The Effect of Conserved Histidine on the Proximity of Fe-S Clusters in Adenosine-5′-Phosphosulfate Reductases from Pseudomonas aeruginosa and Enteromorpha intestinalis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid, Site-Directed Mutagenesis, and Recombinant Protein Expressions
2.2. Recombinant Protein Expression and Purification
2.3. Iron Contents
2.4. Sulfide Contents
2.5. Enzyme Activity Assay
2.6. Binding Assay
2.7. Statistical Analysis
3. Results and Discussion
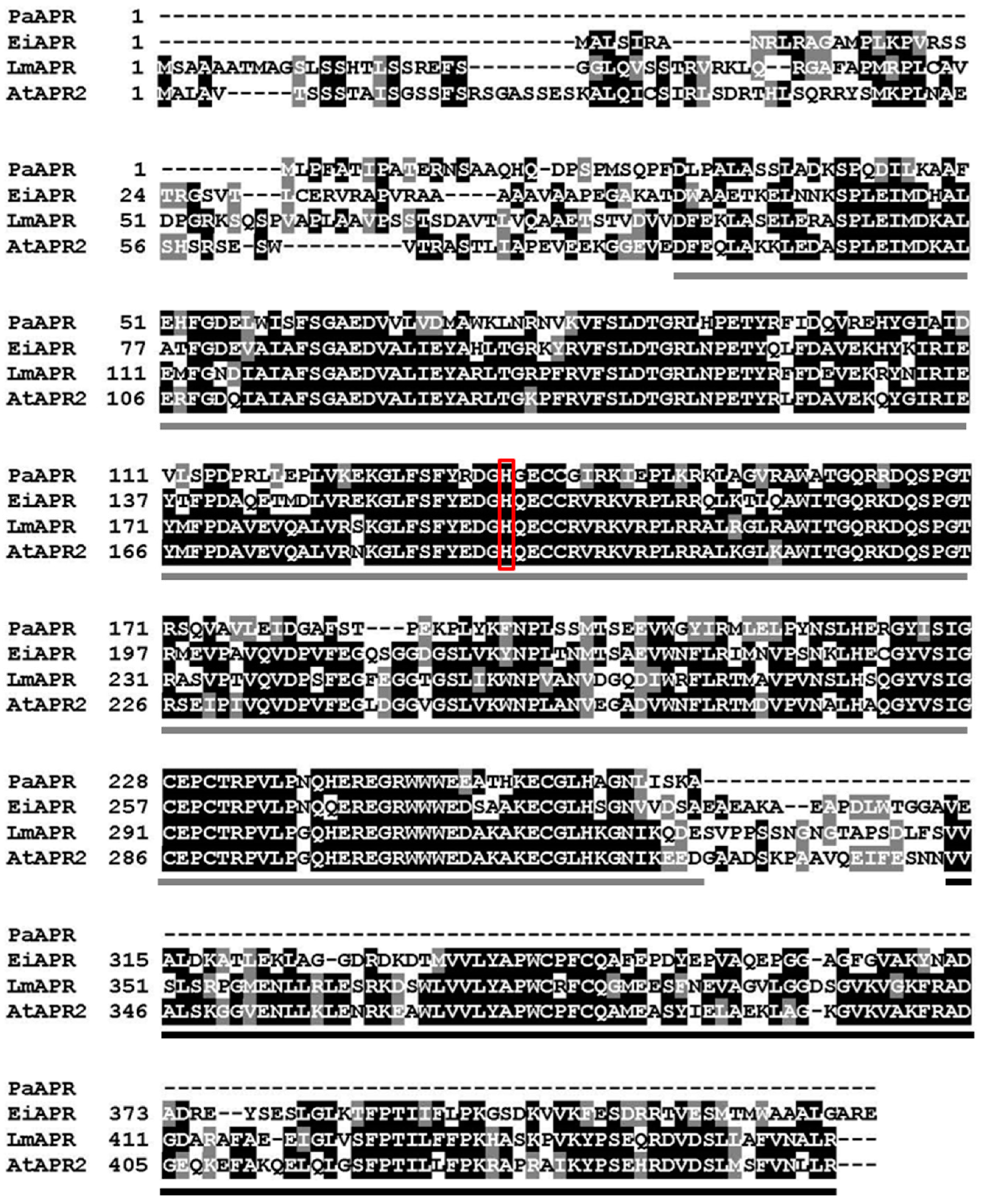
3.1. Molecular Cloning and Protein Expression and Purification
3.2. Protein Spectra, and Iron and Sulfur Contents
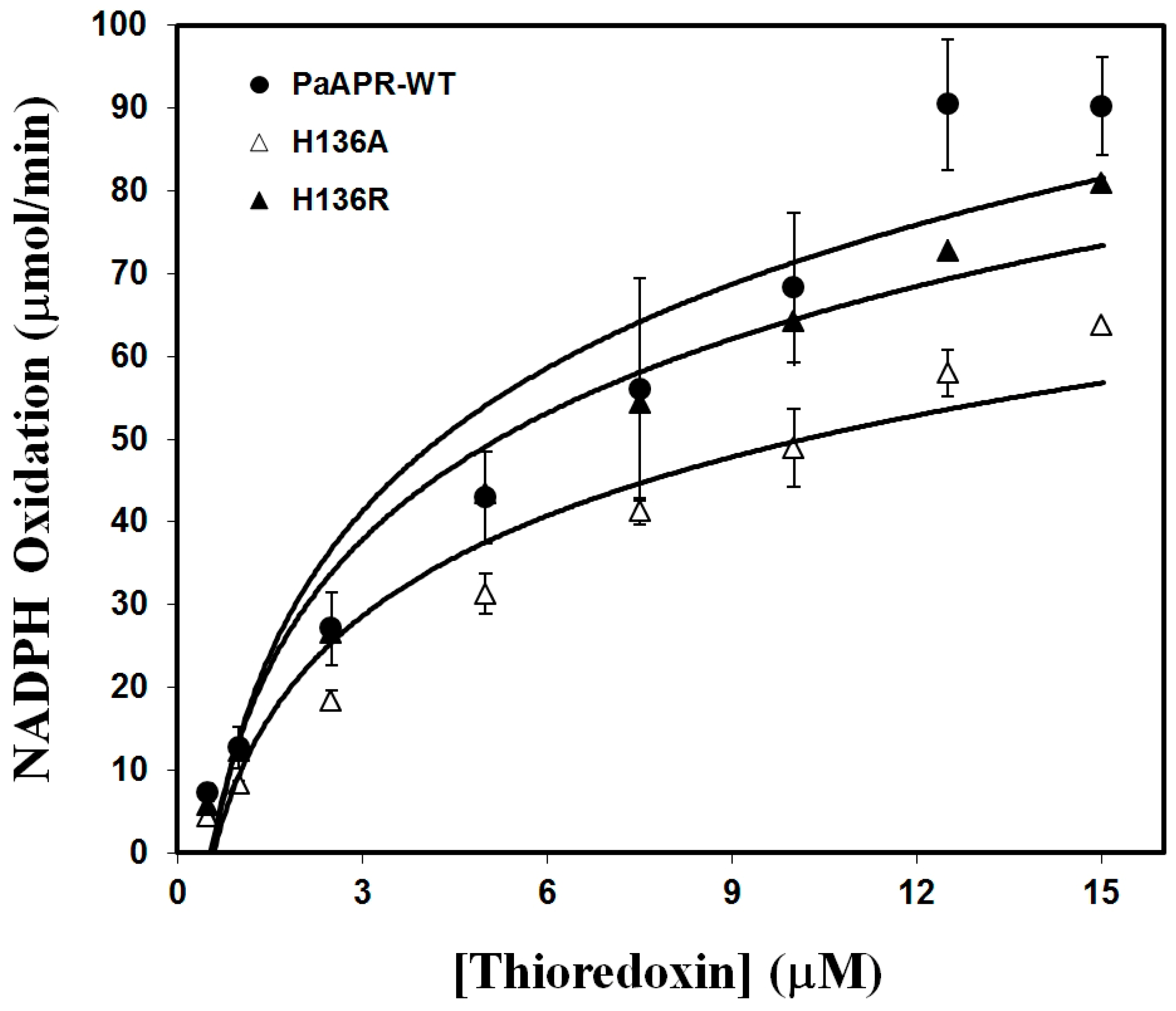
3.3. PaAPR and Variant Activity and Kinetic Analysis
3.4. EiAPR and Variants Activity and Kinetic Analysis
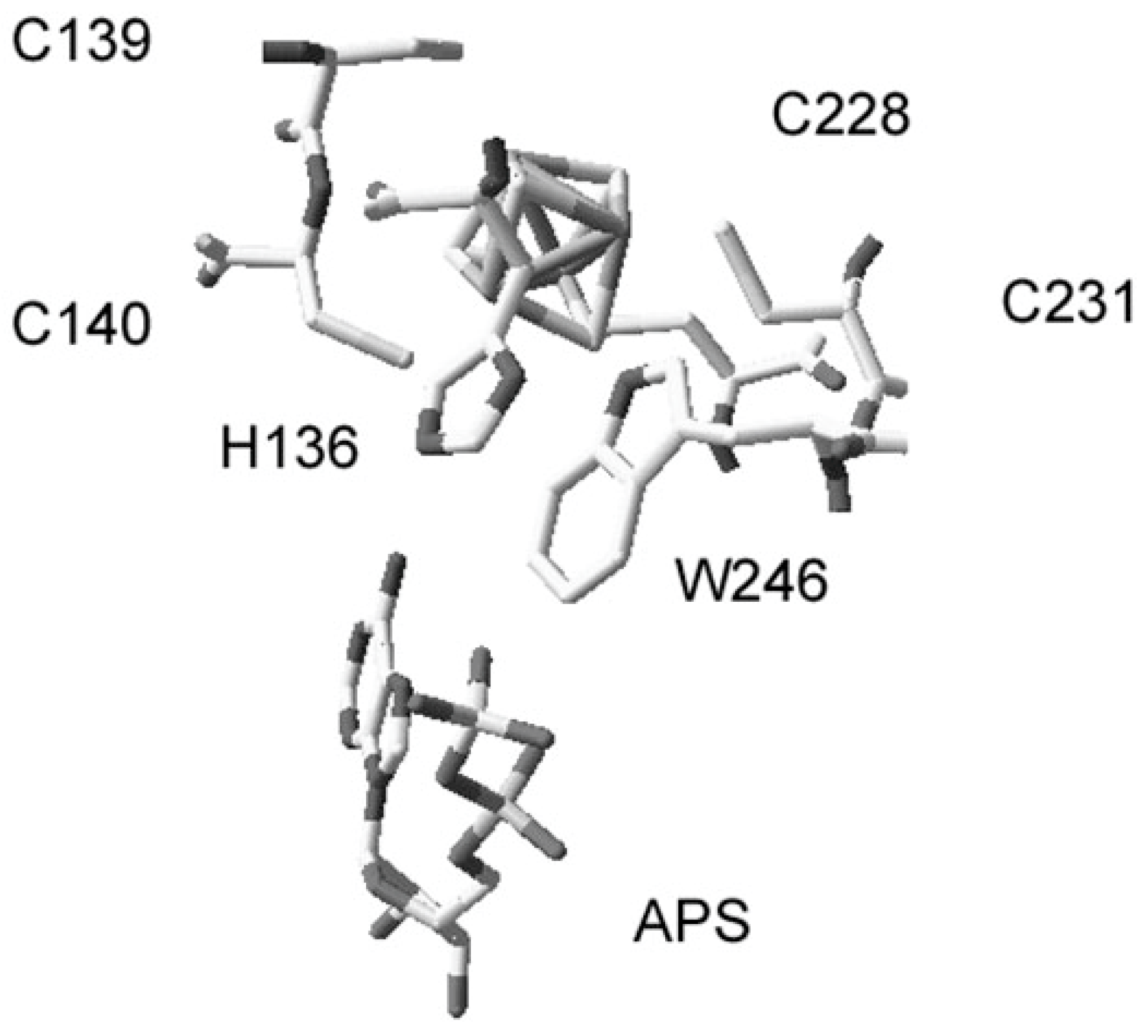
3.5. Histidine Substitutions in APS Reductase: Activity and Binding Impacts
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Takahashi, H.; Kopriva, S.; Giordano, M.; Saito, K.; Hell, R. Sulfur assimilation in photosynthetic organ-isms: Molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 2011, 62, 157–184. [Google Scholar] [CrossRef] [PubMed]
- Sauter, M.; Moffatt, B.; Saechao, M.C.; Hell, R.; Wirtz, M. Methionine salvage and S-adenosylmethionine: Essential links between sulfur, ethylene and polyamine biosynthesis. Biochem. J. 2013, 451, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Calderwood, A.; Kopriva, S. Hydrogen sulfide in plants: From dissipation of excess sulfur to sig-naling molecule. Nitric Oxide. 2014, 41, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Ravilious, G.E.; Jez, J.M. Structural biology of plant sulfur metabolism: From assimilation to biosynthesis. Nat. Prod. Rep. 2012, 29, 1138–1152. [Google Scholar] [CrossRef] [PubMed]
- Saito, K. Sulfur assimilatory metabolism. The long and smelling road. Plant Physiol. 2004, 136, 2443–2450. [Google Scholar] [CrossRef]
- Brunold, C. Reduction of sulfate to sulfide. In Sulfur Nutrition and Sulfur Assimilation in Higher Plants: Fundamental, Environmental and Agricultural Aspects; Rennenberg, H., Brunold, C., De Kok, L.J., Stulen, I., Eds.; SPB Academic Publishing: The Hague, Germany, 1990; pp. 13–31. ISBN 90-5103-038-X. [Google Scholar]
- Kopriva, S.; Buchert, T.; Fritz, G.; Suter, M.; Benda, R.; Schunemann, V.; Koprivova, A.; Schurmann, P.; Trautwein, A.X.; Kroneck, P.M.; et al. The presence of an ironsulfur cluster in adenosine 5′-phosphosulfate reductase separates organisms utilizing adenosine 5′-phosphosulfate and phosphoadeno-sine 5′-phosphosulfate for sulfate assimilation. J. Biol. Chem. 2002, 277, 21786–21791. [Google Scholar] [CrossRef]
- Noctor, G.; Arisi, A.M.; Jouanin, L.; Kunert, K.J.; Rennenberg, H.; Foyer, C.H. Glutathione: Biosynthesis, metabolism and relationship to stress tolerance explored in transformed plants. J. Exp. Bot. 1998, 49, 623–647. [Google Scholar] [CrossRef]
- Bick, J.A.; Åslund, F.; Chen, Y.; Leustek, T. Glutaredoxin function for the carboxyl terminal domain of the plant-type 5‘-adenylyl sulfate (APS) reductase. Proc. Natl. Acad. Sci. USA 1998, 95, 8404–8409. [Google Scholar] [CrossRef] [PubMed]
- Lillig, C.H.; Prior, A.; Schwenn, J.D.; Aslund, F.; Ritz, D.; Vlamis-Gardikas, A.; Holmgren, A. New thi-oredoxins and glutaredoxins as electron donors of 3′-phosphoadenylylsulfate reductase. J. Biol. Chem. 1999, 274, 7695–7698. [Google Scholar] [CrossRef]
- Ravilious, G.E.; Nguyen, A.; Francois, J.A.; Jez, J.M. Structural basis and evolution of redox regulation in plant adenosine-5′-phosphosulfate kinase. Proc. Natl. Acad. Sci. USA 2012, 109, 309–314. [Google Scholar] [CrossRef]
- Koprivova, A.; Kopriva, S. Sulfation pathways in plants. Chem. Biol. Interact. 2016, 259, 23–30. [Google Scholar] [CrossRef]
- Weber, M.; Suter, M.; Brunold, C.; Kopriva, S. Sulfate assimilation in higher plants characterization of a stable intermediate in the adenosine 5‘-phosphosulfate reductase reaction. Eur. J. Biochem. 2000, 267, 3647–3653. [Google Scholar] [CrossRef]
- Kim, S.K.; Rahman, A.; Conover, R.C.; Johnson, M.K.; Mason, J.T.; Hirasawa, M.; Moore, M.L.; Leustek, T.; Knaff, D.B. Properties of the cysteine residues and the iron-sulfur cluster of the assimilatory 5′-adenylyl sulfate reductase from Enteromorpha intestinalis. Biochemistry 2006, 45, 5010–5018. [Google Scholar] [CrossRef]
- Kim, S.K.; Rahman, A.; Bick, J.A.; Conover, R.C.; Johnson, M.K.; Mason, J.T.; Hirasawa, M.; Leustek, T.; Knaff, D.B. Properties of the cysteine residues and iron−sulfur cluster of the assimilatory 5‘-adenylylsulfate reductase from Pseudomonas aeruginosa. Biochemistry 2004, 43, 13478–13486. [Google Scholar] [CrossRef]
- Carroll, K.S.; Gao, H.; Chen, H.; Stout, C.D.; Leary, J.A.; Bertozzi, C.R. A conserved mechanism for sulfonucleotide reduction. PLoS Biol. 2005, 3, e250. [Google Scholar] [CrossRef]
- Chartron, J.; Carroll, K.S.; Shiau, C.; Gao, H.; Leary, J.A.; Bertozzi, C.R.; Stout, C.D. Substrate recogni-tion, protein dynamics, and iron-sulfur cluster in Pseudomonas aeruginosa adenosine-5′- phosphsulfate re-ductase. J. Mol. Biol. 2006, 364, 162–169. [Google Scholar] [CrossRef]
- Feliciano, P.R.; Carroll, K.S.; Drennan, C.L. Crystal Structure of the [4Fe-4S] Cluster-Containing Aden-osine-5′-phosphosulfate Reductase from Mycobacterium tuberculosis. ACS Omega 2021, 6, 13756–13765. [Google Scholar] [CrossRef]
- Carroll, K.S.; Gao, H.; Chen, H.; Leary, J.A.; Bertozzi, C.R. Investigation of the iron-sulfur cluster in Mycobacterium tuberculosis APS reductase: Implications for substrate binding and catalysis. Biochemistry. 2005, 44, 14647–14657. [Google Scholar] [CrossRef]
- Kopriva, S.; Buchert, T.; Fritz, G.; Suter, M.; Weber, M.; Benda, R.; Schaller, J.; Feller, U.; Schurmann, P.; Schunemann, V.; et al. Plant adenosine 5′-phosphosulfate re-ductase is a novel iron-sulfur protein. J. Biol. Chem. 2001, 276, 42881–42886. [Google Scholar] [CrossRef]
- Gao, Y.; Schofield, O.; Leustek, T. Characterization of sulfate assimilation in marine algae focusing on the enzyme 5‘-adenylylsulfate (APS) reductase. Plant Physiol. 2000, 123, 1087–1096. [Google Scholar] [CrossRef]
- Chung, J.S.; Noguera-Mazon, V.; Lancelin, J.M.; Kim, S.K.; Hirasawa, M.; Hologne, M.; Leustek, T.; Knaff, D.B. The Interaction Domain on Thioredoxin for Pseudomonas aeruginosa 5′-adenylylsulfate Re-ductase. J. Biol. Chem. 2009, 284, 31181–31189. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Massey, V. Studies on succinic dehydrogenase: VII. Valency state of the iron in beef heart succinic de-hydrogenase. J. Biol. Chem. 1957, 229, 763–770. [Google Scholar] [CrossRef] [PubMed]
- King, T.E.; Morris, R.O. Determination of acid-labile sulfide and sulfhydryl groups. Methods Enzymol. 1967, 10, 634–641. [Google Scholar] [CrossRef]
- Srivastava, A.P.; Knaff, D.B.; Sétif, P. Kinetic studies of a ferredoxin-dependent cyanobacterial nitrate reductase. Biochemistry 2014, 53, 5092–5101. [Google Scholar] [CrossRef]
- Bhave, D.P.; Hong, J.A.; Keller, R.L.; Krebs, C.; Carroll, K.S. Iron-sulfur cluster engineering provides insight into the evolution of substrate specificity among sulfonucleotide reductases. ACS Chem. Biol. 2012, 7, 306–315. [Google Scholar] [CrossRef]

| Gene | Primer | Sequence 5′-3′ | Mutated Codon |
|---|---|---|---|
| PaAPR | H136A For | TTC TAC CGG GAC GGC GCC GGC GAG TGC TGC GGC | CAC→GCC |
| H136A Rev | GCC GCA GCA CTC GCC GGC GCC GTC CCG GTA GAA | ||
| H136R For | TTC TAC CGG GAC GGC CGC GGC GAG TGC TGC GGC | CAC→CGC | |
| H136R Rev | GCC GCA GCA CTC GCC GCG GCC GTC CCG GTA GAA | ||
| EiAPR | H162A For | TTC TAC GAG GAC GGC GCC CAA GAG TGC TGC CGC | CAT→GCC |
| H162A Rev | GCG GCA GCA CTC TTG GGC GCC GTC CTC GTA GAA | ||
| H162R For | TTC TAC GAG GAC GGC CGC CAA GAG TGC TGC CGC | CAT→CGC | |
| H162R Rev | GCG GCA GCA CTC TTG GCG GCC GTC CTC GTA GAA |
| Variant | Sulfide Content (nmol/nmol of Protein) | Iron Content (nmol/nmol of Protein) | |
|---|---|---|---|
| PaAPR | Wild-type | 3.58 ± 0.30 a | 3.95 ± 0.08 a |
| H136A | 2.51 ± 0.06 b | 3.25 ± 0.30 b | |
| H136R | 3.32 ± 0.38 a | 3.93 ± 0.08 a | |
| EiAPR | Wild-type | 3.44 ± 0.63 a | 3.76 ± 0.03 a |
| H162A | 2.14 ± 0.48 b | 2.91 ± 0.33 b | |
| H162R | 3.37 ± 0.20 a | 3.86 ± 0.07 a |
| Variant | Relative Vmax * | Km (μM) | Kd (μM) | |
| (APS) | ||||
| PaAPR | Wild-type | 100.00 ± 11.57 a | 8.33 ± 1.65 a | 20 a |
| H136A | 84.66 ± 3.07 b | 10.55 ± 0.78 b | 70 b | |
| H136R | 102.46 ± 8.60 a | 8.55 ± 2.05 a | 14.8 a |
| Variant | Relative Vmax * | Km (μM) | Kd (μM) | |
| (APS) | ||||
| EiAPR | Wild-type | 100.00 ± 32.68 a | 83.35 ± 23.55 a | 20 a |
| H162A | 48.06 ± 15.68 b | 204.15 ± 5.87 b | 40 b | |
| H162R | 83.18 ± 13.07 a | 183.35 ± 23.55 b | 32 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chung, J.-S.; Kim, S.-K.; Leustek, T. The Effect of Conserved Histidine on the Proximity of Fe-S Clusters in Adenosine-5′-Phosphosulfate Reductases from Pseudomonas aeruginosa and Enteromorpha intestinalis. Microbiol. Res. 2024, 15, 457-467. https://doi.org/10.3390/microbiolres15020031
Chung J-S, Kim S-K, Leustek T. The Effect of Conserved Histidine on the Proximity of Fe-S Clusters in Adenosine-5′-Phosphosulfate Reductases from Pseudomonas aeruginosa and Enteromorpha intestinalis. Microbiology Research. 2024; 15(2):457-467. https://doi.org/10.3390/microbiolres15020031
Chicago/Turabian StyleChung, Jung-Sung, Sung-Kun Kim, and Thomas Leustek. 2024. "The Effect of Conserved Histidine on the Proximity of Fe-S Clusters in Adenosine-5′-Phosphosulfate Reductases from Pseudomonas aeruginosa and Enteromorpha intestinalis" Microbiology Research 15, no. 2: 457-467. https://doi.org/10.3390/microbiolres15020031
APA StyleChung, J.-S., Kim, S.-K., & Leustek, T. (2024). The Effect of Conserved Histidine on the Proximity of Fe-S Clusters in Adenosine-5′-Phosphosulfate Reductases from Pseudomonas aeruginosa and Enteromorpha intestinalis. Microbiology Research, 15(2), 457-467. https://doi.org/10.3390/microbiolres15020031





_Kim.png)

