Multifaceted Applications of Synthetic Microbial Communities: Advances in Biomedicine, Bioremediation, and Industry
Abstract
1. Introduction
2. Definition and Importance of Microbial Communities
2.1. Synthetic Microbial Communities
2.2. Artificial Communities
2.3. Semi-Synthetic Communities
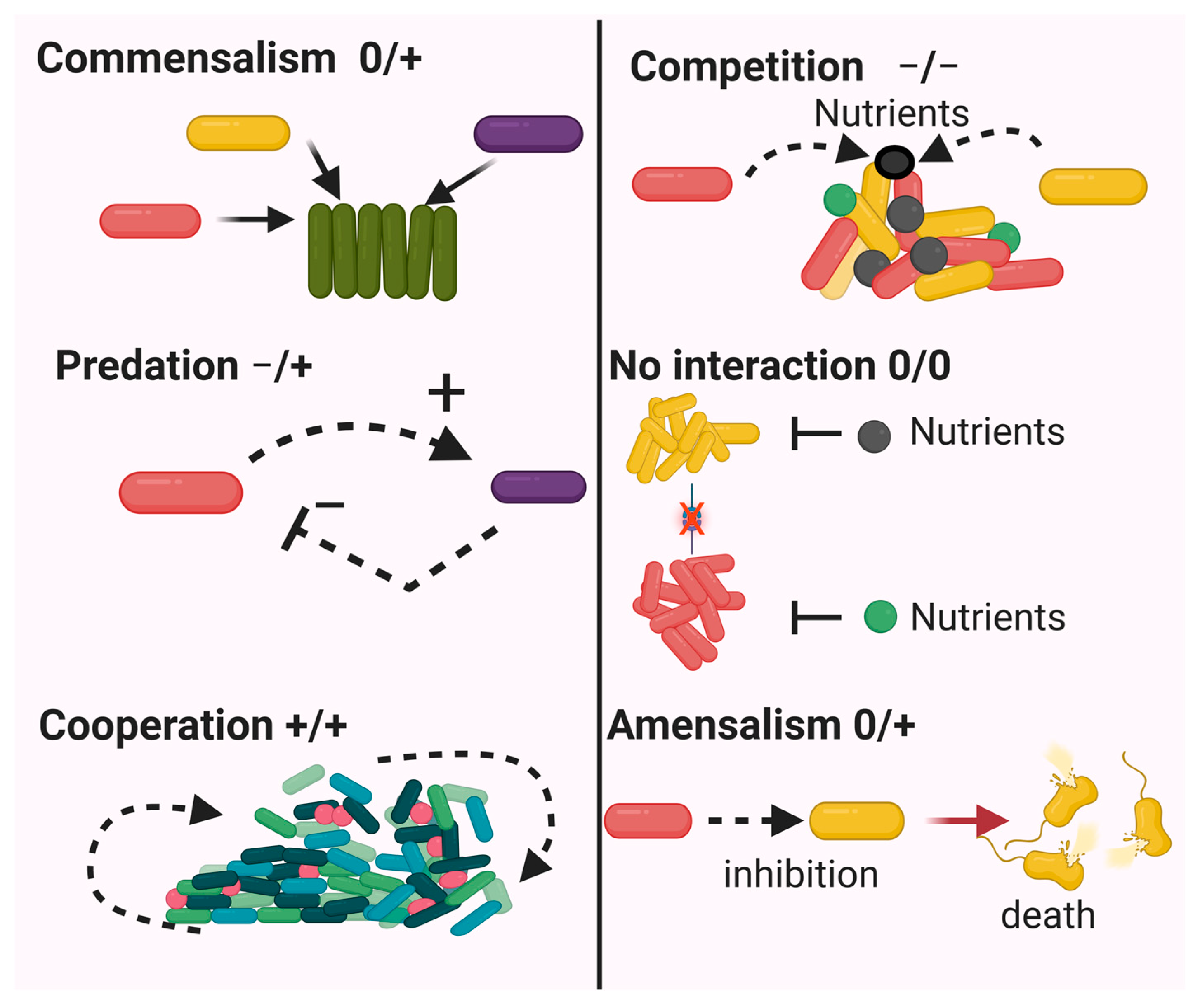
3. Interactions in Microbial Communities
4. Methodologies and Tools in Design of Synthetic Microbial Communities
4.1. Computational Models
4.2. Importance of Genetic Circuits in Microbial Communities
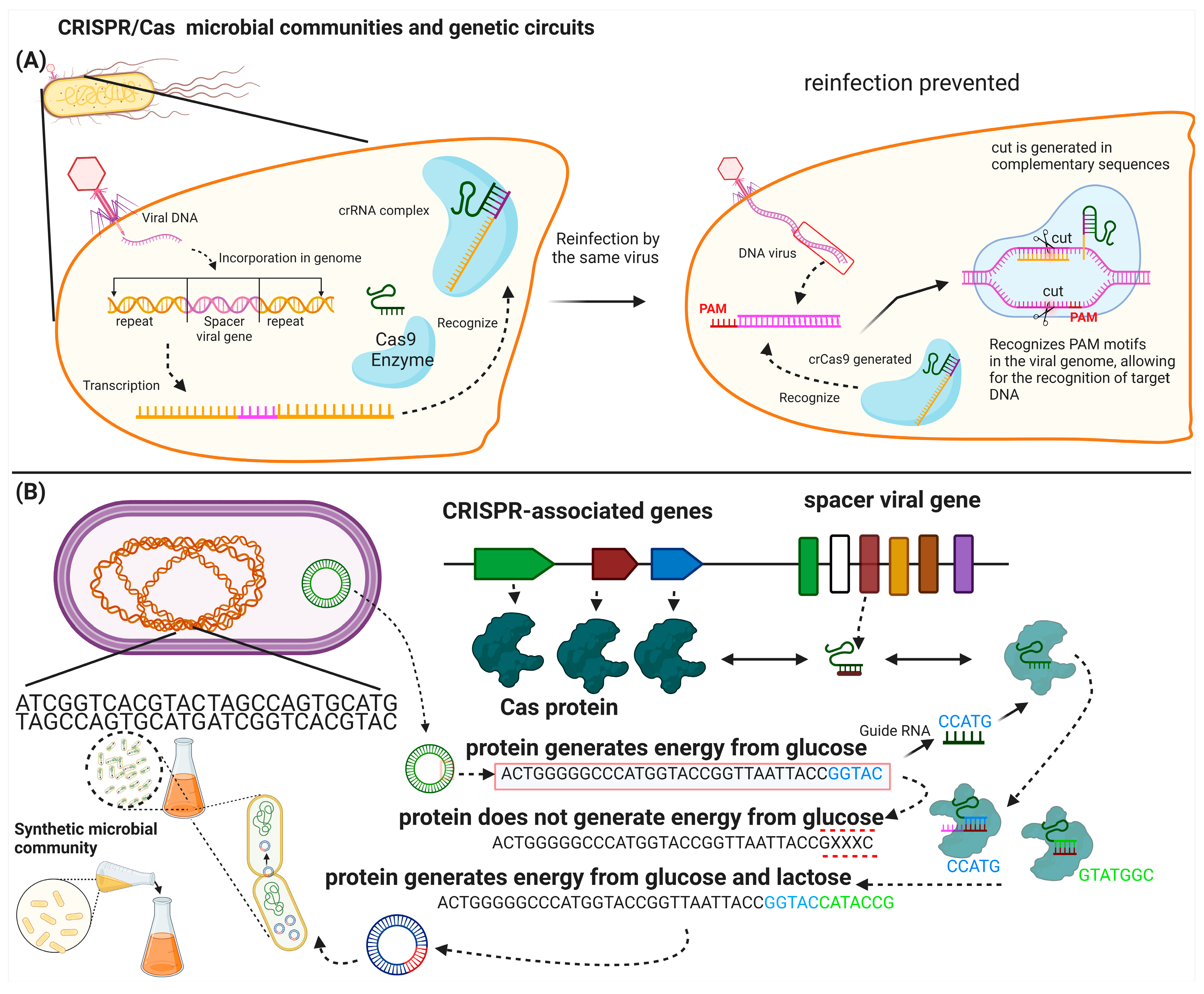
4.3. CRISPR
4.4. Quorum Sensing (QS)
5. Applications of Synthetic Microbial Communities
5.1. Biomedicine and Health
5.2. Bioremediation and Industry
6. Bioethics: Potential Risks in Implementing Synthetic Microbial Communities
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Callieri, C.; Eckert, E.M.; Di Cesare, A.; Bertoni, F. Microbial Communities. In Encyclopedia of Ecology, 2nd ed.; Fath, B., Ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 126–134. [Google Scholar] [CrossRef]
- Gautam, S.S.; Gondwal, M.; Soni, R.; Gautam, B.P.S. Potash biofertilizers: Current development, formulation, and applications. In Trends of Applied Microbiology for Sustainable Economy; Academic Press: Cambridge, MA, USA, 2022; pp. 481–500. [Google Scholar] [CrossRef]
- Ding, Q.; Diao, W.; Gao, C.; Chen, X.; Liu, L. Microbial cell engineering to improve cellular synthetic capacity. Biotechnol. Adv. 2020, 45, 107649. [Google Scholar] [CrossRef] [PubMed]
- Sgobba, E.; Wendisch, V.F. Synthetic microbial consortia for small molecule production. Curr. Opin. Biotechnol. 2020, 62, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Ben Said, S.; Or, D. Synthetic Microbial Ecology: Engineering Habitats for Modular Consortia. Front. Microbiol. 2017, 8, 1125. [Google Scholar] [CrossRef]
- Jagmann, N.; Philipp, B. Design of synthetic microbial communities for biotechnological production processes. J. Biotechnol. 2014, 184, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Johns, N.I.; Blazejewski, T.; Gomes, A.L.; Wang, H.H. Principles for designing synthetic microbial communities. Curr. Opin. Microbiol. 2016, 31, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Großkopf, T.; Soyer, O.S. Synthetic microbial communities. Curr. Opin. Microbiol. 2014, 18, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Letendre, J.H.; Collins, J.J.; Wong, W.W. Synthetic biology in the clinic: Engineering vaccines, diagnostics, and therapeutics. Cell 2021, 184, 881–898. [Google Scholar] [CrossRef]
- Cermak, N.; Datta, M.S.; Conwill, A. Rapid, Inexpensive Measurement of Synthetic Bacterial Community Composition by Sanger Sequencing of Amplicon Mixtures. iScience 2020, 23, 100915. [Google Scholar] [CrossRef]
- Konopka, A. Ecology, microbial. In Encyclopedia of Microbiology; Academic Press: Cambridge, MA, USA, 2009; pp. 91–106. [Google Scholar] [CrossRef]
- Bernstein, H.C.; Carlson, R.P. Microbial Consortia Engineering for Cellular Factories: In vitro to in silico systems. Comput. Struct. Biotechnol. J. 2012, 3, e201210017. [Google Scholar] [CrossRef]
- Blasche, S.; Kim, Y.; Oliveira, A.P.; Patil, K.R. Model microbial communities for ecosystems biology. Curr. Opin. Syst. Biol. 2017, 6, 51–57. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, J.; Wu, Y.; Li, M.; Zhao, Y.; Zhu, H.; Chen, C.; Wang, M.; Chen, B.; Tan, T. Efficient production of chemicals from microorganism by metabolic engineering and synthetic biology. Chin. J. Chem. Eng. 2021, 30, 14–28. [Google Scholar] [CrossRef]
- Aguirre-Cárcer, D. Experimental and computational approaches to unravel microbial community assembly. Comput. Struct. Biotechnol. J. 2020, 18, 4071–4081. [Google Scholar] [CrossRef]
- Mccarty, N.S.; Ledesma-Amaro, R. Synthetic Biology Tools to Engineer Microbial Communities for Biotechnology. Trends Biotechnol. 2019, 37, 181–197. [Google Scholar] [CrossRef]
- Antoniewicz, M.R. A guide to deciphering microbial interactions and metabolic fluxes in microbiome communities. Curr. Opin. Biotechnol. 2020, 64, 230–237. [Google Scholar] [CrossRef]
- Ibáñez, J. La Inteligencia de las Plantas y el Ensamblaje de sus Rizosferas: Conversaciones Mutualistas Entre Raíces y Los Microorganismos del Suelo (Rizosferas) un Universo Invisible Bajo Nuestros Pies. Consejo Superior de Investigaciones Científicas (CSIC). Available online: http://www.madrimasd.org/blogs/universo/2018/04/27/149561 (accessed on 10 July 2024).
- Benigno, O.N.W. Evaluación de Consorcios Microbianos Diseñados Sobre el Desarrollo del Cultivo de Tomate (Solanum lycopersicum) Bajo Invernadero. 2020. Available online: http://dspace.ucuenca.edu.ec/bitstream/123456789/35313/1/Trabajo%20de%20titulacion.pdf (accessed on 26 February 2024).
- Eng, A.; Borenstein, E. Microbial community design: Methods, applications, and opportunities. Curr. Opin. Biotechnol. 2019, 58, 117–128. [Google Scholar] [CrossRef]
- Rojas-Badía, M. Quorum sensing in beneficial plant-bacteria associations. Rev. Colomb. Biotecnol. 2011, 13, 135–143. [Google Scholar]
- Honjo, H.; Iwasaki, K.; Soma, Y.; Tsuruno, K.; Hamada, H.; Hanai, T. Synthetic microbial consortium with specific roles designated by genetic circuits for cooperative chemical production. Metab. Eng. 2019, 55, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson-Palme, J. Microbial model communities: To understand complexity, harness the power of simplicity. Comput. Struct. Biotechnol. J. 2020, 18, 3987–4001. [Google Scholar] [CrossRef] [PubMed]
- Brenner, K.; Karig, D.K.; Weiss, R.; Arnold, F.H. Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium. Proc. Natl. Acad. Sci. USA 2007, 104, 17300–17304. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Qiao, C.; Luan, G.; Luo, Q.; Lu, X. Systematic Identification of Target Genes for Cellular Morphology Engineering in Synechococcus elongatus PCC7942. Front. Microbiol. 2020, 11, 1608. [Google Scholar] [CrossRef]
- García-Jiménez, B.; Torres-Bacete, J.; Nogales, J. Metabolic modelling approaches for describing and engineering microbial communities. Comput. Struct. Biotechnol. J. 2021, 19, 226–246. [Google Scholar] [CrossRef] [PubMed]
- Gallardo-Navarro, O.; Aguilar-Salinas, B.; Rocha, J.; Olmedo-Álvarez, G. Higher-Order Interactions and Emergent Properties of Microbial Communities: The Power of Synthetic Ecology. Heliyon 2024, 10, E33896. [Google Scholar] [CrossRef]
- Bruno, J.F.; Stachowicz, J.J.; Bertness, M.D. Inclusion of facilitation into ecological theory. Trends Ecol. Evol. 2003, 18, 119–125. [Google Scholar] [CrossRef]
- Zhang, H.; Hu, J.; Peng, X.; Zhou, L.; Zhang, T.; Zhang, Y.; Yin, H.; Meng, D. Impacts of ammoniacal odour removal bioagent on air bacterial community. Adv. Biotechnol. 2024, 2, 8. [Google Scholar] [CrossRef]
- Cao, P.; Wei, X.; Wang, G.; Chen, X.; Han, J.; Li, Y. Microbial inoculants and garbage fermentation liquid reduced root-knot nematode disease and As uptake in Panax quinquefolium cultivation by modulating rhizosphere microbiota community. Chin. Herb. Med. 2022, 14, 58–69. [Google Scholar] [CrossRef]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Rev. Microbiol. 2012, 10, 538–550. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, M.; Dong, L.; Zhang, G.; Bai, X.; Wang, J.; Li, Y.; Hu, S.; Yu, X. Interactions between root and rhizosphere microbes mediate phosphorus acquisition in Pinus tabulaeformis. Ind. Crops Prod. 2024, 215, 118624. [Google Scholar] [CrossRef]
- Maqsood, Q.; Sumrin, A.; Waseem, R.; Hussain, M.; Imtiaz, M.; Hussain, N. Bioengineered microbial strains for detoxification of toxic environmental pollutants. Environ. Res. 2023, 227, 115665. [Google Scholar] [CrossRef] [PubMed]
- Requena, T.; Velasco, M. Microbioma humano en la salud y la enfermedad. Rev. Clín. Esp. 2019, 221, 233–240. [Google Scholar] [CrossRef]
- De Lorenzo, V.; Krasnogor, N.; Schmidt, M. For the sake of the Bioeconomy: Define what a Synthetic Biology Chassis is! New Biotechnol. 2021, 60, 44–51. [Google Scholar] [CrossRef]
- Bittihn, P.; Din, M.O.; Tsimring, L.S.; Hasty, J. Rational engineering of synthetic microbial systems: From single cells to consortia. Curr. Opin. Microbiol. 2018, 45, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Kapoore, R.V.; Padmaperuma, G.; Maneein, S.; Vaidyanathan, S. Co-culturing microbial consortia: Approaches for applications in biomanufacturing and bioprocessing. Crit. Rev. Biotechnol. 2021, 42, 46–72. [Google Scholar] [CrossRef] [PubMed]
- Troncoso, C.; Pavez, M.; Santos, A.; Salazar, R.; Barrientos, L. Structural and physiological implications of bacterial cell in antibiotic resistance mechanisms. Int. J. Morphol. 2017, 35, 1214–1223. [Google Scholar] [CrossRef]
- Foresto, E.; Bogino, P.C. Quorum sensing: Un lenguaje común entre bacterias y plantas con importancia en la producción agrícola. Rev. Bol. Biol. 2020, 22, 10–55. [Google Scholar]
- Malla, M.A.; Ansari, F.A.; Bux, F.; Kumari, S. Re-vitalizing wastewater: Nutrient recovery and carbon capture through microbe-algae synergy using omics-biology. Environ. Res. 2024, 259, 119439. [Google Scholar] [CrossRef] [PubMed]
- Trilla Fuertes, L. Biblos-e Archivo. Modelización Computacional de las Alteraciones Metabólicas en Cáncer de Mama. 2019. Available online: https://repositorio.uam.es/handle/10486/687664 (accessed on 30 May 2024).
- Porter, J.R.; Batchelor, E. Using Computational Modeling and Experimental Synthetic Perturbations to Probe Biological Circuits. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2014; pp. 259–276. [Google Scholar] [CrossRef]
- Zomorrodi, A.R.; Segrè, D. Synthetic ecology of microbes: Mathematical models and applications. J. Mol. Biol. 2016, 428, 837–861. [Google Scholar] [CrossRef] [PubMed]
- Hsu, R.H.; Clark, R.L.; Tan, J.W.; Ahn, J.C.; Gupta, S.; Romero, P.A.; Venturelli, O.S. Microbial Interaction Network Inference in Microfluidic Droplets. Cell Syst. 2019, 9, 229–242.e4. [Google Scholar] [CrossRef] [PubMed]
- Glass, D.S.; Riedel-Kruse, I.H. A Synthetic Bacterial Cell-Cell Adhesion Toolbox for Programming Multicellular Morphologies and Patterns. Cell 2018, 174, 649–658.e16. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Riedel-Kruse, I.H. Biofilm lithography enables high-resolution cell patterning via optogenetic adhesin expression. Proc. Natl. Acad. Sci. USA 2018, 115, 3698–3703. [Google Scholar] [CrossRef]
- Gardner, T.S.; Cantor, C.R.; Collins, J.J. Construction of a genetic toggle switch in Escherichia coli. Nature 2000, 403, 339–342. [Google Scholar] [CrossRef]
- Elowitz, M.B.; Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 2000, 403, 335–338. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Yun, H.; Peng, L.; Ji, J.; Wang, W.; Li, X. Deciphering the potential role of quorum quenching in efficient aerobic denitrification driven by a synthetic microbial community. Water Res. 2024, 251, 121162. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.H.; Kim, G.; Seo, S.W. Programable synthetic biology tools for developing microbial cell factories. Curr. Opin. Biotechnol. 2023, 79, 102874. [Google Scholar] [CrossRef]
- Selberg, J.; Gomez, M.; Rolandi, M. The potential for convergence between synthetic biology and bioelectronics. Cell Syst. 2018, 7, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Didovyk, A.; Borek, B.; Hasty, J.; Tsimring, L. Orthogonal Modular Gene Repression in Escherichia coli Using Engineered CRISPR/Cas9. ACS Synth. Biol. 2016, 5, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Barreto, A.C. Quorum Sensing: Sistemas de comunicación bacteriana. Cienc. Actual 2011, 2, 43–50. [Google Scholar]
- Zhang, L.; Zou, W.; Ni, M.; Hu, Q.; Zhao, L.; Liao, X.; Huang, Q.; Zhou, R. Development and application of two inducible expression systems for Streptococcus suis. Microbiol. Spectrum. 2022, 10, e0036322. [Google Scholar] [CrossRef]
- Charubin, K.; Bennett, R.K.; Fast, A.G.; Papoutsakis, E.T. Engineering Clostridium organisms as microbial cell-factories: Challenges & opportunities. Metab. Eng. 2018, 50, 173–191. [Google Scholar] [CrossRef]
- Rivas Lagos, M. Repositorio Académico—Universidad de Chile. Detector de CRISPR en Genoma de Bacterias. 2020. Available online: https://repositorio.uchile.cl/handle/2250/176846 (accessed on 30 May 2024).
- Jeong, S.H.; Lee, H.J.; Lee, S.J. Recent advances in CRISPR-Cas technologies for synthetic biology. J. Microbiol. 2023, 61, 13–36. [Google Scholar] [CrossRef]
- Hutinet, G.; Kot, W.; Cui, L.; Hillebrand, R.; Balamkundu, S.; Gnanakalai, S.; Neelakandan, R.; Carstens, A.B.; Lui, C.F.; Tremblay, D.; et al. 7-Deazaguanine modifications protect phage DNA from host restriction systems. Nat. Commun. 2019, 10, 5442. [Google Scholar] [CrossRef]
- Mojica, F.J.M.; Juez, G.; Rodriguez-Valera, F. Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol. Microbiol. 1993, 9, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Lammoglia-Cobo, M.F.; Lozano-Reyes, R.; García-Sandoval, C.D.; Avilez-Bahena, C.M.; Trejo-Reveles, V.; Muñoz-Soto, R.B.; López-Camacho, C. La revolución en ingeniería genética: Sistema CRISPR/Cas. Investig. Discapacidad. 2016, 5, 116–128, ISSN 2992-779X. [Google Scholar]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Rubin, B.E.; Diamond, S.; Cress, B.F.; Crits-Christoph, A.; Lou, Y.C.; Borges, A.L.; Shivram, H.; He, C.; Xu, M.; Zhou, Z.; et al. Species- and site-specific genome editing in complex bacterial communities. Nat. Microbiol. 2021, 7, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Stephens, K.; Bentley, W.E. Synthetic Biology for Manipulating Quorum Sensing in Microbial Consortia. Trends Microbiol. 2020, 28, 633–643. [Google Scholar] [CrossRef]
- Nealson, K.H.; Hastings, J.W. Bacterial bioluminescence: Its control and ecological significance. Microbiol. Rev. 1979, 43, 496–518. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, K.; Sil, P.C. Effect of diet, pharmaceuticals, and environmental toxicants on gut microbiota imbalance and increased intestinal membrane permeability. In Toxicological Risk Assessment and Multi-System Health Impacts from Exposure; Tsatsakis, A.M., Ed.; Academic Press: Cambridge, MA, USA, 2021; pp. 403–413. [Google Scholar] [CrossRef]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Afzaal, M.; Saeed, F.; Shah, Y.A.; Hussain, M.; Rabail, R.; Socol, C.T.; Hassoun, A.; Pateiro, M.; Lorenzo, J.M.; Rusu, A.V.; et al. Human gut microbiota in health and disease: Unveiling the relationship. Front. Microbiol. 2022, 26, 999001. [Google Scholar] [CrossRef]
- Zhang, J.S.; Chu, C.H.; Yu, O.Y. Oral Microbiome and Dental Caries Development. Dent. J. 2022, 10, 184. [Google Scholar] [CrossRef]
- Van Leeuwen, P.T.; Brul, S.; Zhang, J.; Wortel, M.T. Synthetic microbial communities (SynComs) of the human gut: Design, assembly, and applications. FEMS Microbiol. Rev. 2023, 47, fuad012. [Google Scholar] [CrossRef] [PubMed]
- Donkers, J.M.; van der Vaart, J.I.; van de Steeg, E. Gut-on-a-chip research for drug development: Implications of chip design on preclinical oral bioavailability or intestinal disease studies. Biomimetics 2023, 8, 226. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, W.; Lessing, D.J.; Chu, W. Synthetic microbial consortia for the treatment of Clostridioides difficile infection in mice model. Microb. Biotechnol. 2023, 16, 1985–2006. [Google Scholar] [CrossRef] [PubMed]
- Gurbatri, C.R.; Lia, I.; Vincent, R.; Coker, C.; Castro, S.; Treuting, P.M.; Hinchliffe, T.E.; Arpaia, N.; Danino, T. Engineered probiotics for local tumor delivery of checkpoint blockade nanobodies. Sci. Transl. Med. 2020, 12, eaax0876. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.C.; Clarke, E.J.; Arkin, A.P.; Voigt, C.A. Environmentally controlled invasion of cancer cells by engineered bacteria. J. Mol. Biol. 2006, 355, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Patel, V.K.; Sarim, K.M.; Patel, A.K.; Rout, P.K.; Kalra, A. Synthetic microbial ecology and nanotechnology for the production of Taxol and its precursors: A step towards sustainable production of cancer therapeutics. In Design of Nanostructures for Theranostics Applications; Grumezescu, A.M., Ed.; William Andrew Publishing: Norwich, NY, USA, 2018; pp. 563–587. [Google Scholar] [CrossRef]
- Chen, D.S.; Irving, B.A.; Hodi, F.S. Molecular pathways: Next generation immunotherapy—Inhibiting programmed death ligand 1 and programmed death-1. Clin. Cancer Res. 2012, 18, 6580–6587. [Google Scholar] [CrossRef] [PubMed]
- Buchbinde, A.; Desai, A. CTLA-4 y PD-1 Pathways. Am. J. Clin. Oncol. 2016, 39, 98–106. [Google Scholar] [CrossRef]
- Hu, Y.; Liu, C.; Muyldermans, S. Nanobody-Based Delivery Systems for Diagnosis and Targeted Tumor Therapy. Front. Immunol. 2017, 8, 1442. [Google Scholar] [CrossRef] [PubMed]
- Rylott, E.L.; Bruce, N.C. How synthetic biology can help bioremediation. Curr. Opin. Chem. Biol. 2020, 58, 86–95. [Google Scholar] [CrossRef]
- Fuentes, S.; Méndez, V.; Aguila, P.; Seeger, M. Bioremediation of petroleum hydrocarbons: Catabolic genes, microbial communities, and applications. Appl. Microbiol. Biotechnol. 2014, 98, 4781–4794. [Google Scholar] [CrossRef]
- Schwab, S.; de Souza Pires, A.; Candido, G.Z.; Saggin Júnior, O.J.; Reis, V.M.; Cruz, L.M. Analysis of the endophytic microbiota of roots and culms of two commercial sugarcane cultivars inoculated with a synthetic microbial community. Appl. Soil. Ecol. A Sect. Agric. Ecosyst. Environ. 2024, 195, 105235. [Google Scholar] [CrossRef]
- Chen, J.; Cai, Y.; Wang, Z.; Xu, Z.; Li, J.; Ma, X.; Zhuang, W.; Liu, D.; Wang, S.; Song, A.; et al. Construction of a synthetic microbial community based on multiomics linkage technology and analysis of the mechanism of lignocellulose degradation. Bioresour. Technol. 2023, 389, 129799. [Google Scholar] [CrossRef] [PubMed]
- Revelo-Romo, D.M.; Hurtado Gutiérrez, N.H.; Ruiz Pazos, J.O.; Pabón Figueroa, L.V.; Ordóñez Ordóñez, L.A. Bacterial diversity in the Cr(VI) reducing biocathode of a Microbial Fuel Cell with salt bridge. Rev. Argent. Microbiol. 2019, 51, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Verma, N.; Sharma, R. Bioremediation of Toxic Heavy Metals: A Patent Review. Recent. Pat. Biotechnol. 2017, 11, 171–187. [Google Scholar] [CrossRef]
- Tripathi, S.; Arora, N.; Gupta, P.; Pruthi, P.A.; Poluri, K.M.; Pruthi, V. Microalgae. In Advanced Biofuels; Woodhead Publishing: Sawston, UK, 2019; pp. 97–128. [Google Scholar] [CrossRef]
- Chakravorty, M.; Nanda, M.; Bisht, B.; Sharma, R.K.; Kumar, S.; Mishra, A.; Vlaskin, M.S.; Chauhan, P.K.; Kumar, V. Heavy metal tolerance in microalgae: Detoxification mechanisms and applications. Aquat. Toxicol. 2023, 260, 106555. [Google Scholar] [CrossRef] [PubMed]
- Ardila, L. Medición de la Capacidad de Chlorella Vulgaris y Scenedesmus Acutus para la Remoción de Cromo de Aguas de Curtiembre. Universidad Nacional de Colombia Sede Bogotá Facultad de Ingeniería Departamento de Ingeniería Química y Ambiental. 2012. Available online: https://repositorio.unal.edu.co/handle/unal/20027 (accessed on 26 February 2024).
- Vacca-Jimeno, V.A.; Angulo-Mercado, E.R.; Puentes Ballesteros, D.M.; Torres-Yépez, J.G.; Plaza-Vega, M.E. Using the microalgae Chlorella sp. live suspended in decoloration wastewater from a textile factory. Prospects 2017, 15, 93–99. [Google Scholar] [CrossRef]
- Marrero-Coto, J.; Díaz-Valdivia, A.; Coto-Pérez, O. Mecanismos moleculares de resistencia a metales pesados en las bacterias y sus aplicaciones en la biorremediación. RCCB 2009, 41, 67–78. Available online: https://www.redalyc.org/articulo.oa?id=181221644010 (accessed on 12 February 2024).
- Quiroga, M.V.; Stegen, J.C.; Mataloni, G.; Cowan, D.; Lebre, P.H.; Valverde, A. Microdiverse bacterial clades prevail across Antarctic wetlands. Mol. Ecol. 2024, 33, e17189. [Google Scholar] [CrossRef]
- Abele, D.; Vazquez, S.; Buma, A.; Hernandez, E.; Quiroga, C.; Held, C.; Frickenhaus, S.; Harms, L.; Lopez, J.L.; Helmke, E.; et al. Pelagic and benthic communities of the Antarctic ecosystem of Potter Cove: Genomics and ecological implications. Mar. Genom. 2017, 33, 1–11. [Google Scholar] [CrossRef]
- Roell, M.S.; Zurbriggen, M.D. The impact of synthetic biology for future agriculture and nutrition. Curr. Opin. Biotechnol. 2020, 61, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela-Ruiz, V.; Ayala-Zepeda, M.; Arellano-Wattenbarger, G.L.; Parra-Cota, F.I.; García-Pereyra, G.; Aviña-Martínez, G.N.; de los Santos-Villalobos, S. Las colecciones microbianas y su potencial contribución a la seguridad alimentaria actual y futura. Rev. Latinoam. Rec. Nat. 2018, 14, 18–25, ISSN 2594-0384. [Google Scholar]
- Mardanov, A.V.; Kotlyarov, R.V.; Beletsky, A.V.; Pimenov, N.V.; Nikolaev, Y.A.; Grachev, V.A.; Berestovskaya, Y.Y.; Ravin, N.V. Metagenomic data of the microbial community of lab-scale nitritation-anammox sequencing-batch bioreactor performing nitrogen removal from synthetic wastewater. Data Brief 2019, 27, 104722. [Google Scholar] [CrossRef] [PubMed]
- Schnell, S.; Bak, F.; Pfennig, N. Anaerobic degradation of aniline and dihydroxybenzenes by newly isolated sulfate-reducing bacteria and description of Desulfobacterium anilini. Arch. Microbiol. 1989, 152, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Müller, N. Enhanced aniline degradation by Desulfatiglans anilini in a synthetic microbial community with the phototrophic purple sulfur bacterium Thiocapsa roseopersicina. Syst. Appl. Microbiol. 2019, 42, 125998. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Huang, L.; Wang, W.; Xu, P.; Zanaroli, G.; Tang, H. Maximization of the petroleum biodegradation using a synthetic bacterial consortium based on minimal value algorithm. Int. Biodeterior. Biodegrad. 2020, 150, 104964. [Google Scholar] [CrossRef]
- Silva, R.M.P.; Pozo, M.I.C.; de Oca, J.M.M.; Rodríguez, A.A.; Viñas, M.; Moreno, D.C. Aislamiento y selección de una cepa bacteriana degradadora de hidrocarburos a partir de suelos contaminados con petróleo. Rev. CENIC Cienc. Biol. 2008, 39, 44–51. Available online: https://www.redalyc.org/articulo.oa?id=181214889004 (accessed on 18 November 2023).
- Chen, W.; Li, J.; Sun, X.; Min, J.; Hu, X. High efficiency degradation of alkanes and crude oil by a salt-tolerant bacterium Dietzia species CN-3. Int. Biodeterior. Biodegrad. 2017, 118, 110–118. [Google Scholar] [CrossRef]
- Reyes-Reyes, M.A.; Puentes-Cala, E.A.; Casanova-Montes, E.L.; López-Deluque, F.; Panqueva-Álvarez, J.H.; Castillo-Villamizar, G.A. Immobilization of potentially crude oil degrading bacteria in synthetic and natural organic matrices. Rev. Int. Contam. Ambient. 2018, 34, 597–609. [Google Scholar] [CrossRef]
- Du, R.; Jiang, J.; Qu, G.; Wu, Q.; Xu, Y. Directionally controlling flavor compound profile based on the structure of synthetic microbial community in Chinese liquor fermentation. Food Microbiol. 2023, 114, 104305. [Google Scholar] [CrossRef]
- Jiang, X.; Peng, Z.; Zhang, J. Starting with screening strains to construct synthetic microbial communities (SynComs) for traditional food fermentation. Int. Food Res. 2024, 190, 114557. [Google Scholar] [CrossRef]
- Xin, W.; Zhang, J.; Yu, Y.; Tian, Y.; Li, H.; Chen, X.; Li, W.; Liu, Y.; Lu, T.; He, B.; et al. Root microbiota of tea plants regulate nitrogen homeostasis and theanine synthesis to influence tea quality. Curr. Biol. 2024, 34, 868–880.e6. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Zhao, K.; Tan, X.; Xue, R.; Zeng, Y.; Ratti, C.; Trivedi, P. Perspective on the development of synthetic microbial community (SynCom) biosensors. Trends Biotechnol. 2023, 41, 1227–1236. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Xu, X.; Jin, K.; Liu, Y.; Li, J.; Du, G.; Lv, X.; Liu, L. Genetically encoded biosensors for microbial synthetic biology: From conceptual frameworks to practical applications. Biotechnol. Adv. 2023, 62, 108077. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zheng, X.; Yadav, N.; Saha, S.; Salama, E.-S.; Li, X.; Wang, L.; Jeon, B.-H. Rational management of the plant microbiome for the Second Green Revolution. Plant Commun. 2024, 5, 100812. [Google Scholar] [CrossRef]
- Hao, X.; Wang, X.; Chen, C.; Liu, R.; Yin, Y.; Yao, J.; Xiao, Z.; Liu, X.; Shen, X.; Liu, X. Synthetic bacterial communities reshape microbial communities and enhance nutrient supply in desertified land of Northwest China. Appl. Soil. Ecol. A Sect. Agric. Ecosyst. Environ. 2023, 189, 104972. [Google Scholar] [CrossRef]
- Hays, S.G.; Yan, L.L.; Silver, P.A.; Ducat, D.C. Synthetic photosynthetic consortia define interactions leading to robustness and photoproduction. J. Biol. Eng. 2017, 11, 4. [Google Scholar] [CrossRef]
- Ding, M.-Z.; Song, H.; Wang, E.-X.; Liu, Y.; Yuan, Y.-J. Design and construction of synthetic microbial consortia in China. Synth. Syst. Biotechnol. 2016, 1, 230–235. [Google Scholar] [CrossRef]
- Trump, B.D.; Foran, C.; Rycroft, T.; Wood, M.D.; Bandolin, N.; Cains, M.; Hamilton, K. Development of community of practice to support quantitative risk assessment for synthetic biology products: Contaminant bioremediation and invasive carp control as cases. Environ. Syst. Decis. 2018, 38, 517–527. [Google Scholar] [CrossRef]
- Sivasubramaniam, D.; Franks, A.E. Bioengineering microbial communities: Their potential to help, hinder and disgust. Bioengineered 2016, 7, 137–144. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Contreras-Salgado, E.A.; Sánchez-Morán, A.G.; Rodríguez-Preciado, S.Y.; Sifuentes-Franco, S.; Rodríguez-Rodríguez, R.; Macías-Barragán, J.; Díaz-Zaragoza, M. Multifaceted Applications of Synthetic Microbial Communities: Advances in Biomedicine, Bioremediation, and Industry. Microbiol. Res. 2024, 15, 1709-1727. https://doi.org/10.3390/microbiolres15030113
Contreras-Salgado EA, Sánchez-Morán AG, Rodríguez-Preciado SY, Sifuentes-Franco S, Rodríguez-Rodríguez R, Macías-Barragán J, Díaz-Zaragoza M. Multifaceted Applications of Synthetic Microbial Communities: Advances in Biomedicine, Bioremediation, and Industry. Microbiology Research. 2024; 15(3):1709-1727. https://doi.org/10.3390/microbiolres15030113
Chicago/Turabian StyleContreras-Salgado, Edgar Adrian, Ana Georgina Sánchez-Morán, Sergio Yair Rodríguez-Preciado, Sonia Sifuentes-Franco, Rogelio Rodríguez-Rodríguez, José Macías-Barragán, and Mariana Díaz-Zaragoza. 2024. "Multifaceted Applications of Synthetic Microbial Communities: Advances in Biomedicine, Bioremediation, and Industry" Microbiology Research 15, no. 3: 1709-1727. https://doi.org/10.3390/microbiolres15030113
APA StyleContreras-Salgado, E. A., Sánchez-Morán, A. G., Rodríguez-Preciado, S. Y., Sifuentes-Franco, S., Rodríguez-Rodríguez, R., Macías-Barragán, J., & Díaz-Zaragoza, M. (2024). Multifaceted Applications of Synthetic Microbial Communities: Advances in Biomedicine, Bioremediation, and Industry. Microbiology Research, 15(3), 1709-1727. https://doi.org/10.3390/microbiolres15030113








