Abstract
Endophytic fungus is crucial for maintaining plant health and defense mechanisms, acting as protective barriers against pathogens, and producing medicinally beneficial bioactive compounds. Genome sequencing and metagenomics have significantly enhanced the understanding of fungal diversity and metabolic capabilities, enabling the identification of new genes and substances. Traditional culture-dependent methods have been complemented by culture-independent techniques, offering a more comprehensive view of fungal diversity. Using both culture-dependent and culture-independent techniques, the present research investigation explored the diversity of endophytic fungi encountered in the foliage of Hardwickia binata. The study examined the topographical characteristics and nutritional content of soil samples collected from the locality of the selected plant sample, H. binata, to better comprehend the effects on the plant’s growth. The balanced nutrient constituted approximately a pH of 7.2, which suggested an alkaline nature and promoted plant development. The ratio of nitrogen, phosphorous, and potassium remained 3:1:1. A total of 25 fungal isolates, categorized into 17 morphotypes, were obtained using the culture-dependent approach; Curvularia and Nigrospora emerged as the most common genera. Furthermore, the prediction of the ITS2 secondary structure supports the identification of species, highlighting a wide variety of fungal species present in H. binata. The culture-independent approach generated 69,570 high-quality sequences, identifying 269 Operational Taxonomic Units (OTUs). The dominant Ascomycota phylum, along with various genera, indicated a rich fungal community associated with H. binata. This study advances the understanding of the endophytic fungus communities that are associated with H. binata and the nature of soil ecology. The findings emphasize the significance of holistic techniques in the study of microbial dynamics within plant systems as well as their implications for ecosystem management and plant health.
1. Introduction
Endomycetous fungi thrive in healthy plant tissues and are symbiotically associated with most plants in their natural ecosystems. This allows for the identification of endophytic microbes within every examined plant genus [1,2]. Endophytic microorganisms provide bioactive chemicals that enhance plant development, adaptation, uptake of nutrients, fixation of nitrogen, stress tolerance, and disease control, thereby establishing long-lasting mutually beneficial relationships [3]. Fungal abundance, on the other hand, varies substantially among habitats and ecosystems [4]. Fungi are extremely varied and difficult to predict accurately, which makes academic research particularly challenging. Fungal endophytes work with host plants to regulate development, allowing researchers to assess the diversity and species of fungal communities [5]. Despite the diversity of plant species found in tropical climates, research into strategies for isolating endophytes from these habitats has recently increased [6,7]. Surrounded by a wide variety of native flora and animals, the Nagamalai hills, 12 km south of Madurai city, possess untapped assets. Popular by several names, “Anjan,” or Hardwickia binata Roxb, is a deciduous tree that has been used in traditional medicine to cure a wide range of conditions, including leprosy, dyspepsia, leucorrhea, cancer, diarrhea, and Gram-positive and Gram-negative bacteria [8,9]. As far as we can determine, H. binata, one of the most common woodland plants, has no record of endophytic fungus.
Microbial research, especially on fungi, is challenging because of the wide range of species and the challenge of developing accurate forecasts. However, different researchers have made different estimates about the number of fungal species and the expected diversity of fungal ecosystems on Earth [10]. Analyzing the structure and composition of the microbial communities associated with hosts has been made easier with the gradual development of many approaches [11]. There are two different approaches that have been used to identify endophytic fungi: cultivation-dependent screening, which uses traditional microbiological methods to isolate microbes on solid media, and cultivation-independent screening, which uses metagenomic analysis [12]. Plant endophyte communities can be studied using both direct sequencing and culture-dependent methods. Culture-dependent methods include putting sterile tissue on an agar medium, promoting endophyte development, and keeping cells in pure culture for genetic and morphological identification. Direct sequencing is an additional possible method. A combination of primers is used in PCR to get the necessary sequences from the DNA extracted from the surface-sterilized plant tissue. Sequences having a “known identity”, which is usually (but not always) discovered in a public database, are compared to the obtained sequences in the next phase [9].
From culture-based techniques to molecular biology, endophytic diversity research has progressed, providing new insights. The majority of fungal identification methods require viewing fungi in their natural habitat or grown on growth media, whereas mycological research uses morphological, phylogenetic, and ecological aspects for identification. Although studying fungal variation can greatly benefit from conventional methods, molecular approaches offer more sophisticated methodologies. For taxonomic identification, cultivation is insignificant; however, molecular methods can be beneficial for reliably distinguishing fungi by examining their morphology [13,14]. Mycologists were able to investigate newly obtained as well as previously maintained microbial samples with greater accuracy via a combination of more conventional methods (morphological assessments) with amplification technology. Concerning this, several new taxa have been proposed or established by fungus taxonomists.
A non-cultivational approach to studying genomes from ambient microbes is known as metagenomics [15]. Almost 99% of the microorganisms in an environmental sample may be identified using this method [16]. Interpreting the environment and its inhabitants has been completely transformed by the concept of exploring the entire microflora. The genetic study of environmental samples has emerged as a key technique for obtaining a deeper understanding of the ecological diversity, operational and structural variation, and evolutionary history of the species [17]. A well-known option for high-throughput genome sequencing includes the Illumina MiSeq and HiSeq technologies. The platforms can generate millions of measures within just one cycle and process hundreds of samples simultaneously. These methods have examined fungal diversity in a variety of samples based on recorded data [18]. The main objective of the current study is to comprehend the diversity and cultivation potential of the fungal community in H. binata leaves. To this end, the two approaches: one that is culture-dependent and involves cultivating and identifying endophytic fungi, and another that is culture-independent and involves direct molecular identification of endophytic fungi. The results shed information on the factors influencing endophyte diversity and how to optimize endophyte discovery efficiency.
2. Materials and Methods
2.1. Materials Used
The chemicals and media components used in the current study were obtained from HiMedia, Mumbai, India. The internal transcribed spacer (ITS) primers for molecular identification were obtained from Sigma-Aldrich, Bangalore, India.
2.2. Description of the Site, Selection of Plants, and Soil Analysis
An increasing interest among research organizations in the isolation of endophytes prompted an investigation into the isolation of endophytic fungi in different regions of the Nagamalai hills in Madurai. The research site, located at an altitude of 150 m, and a latitude of 9°54′ N, and a longitude of 78°00′ E, contains a wide variety of plant species distributed across its nearly four kilometers of 1500-foot-tall terrain [19]. The hill’s subhumid eco-climate is slightly dry due to limited drainage capacity, despite its rich flora and fauna. The endophytic fungal diversity analysis focused on Hardwickia binata Roxb, a woodland plant in the Fabaceae family and Caesalpinioideae (Figure 1). The confirmation of the botanical samples’ authenticity was conducted by the Botanical Survey of India, Coimbatore. Following the procedure guidelines indicated by Tamilnadu Agricultural University (TNAU) (https://agritech.tnau.ac.in/agriculture/agri_soil_sampling.html, accessed on 31 March 2022), soil samples were obtained from the H. binata research location. Topographic features were documented. Soil physicochemical parameters were analyzed by sending soil samples to the Agriculture Development, ICAR Krishi Vigyan Kendra, CENDECT, Soil and Water Analysis Center, Theni (Dt), Tamilnadu, India.

Figure 1.
Taxonomic classification of the selected plant.
2.3. Collection of Plant Samples
About sixty completely developed, dark green leaves were hand-picked from the lowest branches of H. binata to directly collect the endophytic fungus populations. The samples were collected according to strict aseptic standards and promptly transported to the laboratory in sterile bags to reduce the chance of contamination. The endophytic fungus identified in the foliage of H. binata was isolated and studied after being properly sterilized and diced into three hundred segments via a clean, sterile blade. The resulting segments were subsequently rinsed under running water.
2.4. Culture-Dependent Approach
2.4.1. Endophytic Fungi Isolation and Morphological Characterization
The leaf segments were rigorously surface sterilized under the [20] methodology to remove any exterior contaminants and make it less difficult to isolate endophytic fungi. The process included providing the foliage sections with a complete rinse with normal water before submerging the leaf bits in a 70% ethanol and 4% NaOCl solution for disinfection. After being air-dried using sterile filter papers, the sterilized foliage segments have been rinsed in sterilized distilled water to remove any remaining moisture residue. Potato dextrose agar plates were used to examine the effectiveness of the surface sterilizing process. Effective sterilization can be determined by the absence of microbial growth. To avoid bacterial and fungal contamination, the sterilized leaf segments were laid out on a medium encompassing potato extract, dextrose, agar-agar, and streptomycin. Subsequent to an incubation period of 7 days, the fungal isolates were subcultured onto new PDA plates at 24 °C following a 10-day period. Pure cultures were kept at 4 °C for further examination and preservation once morphological characterization was completed.
2.4.2. Assembling Sequences and Phylogenetic Tree Construction
The protocol described by [21] was followed for the genomic DNA extraction from fungal mycelia. The isolated DNA was run through agarose gel electrophoresis (0.8%) to determine its purity. Using fungal specific primers (ITS1-F (5′-CTTGGTCATTTAGAGGAAGTAA-3′ and ITS4-R (5′-CTTGGTCATTTAGAGGAAGTAA-3′), the Internal Transcribed Spacer (ITS) region was amplified. A polymerase chain reaction (PCR) was executed in a 25 μL reaction consisting of 1 μL of DNA template, 1.25 μL of forward and reverse primers, and 12.5 μL of 2 × DNA master mix (Amplicon). The PCR amplification was performed using the Prima-96™ Thermocycler instrument from Himedia, Mumbai, India. The reaction conditions included denaturation at 94 °C for 4 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 58.2 °C for 1 min, and extension at 72 °C for 2 min. Finally, there was a 7 min extension at 72 °C. Confirmation of the amplicons was executed on 1% agarose gels, followed by purification and sanger sequencing performed by Bio kart India Pvt. Ltd., located in Bangalore, India. Analyzed using NCBI and BLAST, the amplified sequence yielded valuable insights. Additionally, a phylogenetic tree was meticulously crafted using MEGA X [22].
2.4.3. ITS2 Secondary Structure Prediction
By using GenBank annotation or Hidden Markov models (HMMs) executed via the web server (http://its2.bioapps.biozentrum.uni-wuerzburg.de, accessed on 15 April 2022), the ITS2 area was determined and delineated [23]. Subsequently, the BLAST algorithm was used to do sequence similarity searches. A comparative analysis was conducted between each consensus sequence and published sequences obtained from reference species in various culture collections. The phylogenetic analysis only used sequences that were associated with verified species. The phylogenetic analysis used the ITS2 sequences acquired from reference species with the best coverage and identity using the BLAST search. The ribotypes of the endophytic isolates and the reference taxa were annotated using the ITS2 Database [24]. To generate synchronous alignment of sequence and secondary structures, 4SALE V1.7.1 was used with the ITS2 sequence and secondary structure for all of the sequences. The present study has modeled the genera-specific consensus secondary structure and analyzed compensatory base change (CBC). As mentioned earlier, the phylogenetic tree was rebuilt using the Phangorn program, which is part of the R framework. Using the Phangorn tool in the R environment, the phylogenetic tree was recreated. The General Time Reversible (GTR+I+G) substitution model was used to build the tree using the maximum likelihood (ML) approach. Phylogenetic tree branch support values were computed using 1000 repetitions.
2.5. Culture-Independent Approach
DNA Extraction and Metagenomic Analysis
The genomic DNA was extracted from the homogenized leaf samples using the QIAGEN soil kit. Afterwards, the concentration and purity of DNA were evaluated using a 1% agarose gel. Then, PCR amplification was carried out with specific primers (ITS1: 5′TTGGTCATTTAGAGGAAGTAA3′ and ITS2: 5′GCTGCGTTCTTCATCGATGC3′) with unique barcodes to amplify the nuclear ribosomal Internal Transcribed Spacer (ITS) region. PCR reactions were performed using TAQ Master Mix, with cycling conditions consisting of 30 cycles at 95 °C for 45 s, 56 °C for 45 s, and 72 °C for 60 s, followed by a final extension at 72 °C for 10 min. Products of around 400–450 bp were examined for further analysis. Subsequently, the PCR products were quantified by measuring the 260/280 ratio using Nano Drop readings. To remove any remaining primers, the purified products were treated with Ampure beads. After conducting an additional 8 cycles of PCR, Illumina barcoded adapters were used to prepare the sequencing libraries. The libraries were then purified and quantitated using the Qubit dsDNA High Sensitivity assay kit. Sequencing was performed using the Illumina Miseq platform and a 2X00PE sequencing kit, resulting in the production of 250 bp paired-end reads for subsequent analysis. The culture-independent documentation was performed according to the standard procedure followed by Biokart Pvt. Ltd., Bangalore, India.
2.6. Data Analysis in the Pipeline
Paired-end reads were matched to samples using their distinct barcodes, and then the barcode regions and primer sequence were removed. Paired-end reads were combined, and splicing sequences were acquired. Comprehensive filtering was conducted on the splicing sequences to ensure the acquisition of pristine, high-quality tags [25]. The tags were compared with a reference database to detect and remove any chimeric sequences. Sequence analyses of the clean tags were conducted using the QIIME2 pipeline, a powerful tool for analyzing genetic data. Sequences that shared a similarity of at least 97% were grouped together into Operational Taxonomic Units (OTUs). Sequences from each OTU were carefully examined for additional annotation. For taxonomic assignment of the representative sequences, the Unite Database (https://unite.ut.ee/, accessed on 30 April 2022) [26] was used with the blast algorithm. Individual data points were eliminated from the dataset. All further analyses were conducted using this normalized dataset.
3. Results
3.1. Soil Analysis
The soil sample’s nutritional content and topographical characteristics were examined in the investigation (Table 1). The findings showed that the soil at the H. binata location had a pH of 7.2, which is somewhat alkaline. The soil seems to be rich in organic matter, as shown by the measured organic carbon content of 7.4%. A modest amount of salt was indicated by an electrical conductivity of around 0.65 dS/m. The soil had a balanced nutritional profile that was good for plant development, with a nitrogen-phosphorus-potassium (NPK) ratio of 3:1:1. In addition, the amounts of iron (Fe), manganese (Mn), zinc (Zn), copper (Cu), and boron (B) were determined to be 2.34, 1.74, 1.35, 1.5, and 0.35 units, respectively, when testing the soil for micro-nutrient availability. These results shed light on the soil’s fertility and its ability to sustain plant life, in particular the development of H. binata.

Table 1.
Summary of soil nutrients at sampling locations of the study.
3.2. Culture-Dependent
Cultured Endophytic Fungus: Diversity and Community Structure
The investigation into endophytic fungal diversity within the foliage of H. binata employed a culture-dependent approach, yielding 60 fungal isolates from symptomless leaves. Subsequent morphological analysis revealed 25 distinct isolates, which were further categorized into 17 morphotypes based on cultural traits (Figure 2). This classification served to streamline the dataset by eliminating redundant cultures, enhancing the precision of subsequent analyses. Morphological and phylogenetic analyses, utilizing subsets of ITS sequence data, provided insights into the taxonomic composition of endophytic isolates sourced from H. binata. Genus-level classification revealed enrichment in taxa such as Fusarium 8%, Phoma 4%, Didymella 4%, Talaromyces 4%, Sphaeropsis 4%, Curvularia 16%, Rhytidhysteron 8%, Diaporthe 4%, Phyllosticta 4%, Aspergillus 8%, Sordaria 4%, Nigrospora 12%, Xylaria 4%, Pestalotiopsis 4%, Daldinia 4%, and Pseudopithomyces 4%. These taxa span diverse fungal classes, including Sordariomycetes, Dothidiomycetes, Ascomycetes, Eurotiomycetes of the phylum Ascomycota, and Agaricomycetes of the phylum Basidiomycota. The taxonomic distribution of morphotypes isolated from the plant samples is detailed in Figure 3. Among these, the genus Curvularia and Nigrospora emerge as the predominant genera, constituting 16% and 12%, respectively, of the isolated endophytic fungi. These morphotypes were further identified by the use of ITS2 secondary structure prediction.
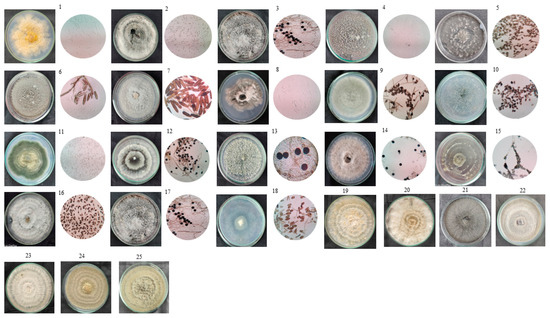
Figure 2.
Macroscopic and microscopic view of endophytic fungal communities isolated from the plant H. binata through the culture-dependent approach (40× magnification). 1-Fusarium hainanense, 2-Phoma macrostoma, 3-Didymella macrostroma, 4-Sphaeropsis visci, 5-Curvularia geniculate, 6-Rhytidhysterom rufulum, 7-Rhytidhysterom rufulum, 8-Fusarium incarnatum, 9-Curvularia sp., 10-Coprinellus radians, 11-Aspergillus sydowii, 12-Nigrospora sphaerica., 13-Nigrospora oryzae, 14-Nigrospora sp., 15-Aspergillus eburneocremeus, 16-Pestalotiopsis trachicarpicola, 17-Daldinia sp., 18-Pseudopitomyces karoo, 19-Curvularia lunata, 20-Curvularia americana, 21-Diapothe biconispora, 22-Talaromyces verraculosus, 23-Xylaria psidii, 24-Sordaria fimicola, 25-Phyllosticta paracaoitalens.
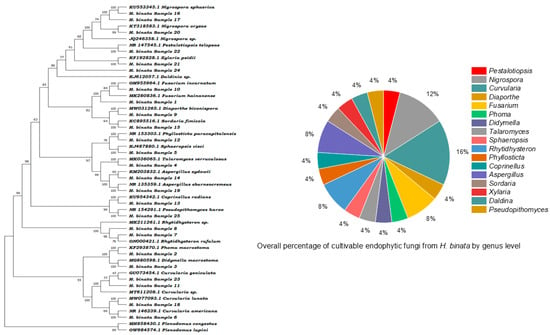
Figure 3.
Phylogenetic relationships between reference strains and representative isolates as determined by the ITS region (outgroup as Plenodomus). Species distribution along genus lines as determined by the culture-dependent approach.
3.3. Prediction of ITS 2 Secondary Structure
Comparative analysis revealed a similarity in taxonomy preferences between the Internal Transcribed Spacer (ITS) and ITS2 regions in fungi. Notably, ITS2 exhibited promising potential as a standalone marker for species delimitation within fungal communities. Utilizing sequences retrieved from the ITS2 database, phylogenetic reconstruction was conducted to ascertain fungal species diversity. Within H. binata, 17 morphotypes underwent ITS2 secondary structure prediction (Figure 4), exhibiting lengths ranging from 155 to 207 bp. Notably, Fusarium (156 bp), Rhytidhysteron (158 bp), and Curvularia (171 bp) presented with three helices, while others, including Phoma (160 bp), Didymella (160 bp), Talaromyces (160), Sphaeropsis (165 bp), Diaporthe (162 bp), Phyllosticta (167 bp), Coprinellus (207 bp), Aspergillus (170 bp), Sordaria (155 bp), Nigrospora (159 bp), Xylaria (164 bp), Pestalotiopsis (170 bp), Daldinia (167 bp), and Pseudopithomyces (161 bp), formed four helices with elongated third helices. Compensatory Base Changes (CBC) analysis indicated the absence of CBCs among isolates like Fusarium, Rhytidhysteron, Curvularia, and others (Supplementry Figure S1 and Table S1). Phylogenetic analysis coupled with ITS2 secondary structure prediction led to the identification of Curvularia lunata, Nigrospora lacticolonia, and Daldinia eschscholtzii, alongside the recognition of 25 isolates encompassing diverse fungal species within H. binata Table 2. By analyzing the RNA molecules, the distinct genetic structures in the conserved nuclear region known as the ITS2 secondary structures may be able to help distinguish between the endophytic fungal individuals at the genotypic level.
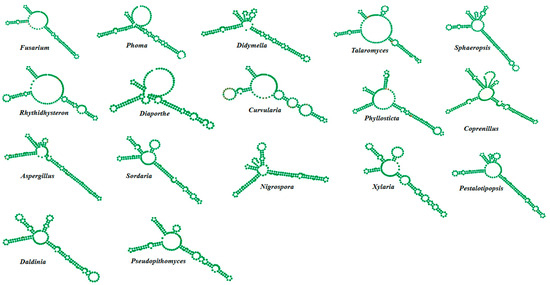
Figure 4.
Isolated fungi from the culture-dependent method: a consensus model for their ITS2 secondary structure.

Table 2.
Endophytic fungal population structure as assessed using a culture-dependent technique.
3.4. Culture-Independent
Examination of Endophytic Fungal Community Structures Using a Culture-Independent Approach
A total of 69,570 high-quality sequences were obtained from Illumina Miseq sequencing analysis after quality control and filtering. A total of 269 OTUs were generated from the obtained sequences, with a GC content of 56.5%. After chimeric removal and filtering, the ITS libraries seem to have covered a significant portion of the fungal variety in the samples utilized in this research. This indicated that the sequence data correctly mirrored the structural makeup of the endophytic fungus samples. Utilizing the UNITE fungal ITS database, which is validated by QIIME, we classified every OTU to the species level, exposing variations in the mycobiome.
3.5. Examining Operational Taxonomic Units
Figure 5 shows the fraction of fungal genera’ abundances at the phylum and genus levels. The fungal community presented in H. binata was identified through a culture-independent method and attributed to 10 phyla, 37 classes, 79 orders, 133 families, and 189 genera. The phyla include Ascomycota (71.3%), Basidiomycota (17.8%), Mortierellomycota (2.2%), Mucromycota (1.4%), Olpidiomycota (1.4%), Neocallimastigomycota (1.1%), Rozellomycota (1.1%), Glomeromycota (1.1%), and Zoopagomycota (0.3%). At the genus level, Tilletia (26%) was the most dominant, followed by Ramichloridium (23%), Dissoconium (13%), Aspergillus (10%), Penicillium (6%), Cladosporium (5%), Pichia (4%), Talaromyces (4%), and Fusarium (4%). These results suggest that the abundance of the culturable endophytic communities was similar. Furthermore, a wide range of microfungi were included in all OTUs with a sequence similarity of ≥97% that had sequences that were made publicly available with species identification (Supplementary Table S2). The Ascomycota phylum was found to be predominant, and one percent of the fungi belong to unidentified and unclassified phyla. As such, the sequences are not easily allocated to any recognized category using the present taxonomy references database.
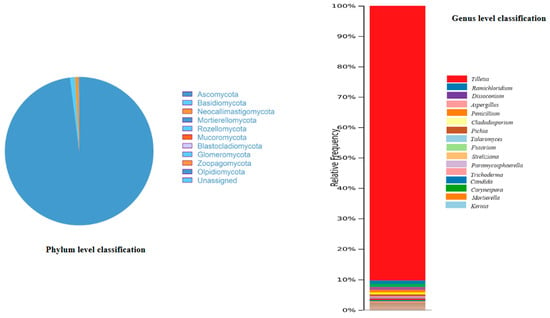
Figure 5.
Phylum- and genus-level endophytic fungal community composition and relative abundance (column length reflects genus percentage and column color reflects fungal genus color) in the sample. For sequences that could not be categorized well into any existing category, the label “unassigned” was applied.
3.6. Rarefaction Analysis and Richness Estimates
As the curves began to approach saturation, suggesting that the chosen sequence data accurately matched the fungal abundance of these samples, the rarefaction curves demonstrated that the sequencing diligence was more comprehensive in covering the fungal variety (Figure 6). To put it another way, the rarefaction curve indicated that almost every species in the community of the samples was covered by the sequencing data. Conversely, the endophytic microbial component distributions in H. binata were found at several taxonomic levels, and the endophytic community complexity was estimated using various fungal diversity indices, such as Shannon and Simpson (Table 3).
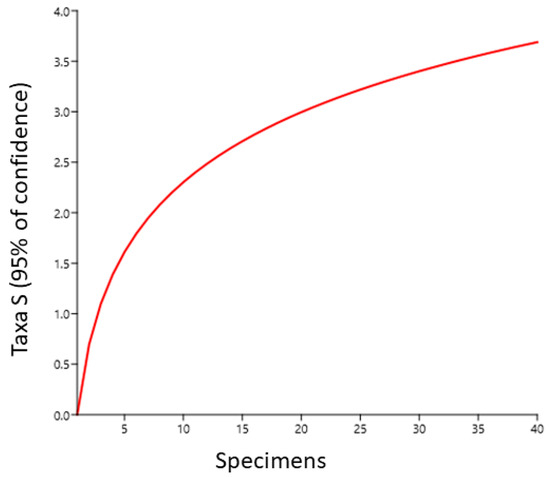
Figure 6.
Endophytic fungal rarefaction curves of the plant sample.

Table 3.
The community composition and richness of endophytic fungal isolates from the leaves of H. binata.
3.7. Culture-Dependent versus Culture-Independent
Culture-dependent and culture-independent methods are employed in studying endophytic fungal diversity, each offering unique insights into community structure and composition. Comparing the number of independent isolated fungus species (25 total) obtained in this work to the total number of OTUs (269 total) found via sequencing would seem to validate this approach. However, several of the proportionately dominant taxa revealed by the sequencing method were really represented by isolates. In general, the ITS sequences of mycobial endophytes found using both techniques yielded a direct comparison that showed 97% similarity or higher between all fungal taxa found using the conventional methods and even fungal OTUs found using the culture-independent methods.
The fungal community analysis using a culture-independent and culture-dependent approach revealed a dominant Ascomycota phylum, accounting for 71.3% of the community. A small fraction, approximately 1%, could not be classified under known phyla and were categorized under the “uncultured” group. This highlights the complexity and diversity of the fungal community and the limitations of current taxonomic classifications in fully characterizing microbial populations. Both methods identified a number of frequently found fungus taxa, including Aspergillus, Nigrospora, and Fusarium. In the mycobiome, there are several highly or often discovered genera that were missed using the conventional approach. These include Dissoconium, Pichia, Lasiodiplodia, Cladosporium, Tilletia, Ramichloridium, and Trichothecium. The common and distinct fungi among the samples were determined using a Venn diagram. A total of 189 fungal taxa have been identified in the comparative study using culture-independent approaches. Furthermore, 17 fungal genera were discovered using culture-dependent approaches. Remarkably, the overlap between the two methods revealed that 17 fungal genera were shared by both datasets. This convergence highlights the value of using alternative methodologies to thoroughly investigate fungal communities by indicating a degree of consistency between culture-independent and culture-dependent methods in capturing a fraction of the fungal diversity (Figure 7). Comparing the two approaches, the culture-dependent method provided detailed insights into cultivable fungal species, allowing for precise morphological characterization and taxonomic classification. However, it may underestimate fungal diversity due to limitations in cultivability. On the other hand, the culture-independent approach offered a broader view of fungal diversity by detecting non-cultivable taxa, albeit with challenges in taxonomic assignment. Integrating both methods can enhance the comprehensive analysis of endophytic fungal communities, providing a more holistic understanding of their structure and composition.
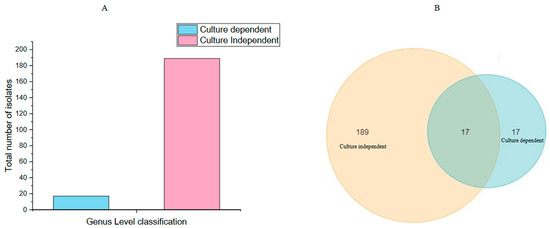
Figure 7.
A column graph and a Venn diagram depicting the total count of endophytic fungi obtained through culture-dependent and culture-independent methods. (A) Show the total number of fungal isolates from H. binata that were acquired by culture-dependent and culture-independent methods. (B) Genomic level comparison utilizing culture-dependent and culture-independent methods.
4. Discussion
Endophytic fungi are commonplace within numerous plant tissues and are essential to the health and defense systems of plants. Microbial colonization of plant tissues acts as a barrier to prevent pathogen invasion, increasing the resistance of the host plant [27,28,29]. Moreover, endophytic fungi are renowned for their production of bioactive metabolites with diverse pharmaceutical properties, including antiviral and anticarcinogenic activities [30,31]. Recent advancements in genomics and metagenomics have revolutionized our understanding of fungal diversity and metabolic potential, facilitating the discovery of novel genes and metabolic compounds [32,33].
Traditionally, culture-dependent methods have been employed to isolate and identify endophytic fungi based on morphological and phenotypic characteristics [34,35]. However, limitations inherent to these methods have spurred the development of culture-independent techniques, such as metagenomics, enabling the direct examination of microbial communities from environmental samples [36,37]. By integrating both culture-dependent and culture-independent approaches, researchers can achieve a more comprehensive understanding of fungal diversity, revealing species that may elude traditional cultivation methods [38]. This integrated methodology not only enhances fungal species identification but also facilitates in-depth investigations into their ecological roles and potential applications across various industries, including biotechnology, agriculture, and medicine.
The study’s findings shed light on important soil properties and nutrient content for H. binata’s development. A soil with a high organic carbon content and a slightly alkaline pH is ideal for plant development. Soil micronutrient availability and a balanced nitrogen-phosphorus-potassium (NPK) ratio suggest the soil may maintain healthy growth despite moderate saline levels. Results from a study by [39] were similar. The two locations that were examined are located in an area that has a history of Pb-Zn mining dating back over 300 years. Pb, Zn, and Cd were significantly present in the soils and plants of both locations throughout the sample period. Soils from the slag heap had a greater level of HMs than those from the wasteland, on average. Soil organic matter, total nitrogen, phosphorus, potassium, accessible potassium, and hydrolyzable nitrogen were all substantially reduced in the slag heap soils compared to the wasteland soils, with the exception of available phosphorus. To maximize H. binata’s development in the research region, however, more research into its unique needs with regard to soil salinity, pH, and nutrient levels is required. Insightful management techniques may be built upon these results, which add to our knowledge of soil fertility and help ensure the long-term viability of H. binata populations.
The study represents a pioneering investigation into the endophytic fungal population inhabiting H. binata leaves, utilizing both traditional culture-dependent methods and modern culture-independent techniques. Precise taxonomic identifications were derived from DNA analysis, enabling a comprehensive comparison of taxa obtained from both methods using the same samples. This study marks the first report on the identification and isolation of endophytic fungi within H. binata. The culture-dependent method yielded 60 fungal isolates from symptomless leaves, subsequently identified into 17 morphotypes encompassing diverse taxa, including Fusarium, Phoma, Didymella, Talaromyces, Sphaeropsis, Curvularia, Rhytidhysteron, Diaporthe, Phyllosticta, Aspergillus, Sordaria, Nigrospora, Xylaria, Pestalotiopsis, Daldinia, and Pseudopithomyces genera. Comparable findings were noted in previous research by [40], who isolated 15 endophytic fungi from mature E. pachyclada leaves, identifying species such as Penicillium crustosum, Penicillium chrysogenum, Penicillium commune, Penicillium corylophilum, Alternaria infectoria, Alternaria alternata, Alternaria tenuissima, Aspergillus flavus, and Aspergillus niger. Additionally, previous studies by [41,42] have identified endophytic fungi from medicinal plants and explored the importance of molecular phylogeny in fungal taxonomy, highlighting the challenges associated with DNA barcoding methods such as ITS sequencing due to sequence diversity within and between species. For phylogenetic and taxonomic placement analyses of fungi, the protocol of choice has been the sequencing analysis of the ITS rRNA region. However, building a reliable and repeatable phylogenetic connection is hampered by the sequence diversity of the ITS1 region across species belonging to the same genus [43]. For fungus, the internal transcribed spacer region is the most chosen DNA barcode. But in certain fungal groupings, the inter- and intra-specific distances in ITS sequences differ greatly; as a result, it is not a completely trustworthy method for distinguishing between species [44].
A common core secondary structure of the nuclear ribosomal internal transcribed spacer 1 (ITS1) region in fungi was proposed by Koetschan et al. [45], using a helix-wise folding approach in conjunction with ITS1 sequence annotation based on the Hidden Markov Model. This innovative technique facilitated automated sequence-structure-based taxonomy of ITS1 for environmental fungi and incorporated structural data into phylogenetic analysis. Their findings suggested that ITS2 secondary structure prediction could serve as a valuable tool for species delimitation within fungal communities. Comparative analysis indicated similar taxonomy preferences between ITS and ITS2 regions in fungi, with ITS2 demonstrating potential as a standalone marker. Phylogenetic reconstruction utilizing ITS2 sequences enabled the identification of fungal species diversity within H. binata, revealing insights into genetic diversity and evolutionary relationships among fungal isolates. Specific fungal species, including Curvularia lunata, Nigrospora lacticolonia, and Daldinia eschscholtzii, were identified, along with 25 isolates encompassing diverse fungal species within H. binata. Additionally, [24] discovered distinct helix patterns in isolates from various genera, with some exhibiting a 3-helix pattern and others a 4-helix pattern, highlighting structural variations across species. The ITS2 ribotype findings we obtained are consistent with previous research [22,46] on the genera Colletotrichum, Fusarium, Xylaria, Aerobasidium, Diaporthe, and Aspergillus. Ref. [47] explored the utility of ITS2 molecular morphometrics for delineating Ascomycota species, emphasizing the effectiveness of this approach in elucidating fungal species boundaries and potentially easing challenges associated with molecular characterization. Integration of secondary structure information with primary sequencing data significantly enhances species identification within plant genera and sheds light on their evolutionary history. Furthermore, phylogenetic analysis of ITS2 at higher taxonomic levels leverages the conservative secondary structure qualities of ITS2 and aids in nucleotide sequence evolutionary studies, contributing to improved structure prediction and taxonomic understanding.
The diversity of fungal communities in various habitats, such as soils, plants, and environmental samples, has been thoroughly investigated using cultivation-independent methods [48,49,50]. When studying fungal species diversity, high-throughput sequencing technologies were able to identify more fungi than the culture-based technique would have in the past. As much as one percent of all microorganisms may be cultivated, according to research by [51]. This study employed a metagenomics approach to comprehensively characterize the endophytic fungal community inhabiting H. binata, leveraging the power of culture-independent methods to elucidate its taxonomic composition and diversity. Through high-throughput Illumina MiSeq sequencing analysis, a substantial dataset comprising 69,570 high-quality sequences was generated, leading to the identification of 269 Operational Taxonomic Units (OTUs). The observed GC content of 56.5% further confirmed the typical fungal DNA composition, enhancing the reliability of the obtained results. The utilization of paired-end Illumina sequencing in similar studies, such as that by [52], has provided comparable insights into fungal diversity within endophytic communities, reinforcing the robustness of the methods employed. Classification of OTUs to the species level using the UNITE fungal ITS database revealed a rich and diverse fungal community associated with H. binata, spanning 10 phyla, 37 classes, 79 orders, 133 families, and 189 genera. The dominance of the Ascomycota phylum, followed by Basidiomycota, underscores the complexity of the fungal community inhabiting H. binata. These findings align with similar research on endophytic fungal communities in related plant species, as demonstrated by [53], analyzed the endophytic fungal assemblages in H. contortus shoots and roots using a multiscale approach. H. contortus’s endophytic fungal flora includes 4 phyla, 11 classes, 17 orders, 21 families, and 18 genera in the shoot sample and 10 classes, 18 orders, 22 families, and 20 genera in the root sample. This suggests that both sections are host to a diverse range of fungal endophytic species. At the genus level, Tilletia emerged as the most dominant genus, followed by Ramichloridium, Dissoconium, Aspergillus, Penicillium, Cladosporium, Pichia, Talaromyces, and Fusarium. The presence of these genera indicates a diverse assemblage of endophytic fungi colonizing H. binata leaves. Moreover, the detection of OTUs with high sequence similarity to publicly available sequences further validates the taxonomic assignments and highlights the presence of various microfungi within the endophytic community. However, because of advancements in high-throughput sequencing techniques, an increasing number of endophytic fungi have been identified, bringing the total number of fungal species to 5.1 million. The identification and traits of the majority of fungi are still unknown, nevertheless, since only around 10,000 species have been studied and categorized [54]. In a comparable way, our research [55] found that ten of the twenty-two most common fungal OTUs could be assigned to specific genera. Phyllosticta, Talaromyces, Aspergillus, Aporospora, Cladosporium, Alternaria, Fusarium, Penicillium, and Allophaeosphaeria were among the fungi that were identified. The majority of these endophytic fungi, like Fusarium, are often found in nature and may be either harmful pathogens or endophytes of many different types of crops [56].
Our use of Illumina sequencing has allowed us to get some insight into the fungal endophytes in this medicinal plant, and we have also detected a number of fungi that have not yet been classified or recognized, suggesting the presence of novel microbial resources. Rarefaction analysis and richness estimates provided insights into the comprehensiveness of the sequencing effort, demonstrating saturation of rarefaction curves and indicating adequate coverage of fungal diversity within the sampled H. binata population. The results were in line with those of the study by [57], which established that all samples had reached an equilibrium level in terms of sequencing depth and that taxonomic designations were accurate. Based on the numbers of OTUs, diversity, and coverage indices, [2] demonstrated that fungal sequence diversity was compared in each sample. Fungal communities in the subterranean regions (rhizosphere and root) are exceedingly diverse, according to the diversity index that was calculated for each possible combination of microhabitats. Furthermore, research showed that the fungal endophyte biodiversity in the above-ground components (leaves and stems) is not very great. Endophytic fungal species were most reliably identified in the root sample, rhizosphere, leaves, and stems, according to the Chao 1 and Shannon indices. The presence of a varied array of fungal species in the rhizosphere was shown using Simpson’s diversity index. It is clear that the stems have the lowest diversity indices throughout all parts of the sample, which indicates that the community’s phylotypes are most unevenly distributed. Based on this, our study also estimated the fungal diversity indices, such as the Shannon and Simpson indices, which underscore the complexity of the endophytic microbial community inhabiting H. binata, indicating a diverse assemblage of fungal taxa.
The investigation of endophytic fungal diversity often employs both culture-dependent and culture-independent methods, each offering distinct advantages in revealing community structure and composition. In this study, comparing the results obtained from these two approaches provides valuable insights into the fungal community associated with the host plant, H. binata. The culture-dependent method yielded a total of 25 independent, isolated fungal species, whereas the culture-independent method identified 269 Operational Taxonomic Units (OTUs) through sequencing. This discrepancy underscores the complementary nature of these techniques, with culture-dependent methods providing detailed insights into cultivable fungal species, including precise morphological characterization and taxonomic classification. According to research by [58], most genera found using the culture-independent approach are saprotrophs, while most genera found using the conventional method are phytopathogens. Although in six instances the identification is only up to the genus level owing to a data shortage, they were able to identify several taxa to the species level using the conventional strategy that combined morphological and genetic methods. However, it is important to note that several proportionately dominant taxa revealed by sequencing were also represented by isolates, indicating a degree of consistency between the two methods in capturing fungal diversity.
The fungal community analysis revealed a dominant Ascomycota phylum, comprising 71.3% of the community, with approximately 1% remaining unclassified under known phyla. This highlights both the complexity and diversity of the fungal community associated with H. binata, as well as the limitations of current taxonomic classifications in fully characterizing microbial populations. Both methods identified commonly found fungal taxa such as Aspergillus, Nigrospora, and Fusarium. However, the culture-independent approach unveiled additional frequently encountered genera that were missed using the conventional approach, including Dissoconium, Pichia, Lasiodiplodia, Cladosporium, Tilletia, Ramichloridium, and Trichothecium. The comparison of common and distinct fungi among the samples using a Venn diagram revealed a total of 189 fungal taxa identified through culture-independent approaches, with 17 fungal genera discovered using culture-dependent methods. Remarkably, 17 fungal genera were shared by both datasets, indicating a degree of consistency between culture-independent and culture-dependent methods in capturing a fraction of fungal diversity. Our findings were in line with those of [52], who also found that most of the OTUs were consistent between culture-dependent and culture-independent methods of detection, with the exception of Aspergillus (which was more common in the former but less so in the latter) and Cadophora sp. (a fungal taxon that was commonly found in the latter but not in the former). Because Aspergillus sp. may grow rapidly, even at low concentrations in the tissue samples, this finding might be due to their presence. So, they may proliferate rapidly and yield many isolates in the culture-dependent method, but they would go undetected in the next generation of sequencing.
Combining effective high-resolution phylogenetic fingerprinting with high-throughput culture methods allows us to recover previously unknown species of microorganisms, allowing us to discover and get certain unique or difficult-to-cultivate microbes [59]. The functional information of the fungal OTUs derived from culture-independent approaches may be better understood by comparing conventional and culture-independent data [58]. Integrating both culture-dependent and culture-independent methods enhances the comprehensive analysis of endophytic fungal communities. While culture-dependent methods provide detailed insights into cultivable species, they may underestimate fungal diversity due to limitations in culturability. Conversely, culture-independent approaches offer a broader view of fungal diversity by detecting non-cultivable taxa, albeit with challenges in taxonomic assignment. By combining these methodologies, researchers can gain a more holistic understanding of endophytic fungal community structure and composition, contributing to our knowledge of fungal ecology and its implications for plant health and ecosystem dynamics. Overall, this study has demonstrated the promise of combining both methods in a single study to connect NGS datasets with morphologically and phylogenetically identified fungal isolates from culture. This complementary approach allows for accurate taxonomic labeling of fungal endophytes found using NGS, which is important because NGS studies without reference cultures can be misleading and a lot of the data from the past needs to be viewed critically.
5. Conclusions
This study explores the diversity of endophytic fungal communities within H. binata leaves using both culture-dependent and culture-independent methods. Culture-dependent methods allowed for the isolation and identification of cultivable fungal species, while culture-independent techniques allowed for the detection of non-cultivable taxa. The study found that endophytic fungi play a crucial role in plant health and defense mechanisms in the presence of the Ascomycota phylum. The identification of bioactive metabolites produced by these fungi also suggests their potential pharmaceutical applications in medicine and biotechnology. The study used high-throughput sequencing technologies to bridge the gap between NGS datasets and taxonomically identified fungal isolates, ensuring accurate taxonomic labeling and enhancing the reliability of fungal community studies. The study also highlighted the complementary nature of these techniques in elucidating fungal ecology. The combined use of culture-dependent and culture-independent methods holds promise for advancing our understanding of fungal ecology and its implications for plant health and ecosystem dynamics. Future research should continue to use both methodologies to unravel the complex relationships between endophytic fungi, their host plants, and the environment.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microbiolres15020053/s1, Figure S1: ITS2 Secondary structure predicted phylogenetic tree of H. binata; Table S1: Compensatory base changes (CBC) analysis in the sequences of H. binata. Table S2: Culture-independent molecular approaches reveal the diversity of fungal endophytic communities in leaves of H. binata.
Author Contributions
M.J.X.S., M.T. and I.M.: investigation, methodology, conceptualization, writing—original draft and editing, P.R. and N.P.: writing—review and editing. M.P.: supervision, conceptualization, writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the “RUSA” Madurai Kamaraj University, Madurai, India.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data supporting the findings are provided in the Supplementary File.
Acknowledgments
The authors acknowledge “DST-PURSEand RUSA” Madurai Kamaraj University, Madurai, India for the instrument facility.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. No conflict of interest exists in the submission of this manuscript, and manuscript is approved by all authors for publication.
References
- Hyde, K.D.; Soytong, K. The fungal endophyte dilemma. Fungal Divers. 2018, 33, e173. [Google Scholar]
- Jin, H.; Yang, X.; Lu, D.; Li, C.; Yan, Z.; Li, X.; Zeng, L.; Qin, B. Phylogenic diversity and tissue specificity of fungal endophytes associated with the pharmaceutical plant, Stellera chamaejasme L. revealed by a cultivation-independent approach. Antonie Van Leeuwenhoek 2015, 108, 835–850. [Google Scholar] [CrossRef]
- Fadiji, A.E.; Ayangbenro, A.S.; Babalola, O.O. Metagenomic profiling of the community structure, diversity, and nutrient pathways of bacterial endophytes in maize plant. Antonie Van Leeuwenhoek 2020, 113, 1559–1571. [Google Scholar] [CrossRef]
- Wu, B.; Hussain, M.; Zhang, W.; Stadler, M.; Liu, X.; Xiang, M. Current insights into fungal species diversity and perspective on naming the environmental DNA sequences of fungi. Mycology 2019, 10, 127–140. [Google Scholar] [CrossRef]
- Verma, V.C.; Singh, S.K.; Kharwar, R.N. Histological Investigation of Fungal Endophytes in Healthy Tissues of Azadirachta indica A. Juss. Agric. Nat. Resour. 2012, 46, 229–237. [Google Scholar]
- Arnold, A.E.; Henk, D.A.; Eells, R.L.; Lutzoni, F.; Vilgalys, R. Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR. Mycologia 2007, 99, 185–206. [Google Scholar] [CrossRef]
- Suryanarayanan, T.S.; Murali, T.S.; Thirunavukkarasu, N.; Rajulu, G.; Venkatesan, G.; Sukumar, R. Endophytic fungal communities in woody perennials of three tropical forest types of the Western Ghats, southern India. Biodivers. Conserv. 2011, 20, 913–928. [Google Scholar] [CrossRef]
- Shingade, S.P.; Kakde, R.B. A Review on “Anjan” Hardwickia binata Roxb: Its Phytochemical Studies, Traditional uses and Pharmacological activities. Pharmacogn. Rev. 2021, 15, 65. [Google Scholar] [CrossRef]
- Høyer, A.K.; Hodkinson, T.R. Hidden fungi: Combining culture-dependent and-independent DNA barcoding reveals inter-plant variation in species richness of endophytic root fungi in Elymus repens. J. Fungi 2021, 7, 466. [Google Scholar] [CrossRef]
- Hawksworth, D.L.; Lücking, R. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol. Spectr. 2017, 5, 1–17. [Google Scholar] [CrossRef]
- Miguel, P.S.B.; de Oliveira, M.N.V.; Delvaux, J.C.; de Jesus, G.L.; Borges, A.C.; Tótola, M.R.; Costa, M.D. Diversity and distribution of the endophytic bacterial community at different stages of Eucalyptus growth. Antonie Van Leeuwenhoek 2016, 109, 755–771. [Google Scholar] [CrossRef]
- Mashiane, R.A.; Ezeokoli, O.T.; Adeleke, R.A.; Bezuidenhout, C.C. Metagenomic analyses of bacterial endophytes associated with the phyllosphere of a Bt maize cultivar and its isogenic parental line from South Africa. World J. Microbiol. Biotechnol. 2017, 33, 1–12. [Google Scholar] [CrossRef]
- Bálint, M.; Bahram, M.; Eren, A.M.; Faust, K.; Fuhrman, J.A.; Lindahl, B.; Tedersoo, L. Millions of reads, thousands of taxa: Microbial community structure and associations analyzed via marker genes. FEMS Microbiol. Rev. 2016, 40, 686–700. [Google Scholar] [CrossRef]
- Peršoh, D. Plant-associated fungal communities in the light of meta’omics. Fungal Divers. 2015, 75, 1–25. [Google Scholar] [CrossRef]
- Ghosh, S.; Das, A.P. Metagenomic insights into the microbial diversity in manganese-contaminated mine tailings and their role in biogeochemical cycling of manganese. Sci. Rep. 2018, 8, 8257. [Google Scholar] [CrossRef]
- Alves, L.D.F.; Westmann, C.A.; Lovate, G.L.; de Siqueira, G.M.V.; Borelli, T.C.; Guazzaroni, M.E. Metagenomic approaches for understanding new concepts in microbial science. Int. J. Genom. 2018, 2018, 2312987. [Google Scholar] [CrossRef]
- Shokralla, S.; Spall, J.L.; Gibson, J.F.; Hajibabaei, M. Next-generation sequencing technologies for environmental DNA research. Mol. Ecol. 2012, 21, 1794–1805. [Google Scholar] [CrossRef]
- Al-Bulushi, I.M.; Bani-Uraba, M.S.; Guizani, N.S.; Al-Khusaibi, M.K.; Al-Sadi, A.M. Illumina MiSeq sequencing analysis of fungal diversity in stored dates. BMC Microbiol. 2017, 17, 72. [Google Scholar] [CrossRef]
- Mani, I.; Thangavel, M.; Surendrababu, A.; Sneha, M.; Rajagopal, R.; Alfarhan, A.; Pandi, M. Unveiling the Bioprospecting Efficacy and Textile Dyeing of a Novel Endophytic Mycobial Red Pigment. Indian J. Microbiol. 2024, 1–17. [Google Scholar] [CrossRef]
- Kjer, J.; Debbab, A.; Aly, A.H.; Proksch, P. Methods for isolation of marine-derived endophytic fungi and their bioactive secondary products. Nat. Protoc. 2010, 5, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Cenis, J.L. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res. 1992, 20, 2380. [Google Scholar] [CrossRef] [PubMed]
- GokulRaj, K.; Sundaresan, N.; Ganeshan, E.J.; Rajapriya, P.; Muthumary, J.; Sridhar, J.; Pandi, M. Phylogenetic reconstruction of endophytic fungal isolates using internal transcribed spacer 2 (ITS2) region. Bioinformation 2014, 10, 320. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Schleicher, T.; Schultz, J.; Müller, T.; Dandekar, T.; Wolf, M. 5.8 S-28S rRNA interaction and HMM-based ITS2 annotation. Gene 2009, 430, 50–57. [Google Scholar] [CrossRef]
- Del Carmen Flores-Vallejo, R.; Folch-Mallol, J.L.; Sharma, A.; Cardoso-Taketa, A.; Alvarez-Berber, L.; Villarreal, M.L. ITS2 ribotyping, in vitro anti-inflammatory screening, and metabolic profiling of fungal endophytes from the Mexican species Crescentia alata Kunth. S. Afr. J. Bot. 2020, 134, 213–224. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.; Bahram, M.; Larsson, K.H. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef]
- Nair, D.N.; Padmavathy, S.J.T.S.W.J. Impact of endophytic microorganisms on plants, environment and humans. Sci. World J. 2014, 2014, 250693. [Google Scholar] [CrossRef]
- Dos Reis, J.B.A.; Lorenzi, A.S.; do Vale, H.M.M. Methods used for the study of endophytic fungi: A review on methodologies and challenges, and associated tips. Arch. Microbiol. 2022, 204, 675. [Google Scholar] [CrossRef]
- Verma, V.; Srivastava, A.; Garg, S.K.; Singh, V.P.; Arora, P.K. Incorporating omics-based tools into endophytic fungal research. Biotechnol. Notes 2023, 5, 1–7. [Google Scholar] [CrossRef]
- Hirakue, A.; Sugiyama, S. Relationship between foliar endophytes and apple cultivar disease resistance in an organic orchard. Biol. Control 2018, 127, 139–144. [Google Scholar] [CrossRef]
- Adeleke, B.S.; Babalola, O.O. Pharmacological potential of fungal endophytes associated with medicinal plants: A review. J. Fungi 2021, 7, 147. [Google Scholar] [CrossRef]
- Ahrendt, S.R.; Quandt, C.A.; Ciobanu, D.; Clum, A.; Salamov, A.; Andreopoulos, B.; Grigoriev, I.V. Leveraging single-cell genomics to expand the fungal tree of life. Nat. Microbiol. 2018, 3, 1417–1428. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Bahram, M.; Sánchez-Castro, I.; Dai, D.Q.; Ariyawansa, K.G.; Jayalal, U.; Tedersoo, L. Current insight into culture-dependent and culture-independent methods in discovering Ascomycetous Taxa. J. Fungi 2021, 7, 703. [Google Scholar] [CrossRef]
- Yao, Y.Q.; Lan, F.; Qiao, Y.M.; Wei, J.G.; Huang, R.S.; Li, L.B. Endophytic fungi harbored in the root of Sophora tonkinensis Gapnep: Diversity and biocontrol potential against phytopathogens. MicrobiologyOpen 2017, 6, e00437. [Google Scholar] [CrossRef]
- Gong, A.; Zhou, T.; Xiao, C.; Jiang, W.; Zhou, Y.; Zhang, J.; Zhang, C. Association between dipsacus saponin VI level and diversity of endophytic fungi in roots of Dipsacus asperoides. World J. Microbiol. Biotechnol. 2019, 35, 1–14. [Google Scholar] [CrossRef]
- Rojas, E.C.; Sapkota, R.; Jensen, B.; Jørgensen, H.J.; Henriksson, T.; Jørgensen, L.N.; Collinge, D.B. Fusarium head blight modifies fungal endophytic communities during infection of wheat spikes. Microb. Ecol. 2020, 79, 397–408. [Google Scholar] [CrossRef]
- Ruiz Gómez, F.J.; Navarro-Cerrillo, R.M.; Pérez-de-Luque, A.; Oβwald, W.; Vannini, A.; Morales-Rodríguez, C. Assessment of functional and structural changes of soil fungal and oomycete communities in holm oak declined dehesas through metabarcoding analysis. Sci. Rep. 2019, 9, 5315. [Google Scholar] [CrossRef]
- Forbes, J.D.; Knox, N.C.; Ronholm, J.; Pagotto, F. Metagenomics: The next culture-independent game changer. Front. Microbiol. 2017, 8, 261928. [Google Scholar] [CrossRef]
- Parmar, S.; Li, Q.; Wu, Y.; Li, X.; Yan, J.; Sharma, V.K.; Li, H. Endophytic fungal community of Dysphania ambrosioides from two heavy metal-contaminated sites: Evaluated by culture-dependent and culture-independent approaches. Microb. Biotechnol. 2018, 11, 1170–1183. [Google Scholar] [CrossRef]
- Khalil, A.M.A.; Hassan, S.E.D.; Alsharif, S.M.; Eid, A.M.; Ewais, E.E.D.; Azab, E.; Fouda, A. Isolation and characterization of fungal endophytes isolated from medicinal plant Ephedra pachyclada as plant growth-promoting. Biomolecules 2021, 11, 140. [Google Scholar] [CrossRef]
- Amirita, A.; Sindhu, P.; Swetha, J.; Vasanthi, N.S.; Kannan, K.P. Enumeration of endophytic fungi from medicinal plants and screening of extracellular enzymes. World J. Sci. Technol. 2012, 2, 13–19. [Google Scholar]
- Rampersad, S.N. ITS1, 5.8 S and ITS2 secondary structure modelling for intra-specific differentiation among species of the Colletotrichum gloeosporioides sensu lato species complex. SpringerPlus 2014, 3, 684. [Google Scholar] [CrossRef]
- Kapoor, N.; Gambhir, L.; Saxena, S. Secondary structure prediction of ITS rRNA region and molecular phylogeny: An integrated approach for the precise speciation of Muscodor species. Ann. Microbiol. 2018, 68, 763–772. [Google Scholar] [CrossRef]
- Prahl, R.E.; Khan, S.; Deo, R.C. The role of internal transcribed spacer 2 secondary structures in classifying mycoparasitic Ampelomyces. PLoS ONE 2021, 16, e0253772. [Google Scholar] [CrossRef]
- Koetschan, C.; Kittelmann, S.; Lu, J.; Al-Halbouni, D.; Jarvis, G.N.; Müller, T.; Janssen, P.H. Internal transcribed spacer 1 secondary structure analysis reveals a common core throughout the anaerobic fungi (Neocallimastigomycota). PLoS ONE 2014, 9, e91928. [Google Scholar] [CrossRef]
- Ahvenniemi, P.; Wolf, M.; Lehtonen, M.J.; Wilson, P.; German-Kinnari, M.; Valkonen, J.P. Evolutionary diversification indicated by compensatory base changes in ITS2 secondary structures in a complex fungal species, Rhizoctonia solani. J. Mol. Evol. 2009, 69, 150–163. [Google Scholar] [CrossRef]
- Sundaresan, N.; Sahu, A.K.; Jagan, E.G.; Pandi, M. Evaluation of ITS2 molecular morphometrics effectiveness in species delimitation of Ascomycota–A pilot study. Fungal Biol. 2019, 123, 517–527. [Google Scholar] [CrossRef]
- Pang, K.L.; Guo, S.Y.; Chen, I.A.; Burgaud, G.; Luo, Z.H.; Dahms, H.U.; Cha, H.J. Insights into fungal diversity of a shallow-water hydrothermal vent field at Kueishan Island, Taiwan by culture-based and metabarcoding analyses. PLoS ONE 2019, 14, e0226616. [Google Scholar] [CrossRef]
- Chi, W.C.; Chen, W.; He, C.C.; Guo, S.Y.; Cha, H.J.; Tsang, L.M.; Pang, K.L. A highly diverse fungal community associated with leaves of the mangrove plant Acanthus ilicifolius var. xiamenensis revealed by isolation and metabarcoding analyses. PeerJ 2019, 7, e7293. [Google Scholar] [CrossRef]
- Donovan, P.D.; Gonzalez, G.; Higgins, D.G.; Butler, G.; Ito, K. Identification of fungi in shotgun metagenomics datasets. PLoS ONE 2018, 13, e0192898. [Google Scholar] [CrossRef]
- Sugiyama, A.; Vivanco, J.M.; Jayanty, S.S.; Manter, D.K. Pyrosequencing assessment of soil microbial communities in organic and conventional potato farms. Plant Dis. 2010, 94, 1329–1335. [Google Scholar] [CrossRef]
- Dissanayake, A.J.; Purahong, W.; Wubet, T.; Hyde, K.D.; Zhang, W.; Xu, H.; Yan, J. Direct comparison of culture-dependent and culture-independent molecular approaches reveal the diversity of fungal endophytic communities in stems of grapevine (Vitis vinifera). Fungal Divers. 2018, 90, 85–107. [Google Scholar] [CrossRef]
- Nischitha, R.; Shivanna, M.B. Comparative Metagenomic Analyses of Endophytic Fungi Assemblages in Shoot and Root Regions of Heteropogon contortus. Proc. Natl. Acad. Sci. USA 2024, 94, 161–167. [Google Scholar] [CrossRef]
- Kaul, S.; Sharma, T.; Dhar, M.K. “Omics” tools for better understanding the plant–endophyte interactions. Front. Plant Sci. 2016, 7, 183603. [Google Scholar] [CrossRef]
- Zhang, Q.; Xue, X.Z.; Miao, S.M.; Cui, J.L.; Qin, X.M. Differential relationship of fungal endophytic communities and metabolic profiling in the stems and roots of Ephedra sinica based on metagenomics and metabolomics. Symbiosis 2020, 81, 115–125. [Google Scholar] [CrossRef]
- Macías-Rubalcava, M.L.; Sánchez-Fernández, R.E.; Roque-Flores, G.; Lappe-Oliveras, P.; Medina-Romero, Y.M. Volatile organic compounds from Hypoxylon anthochroum endophytic strains as postharvest mycofumigation alternative for cherry tomatoes. Food Microbiol. 2018, 76, 363–373. [Google Scholar] [CrossRef]
- Sumbula, V.; Kurian, P.S.; Girija, D.; Cherian, K.A. Impact of foliar application of fungicides on tomato leaf fungal community structure revealed by metagenomic analysis. Folia Microbiol. 2022, 67, 103–108. [Google Scholar] [CrossRef]
- Jayawardena, R.S.; Purahong, W.; Zhang, W. Biodiversity of fungi on Vitis vinifera L. revealed by traditional and high-resolution culture-independent approaches. Fungal Divers. 2018, 90, 1–84. [Google Scholar] [CrossRef]
- Pei, C.; Mi, C.; Sun, L.; Liu, W.; Li, O.; Hu, X. Diversity of endophytic bacteria of Dendrobium officinale based on culture-dependent and culture-independent methods. Biotechnol. Biotechnol. Equip. 2017, 31, 112–119. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).