Biodegradation of Free Cyanide by a New Isolated Alkaliphilic Bacillus licheniformis Strain
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation Sources of Microorganisms
2.2. Microbial Enrichment Cultures
2.3. Microbial Isolation and Morphological Characterization
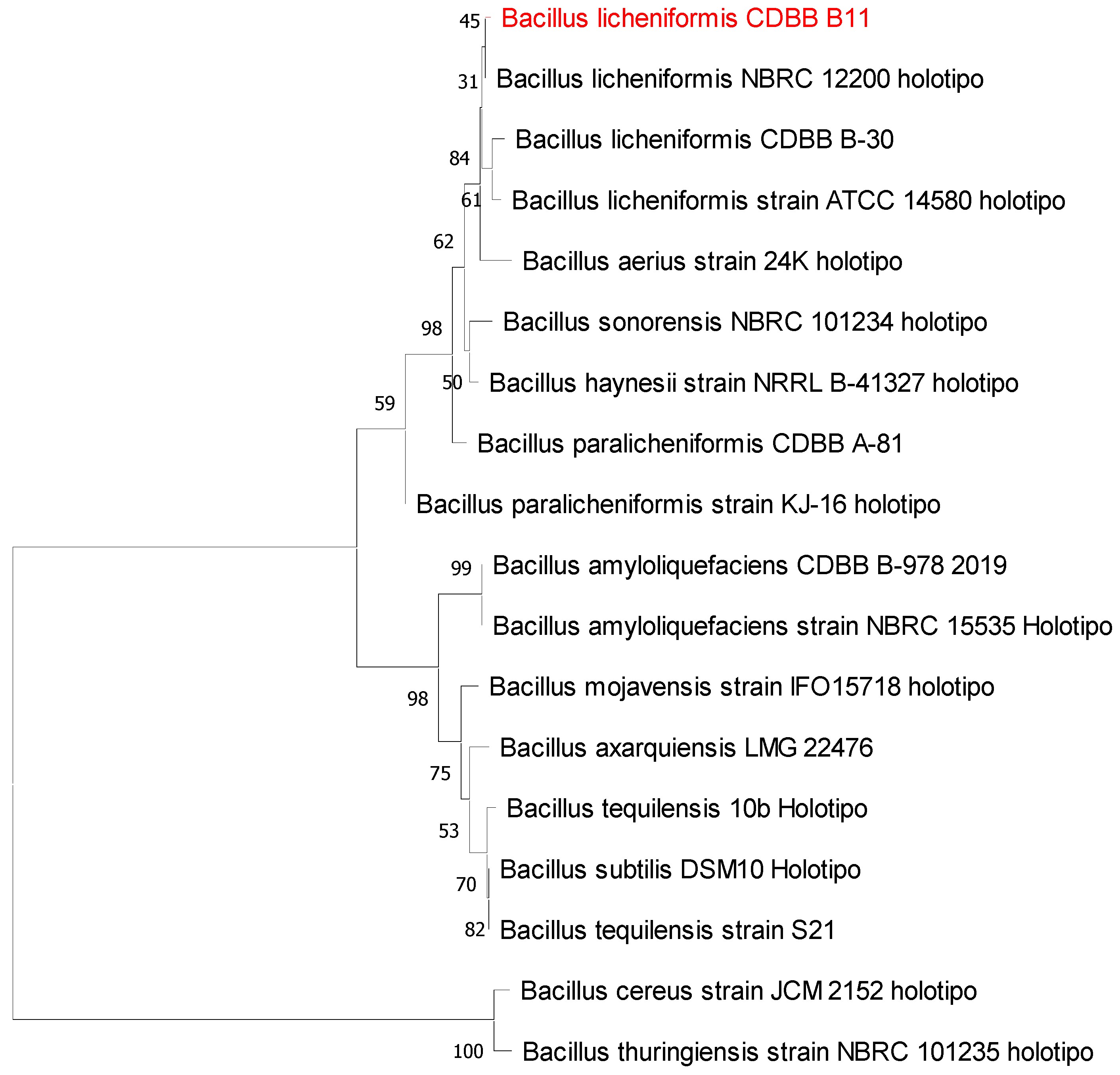
2.4. Molecular Identification of the Selected Bacterial Isolate
2.5. Effect of Carbon Source (Acetate) Concentration on Free-Cyanide Biodegradation by the Selected Bacterial Strain
2.6. Kinetic Study of Cell Growth, Cyanide Biodegradation, and Ammonia Accumulation
2.7. Evaluation of Kinetic Parameters of Cell Growth, Cyanide Biodegradation, and Ammonia Accumulation
2.8. Statistical Analysis
2.9. Analytical Methods
3. Results and Discussion
3.1. Screening and Selection of Cyanide-Degrading Microbial Cultures
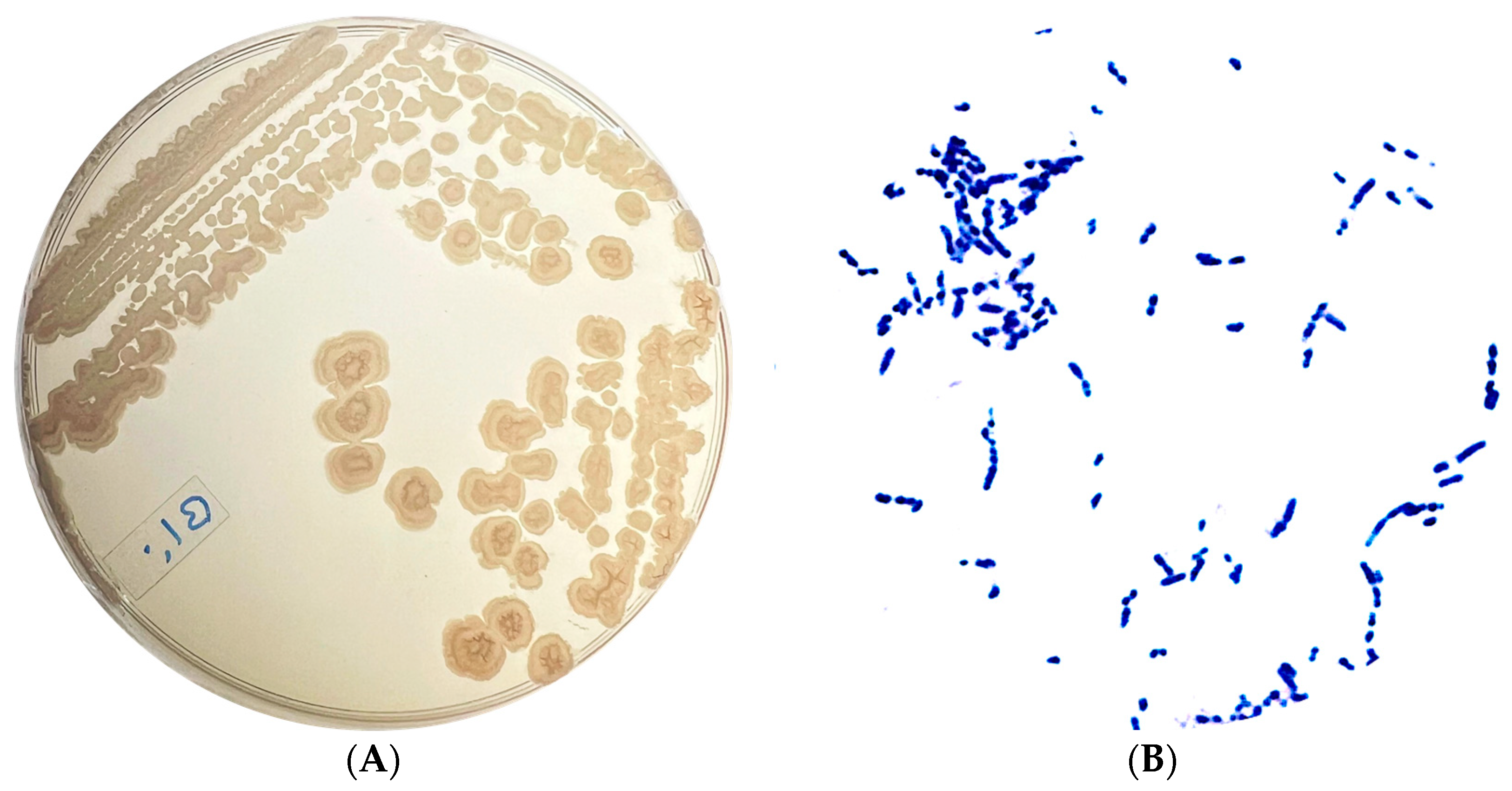
3.2. Isolation, Morphological Characterization, and Molecular Identification of Selected Cyanide-Degrading Bacterium
3.3. Effect of Acetate Concentration on Cyanide Biodegradation by B. licheniformis CDBB B11
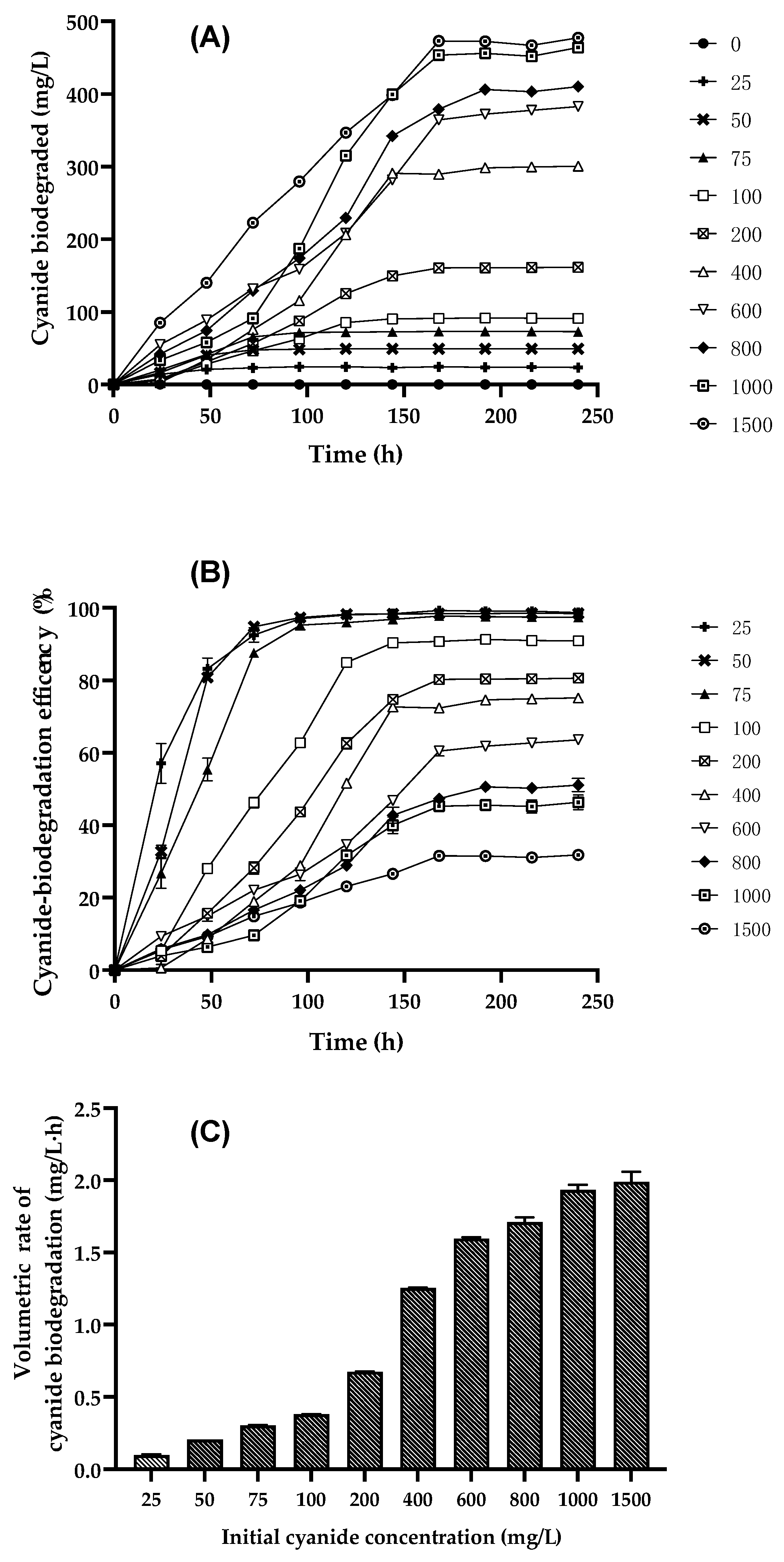
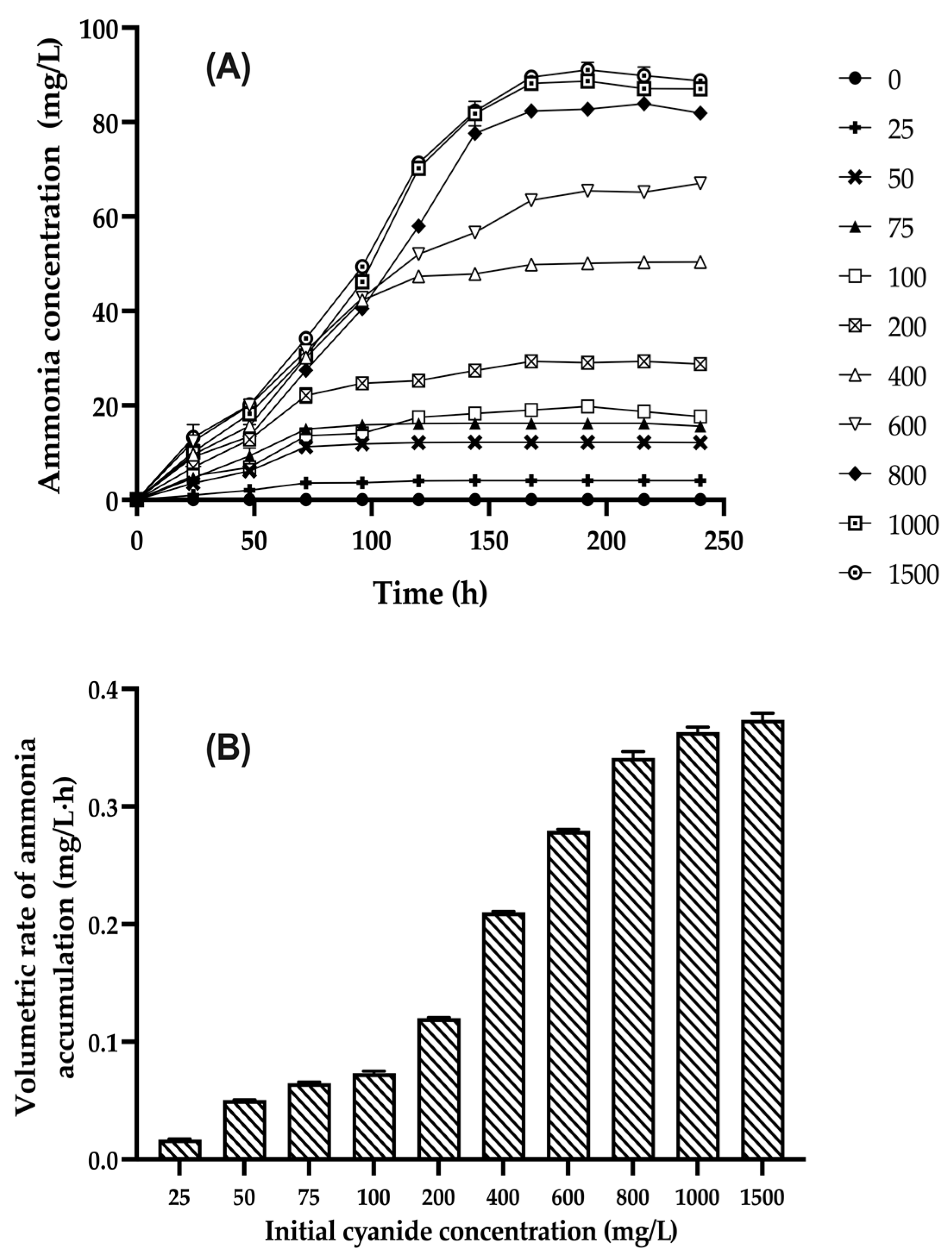
3.4. Kinetic Study of Cell Growth, Cyanide Biodegradation, and Ammonia Accumulation by Bacillus licheniformis CDBB B11 at Different Initial Free-Cyanide Concentrations
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dwivedi, N.; Balomajumder, C.; Mondal, P. Applications of microorganisms in biodegradation of cyanide from wastewater. In Advances in Microbial Biotechnology. Current Trends and Future Prospects; Kumar, P., Patra, J.J., Chandra, P., Eds.; Apple Academic Press, Inc.: Oakville, ON, Canada, 2019. [Google Scholar]
- Zuhra, K.; Szabo, C. The two faces of cyanide: An environmental toxin and a potential novel mammalian gasotransmitter. FEBS J. 2022, 289, 2481–2515. [Google Scholar] [CrossRef] [PubMed]
- Alvillo-Rivera, A.; Garrido-Hoyos, S.; Buitrón, G.; Thangarasu-Sarasvathi, P.; Rosano-Ortega, G. Biological treatment for the degradation of cyanide: A review. J. Mater. Res. Technol. 2021, 12, 1418–1433. [Google Scholar] [CrossRef]
- Alvillo-Rivera, A.; Garrido-Hoyos, S.; Buitrón, G. Cyanide treatment of mining tailings using suspended biomass and moving bed biomass reactors. Environ. Sci. Pollut. Res. 2022, 29, 37458–37470. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Lu, Y.; Feng, Z.; Yu, H.; Ma, P.; Zhu, J.; Wang, Y.; Sun, J. Biodegradation of cyanide by a new isolated Aerococcus viridans and optimization of degradation conditions by response surface methodology. Sustainability 2022, 14, 15560. [Google Scholar] [CrossRef]
- Manogaran, M.; Yakasai, H.M.; Karamba, K.I. Kinetics of cyanide degradation by Azotobacter vinelandii. J. Biochem. Microbiol. Biotechnol. 2019, 7, 38–42. [Google Scholar] [CrossRef]
- Guadalima, M.P.G.; Monteros, D.A.N. Evaluation of the rotational speed and carbon source on the biological removal of free cyanide present on gold mine wastewater, using a rotating biological contactor. J. Water Process Eng. 2018, 23, 84–90. [Google Scholar] [CrossRef]
- Cosmos, A.; Erdenekhuyag, B.-O.; Yao, G.; Li, H.; Zhao, J.; Laijun, W.; Luy, X. Principles and methods of bio-detoxification of cyanide contaminants. J. Mater. Cycles Waste Manag. 2020, 22, 939–954. [Google Scholar] [CrossRef]
- Luque-Almagro, V.M.; Cabello, P.; Sáez, L.P.; Olaya-Abril, A.; Moreno-Vivián, C.; Roldán, M.D. Exploring anaerobic environments for cyanide and cyano-derivatives microbial degradation. Appl. Microbiol. Biotechnol. 2018, 102, 1067–1074. [Google Scholar] [CrossRef]
- Razanamahandry, L.C.; Onwordi, C.T.; Saban, W.; Bashir, A.K.H.; Mekuto, L.; Malenga, E.; Manikandan, E.; Fosso-Kankeu, E.; Maaza, M.; Ntwampe, S.K.O. Performance of various cyanide degrading bacteria on the biodegradation of free cyanide in water. J. Hazard. Mater. 2019, 380, 120900. [Google Scholar] [CrossRef]
- Bhattacharya, R.; Flora, S.J.S. Chapter 19—Cyanide toxicity and its treatment. In Handbook of Toxicology of Chemical Warfare Agents; Gupta, R.C., Ed.; Academic Press: San Diego, CA, USA, 2009; pp. 255–270. [Google Scholar] [CrossRef]
- Terada, A.; Komatsu, D.; Ogawa, T.; Flamandita, D.; Sahlan, M.; Nishimura, M.; Yohda, M. Isolation of cyanide-degrading bacteria and molecular characterization of its cyanide-degrading nitrilase. Biotechnol. Appl. Biochem. 2022, 69, 183–189. [Google Scholar] [CrossRef]
- Maduh, E.U.; Turek, J.J.; Borowitz, J.L.; Rebar, A.; Isom, G.E. Cyanide-induced neurotoxicity: Calcium mediation of morphological changes in neuronal cells. Toxicol. Appl. Pharmacol. 1990, 103, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Rosario, C.G.A.; Vallenas-Arévalo, A.T.; Arévalo, S.J.; Espinosa, D.C.R.; Tenório, J.A.S. Biodegradation of cyanide using a Bacillus subtilis isolated from artisanal gold mining tailings. Braz. J. Chem. Eng. 2023, 40, 129–136. [Google Scholar] [CrossRef]
- Sharma, M.; Akhter, Y.; Chatterjee, S. A review on remediation of cyanide containing industrial wastes using biological systems with special reference to enzymatic degradation. World J. Microbiol. Biotechnol. 2019, 35, 70. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.; Park, J.; Park, H.; Lee, J.-C.; Kim, M.-S.; Lee, J. Key microbes and metabolic potentials contributing to cyanide biodegradation in stirred-tank bioreactors treating gold mining effluent. Miner. Process. Extr. Metall. Rev. 2020, 41, 85–95. [Google Scholar] [CrossRef]
- Sáez, L.P.; Cabello, P.; Ibañez, M.I.; Luque-Almagro, V.M.; Roldán, M.D.; Moreno-Vivián, C. Cyanate assimilation by the alkaliphilic cyanide-degrading bacterium Pseudomonas pseudoalcaligenes CECT5344: Mutational analysis of the cyn gene cluster. Int. J. Mol. Sci. 2019, 20, 3008. [Google Scholar] [CrossRef] [PubMed]
- Cabello, P.; Luque-Almagro, V.M.; Olaya-Abril, A.; Sáez, L.P.; Moreno-Vivián, C.; Roldán, M.D. Assimilation of cyanide and cyano-derivatives by Pseudomonas pseudoalcaligenes CECT5344: From omic approaches to biotechnological applications. FEMS Microbiol. Lett. 2018, 365, fny032. [Google Scholar] [CrossRef] [PubMed]
- Luque-Almagro, V.M.; Huertas, M.-J.; Martínez-Luque, M.; Moreno-Vivián, C.; Roldán, M.D.; García-Gil, J.; Castillo, F.; Blasco, R. Bacterial degradation of cyanide and its metal complexes under alkaline conditions. Appl. Environ. Microbiol. 2005, 71, 940–947. [Google Scholar] [CrossRef] [PubMed]
- Patil, Y.B.; Paknikar, K.M. Biodetoxification of silver-cyanide from electroplating industry wastewater. Lett. Appl. Microbiol. 2000, 30, 33–37. [Google Scholar] [CrossRef]
- Hope, K.M.; Knowles, C.J. The anaerobic utilization of cyanide in the presence of sugars by microbial cultures can involve an abiotic process. FEMS Microbiol. Lett. 1991, 80, 217–220. [Google Scholar] [CrossRef][Green Version]
- Hartley, T.F.; Philcox, J.C.; Willoughby, J. Two methods for monitoring the photodecomposition of sodium nitroprusside in aqueous and glucose solutions. J. Pharm. Sci. 1985, 74, 668–671. [Google Scholar] [CrossRef]
- Moradkhani, M.; Yaghmaei, S.; Nejad, Z.G. Biodegradation of cyanide under alkaline conditions by a strain of Pseudomonas putida isolated from gold mine soil and optimization of process variables through response surface methodology. Period. Polytech. Chem. Eng. 2018, 62, 265–273. [Google Scholar] [CrossRef]
- Karamba, K.I.; Shukor, M.Y.; Syed, M.A.; Zulkharnain, A.; Yasid, N.A.; Khalid, A.; Khalil, K.A.; Ahmad, S.A. Isolation, screening and characterisation of cyanide-degrading Serratia marcescens strain aq07. J. Chem. Pharm. Sci. 2015, 8, 401–406. [Google Scholar]
- Ibrahim, S.N.M.M.; Amalia, S.E.; Agatha, B.; Lestari, M.Y.; Saputra, A.D.; Ikmala, N.L.F.; Ryanto, F.H.A.R.; Dewi, P.A.; Anam, M.K.; Nafidiastri, F.A.; et al. Screening and identifying of cellulolytic bacteria from Alas Purwo National Park. AIP Conf. Proc. 2018, 2002, 020064. [Google Scholar] [CrossRef]
- Ni’matuzahroh; Fitriani, N.; Nuswantara, E.N.; Affandi, M.; Prasongsuk, S.; Kurniawan, S.B. Isolation and characterization of Schmutzdecke in slow sand filter for treating domestic wastewater. J. Ecol. Eng. 2022, 23, 76–88. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley & Sons: New York, NY, USA, 1991; pp. 115–147. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Malmir, N.; Fard, N.A.; Aminzadeh, S.; Moghaddassi-Jahromi, Z.; Mekuto, L. An overview of emerging cyanide bioremediation methods. Processes 2022, 10, 1724. [Google Scholar] [CrossRef]
- Hach Company. Hach Water Analysis Handbook, 5th ed.; Hach Company: Loveland, CO, USA, 2008. [Google Scholar]
- Pérez-Domínguez, J.C.; Higuera-Cobos, Ó.F. Comportamiento electroquímico del cianuro. Ing. Desarro. 2008, 24, 63–76. [Google Scholar]
- Tiong, B.; Bahari, Z.M.; Lee, N.; Jaafar, J.; Ibrahim, Z.; Shahir, S. Cyanide degradation by Pseudomonas pseudoalcaligenes strain W2 isolated from mining effluent. Sains Malays. 2015, 44, 233–238. [Google Scholar] [CrossRef]
- Vallenas-Arévalo, A.T.; Rosario, C.G.A.; Espinosa, D.C.R.; Tenório, J.A.S. Bacterial degradation of free cyanide in alkaline medium using Bacillus licheniformis strain. In Energy Technology 2018; Sun, Z., Wang, C., Guillen, D.P., Neelameggham, N.R., Zhang, L., Howarter, J.A., Wang, T., Olivetti, E., Zhang, M., Verhulst, D., et al., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 367–373. [Google Scholar] [CrossRef]
- Mekuto, L.; Ntwampe, S.K.O.; Kena, M.; Golela, M.T.; Amodu, O.S. Free cyanide and thiocyanate biodegradation by Pseudomonas aeruginosa STK 03 capable of heterotrophic nitrification under alkaline conditions. 3 Biotech 2016, 6, 6. [Google Scholar] [CrossRef]
- Logan, N.A.; Vos, P.D. Bacillus. In Bergey’s Manual of Systematics of Archaea and Bacteria, 1st ed.; Whitman, W.B., Rainey, F., Kämpfer, P., Trujillo, M., Chun, J., DeVos, P., Hedlund, B., Dedysh, S., Eds.; John Wiley & Sons, Inc., in Association with Bergey’s Manual Trust: Hoboken, NJ, USA, 2015; pp. 1–163. [Google Scholar] [CrossRef]
- Slepecky, R.A.; Hemphill, H.E. The genus Bacillus−Nonmedical. In The Prokaryotes. A Handbook on the Biology of Bacteria: Bacteria: Firmicutes, Cyanobacteria, 3rd ed.; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; Volume 4, pp. 530–562. [Google Scholar] [CrossRef]
- Salkinoja-Salonen, S.; Vuorio, R.; Andersson, M.A.; Kämpfer, P.; Andersson, M.C.; Honkanen-Buzalski, T.; Scoging, A.C. Toxigenic strains of Bacillus licheniformis related to food poisoning. Appl. Environ. Microbiol. 1999, 65, 4637–4645. [Google Scholar] [CrossRef]
- Arutchelvan, V.; Elangovan, R.; Venkatesh, K.R.; Nagarajan, S. Biodegradation of cyanide using Bacillus megaterium. J. Ind. Pollut. Control 2005, 21, 247–254. [Google Scholar]
- Nwokoro, O.; Dibua, M.E.U. Degradation of soil cyanide by single and mixed cultures of Pseudomonas stutzeri and Bacillus subtilis. Arch. Ind. Hyg. Toxicol. 2014, 65, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Safa, Z.J.; Aminzadeh, S.; Zamani, M.; Motallebi, M. Significant increase in cyanide degradation by Bacillus sp. M01 PTCC 1908 with response surface methodology optimization. AMB Express 2017, 7, 200. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-F.; Xu, X.-M.; Zhu, Q.; Deng, M.-C.; Feng, L.; Peng, J.; Yuan, J.-P.; Wang, J.-H. An effective method for the detoxification of cyanide-rich wastewater by Bacillus sp. CN-22. Appl. Microbiol. Biotechnol. 2014, 98, 3801–3807. [Google Scholar] [CrossRef] [PubMed]
- Mekuto, L.; Jackson, V.A.; Ntwampe, S.K.O. Biodegradation of free cyanide using Bacillus sp. consortium dominated by Bacillus safensis, lichenformis and tequilensis strains: A bioprocess supported solely with whey. J. Bioremediat. Biodegrad. 2013, 18, 2–7. [Google Scholar] [CrossRef]
- Mekuto, L.; Ntwampe, S.K.O.; Jackson, V.A. Biodegradation of free cyanide and subsequent utilisation of biodegradation by-products by Bacillus consortia: Optimisation using response surface methodology. Environ. Sci. Pollut. Res. 2015, 22, 10434–10443. [Google Scholar] [CrossRef] [PubMed]
- Karamba, K.I.; Ahmad, S.A.; Zulkharnain, A.; Yasid, N.A.; Ibrahim, S.; Shukor, M.Y. Batch growth kinetic studies of locally isolated cyanide-degrading Serratia marcescens strain AQ07. 3 Biotech 2018, 8, 11. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Balomajumder, C. Batch growth kinetic studies for elimination of phenol and cyanide using mixed microbial culture. J. Water Process Eng. 2016, 11, 130–137. [Google Scholar] [CrossRef]
- Akinpelu, E.A.; Ntwampe, S.K.O.; Mpongwana, N.; Nchu, F.; Ojumu, T.V. Biodegradation kinetics of free cyanide in Fusarium oxysporum-beta vulgaris waste-metal (As, Cu, Fe, Pb, Zn) cultures under alkaline conditions. BioResources 2016, 11, 2470–2482. [Google Scholar] [CrossRef]
- Cabuk, A.; Unal, A.T.; Kolankaya, N. Biodegradation of cyanide by a white rot fungus, Trametes versicolor. Biotechnol. Lett. 2006, 28, 1313–1317. [Google Scholar] [CrossRef]
- Alvarado-López, M.J.; Garrido-Hoyos, S.E.; Raynal-Gutiérrez, M.E.; El-Kassis, E.G.; Luque-Almagro, V.M.; Rosano-Ortega, G. Cyanide biodegradation by a native bacterial consortium and its potential for goldmine tailing biotreatment. Water 2023, 15, 1595. [Google Scholar] [CrossRef]
- Maiga, Y.; Young, S.; Orner, K.D.; Mihelcic, J.R.; Harwood, V.J.; Ouattara, A.S. Isolation and assessment of cyanide biodegradation potential of indigenous bacteria from contaminated soil. J. Environ. Prot. 2022, 13, 716–731. [Google Scholar] [CrossRef]
- Mirizadeh, S.; Yaghmaei, S.; Nejad, Z.G. Biodegradation of cyanide by a new isolated strain under alkaline conditions and optimization by response surface methodology (RSM). J. Environ. Health Sci. Eng. 2014, 12, 85. [Google Scholar] [CrossRef] [PubMed]
- Sirianuntapiboon, S.; Chuamkaew, C. Packed cage rotating biological contactor system for treatment of cyanide wastewater. Bioresour. Technol. 2007, 98, 266–272. [Google Scholar] [CrossRef]
- Supromin, N.; Potivichayanon, S.; Toensakes, R. Degradation of metal cyanide from real electroplating wastewater by mixed culture of SUTS 1 and SUTS 2. In Proceedings of the 3rd International Conference on Biological, Chemical and Environmental Sciences (BCES-2015), Kuala Lumpur, Malaysia, 21–22 September 2015. [Google Scholar] [CrossRef]
- Kao, C.M.; Lin, C.C.; Liu, J.K.; Chen, Y.L.; Wu, L.T.; Chen, S.C. Biodegradation of the metal-cyano complex tetracyanonickelate (II) by Klebsiella oxytoca. Enzym. Microb. Technol. 2004, 35, 405–410. [Google Scholar] [CrossRef]
- Suh, Y.-J.; Park, J.M.; Yang, J.-W. Biodegradation of cyanide compounds by Pseudomonas fluorescens immobilized on zeolite. Enzym. Microb. Technol. 1994, 16, 529–533. [Google Scholar] [CrossRef]

| Isolation Source | Geographical Coordinates | |
|---|---|---|
| 1 | Soil contaminated with wastewater from pulp and paper industries (San Rafael, Mexico State, Mexico) | 19°12′34.3″ N 98°45′18.9″ W |
| 2 | Agrochemical-contaminated soil [1] (San Rafael, Mexico State, Mexico) | 19°11′40.3″ N 98°45′23.3″ W |
| 3 | Industrial wastewater-contaminated soil [1] (Ixtapaluca, Mexico State, Mexico) | 19°17′20.3″ N 98°52′58.4″ W |
| 4 | Industrial wastewater-contaminated soil [2] (Chalco, Mexico State, Mexico) | 19°16′09.9″ N 98°51′33.2″ W |
| 5 | Agrochemical-contaminated soil [3] (Chalco, Mexico State, Mexico) | 19°14′09.9″ N 98°53′12.6″ W |
| 6 | Surface-water source (La Compañia River) that receives domestic and industrial effluents (Chalco, Mexico State, Mexico) | 19°16′28.7″ N 98°51′51.6″ W |
| 7 | Heavy metal and hydrocarbon-contaminated soil from a manganese mine (Tianguistengo, Hidalgo, Mexico) | 20°45′00″ N 98°40′00″ W |
| 8 | Heavy metal and hydrocarbon-contaminated soil from an ex-refinery (Azcapotzalco, Mexico City, Mexico) | 19°29′28.73″ N 99°11′25.04″ W |
| 9 | Hydrocarbon-contaminated soil (Tamalín, Veracruz, Mexico) | 21°20′21.08″ N 97°48′44.13″ W |
| 10 | Heavy metal and hydrocarbon-contaminated swamp [1] (Minatitlán, Veracruz, Mexico) | 17°59′03.4″ N 94°29′00.2″ W |
| 11 | Heavy metal and hydrocarbon-contaminated swamp [2] (Minatitlán, Veracruz, Mexico) | 17°59′03.4″ N 94°29′00.2″ W |
| 12 | Tailings from the gold and silver mining industry (Ocampo, Chihuahua, Mexico) | 28°11′34.8″ N 108°24′14.7″ W |
| 13 | Activated sludge (Ixtapaluca, Mexico State, Mexico) | 19°17′34.3″ N 98°53′41.0″ W |
| Enrichment Culture | Initial Cyanide Concentration (mg/L) | ||||||
|---|---|---|---|---|---|---|---|
| 20 | 50 | 100 | 200 | 500 | 1000 | 1500 | |
| B1 | + | + | + | + | + | + | + |
| B2 | + | − | − | − | − | − | − |
| B3 | + | + | − | − | − | − | − |
| B4 | + | + | + | + | + | + | + |
| B5 | + | − | − | − | − | − | − |
| B6 | + | + | + | − | − | − | − |
| B7 | + | + | + | − | − | − | − |
| B8 | + | + | + | + | + | + | + |
| B9 | + | − | − | − | − | − | − |
| B10 | + | − | − | − | − | − | − |
| B11 | + | + | + | + | + | + | + |
| B12 | + | + | + | + | + | + | + |
| B13 | + | + | + | − | − | − | − |
| Enrichment Culture | Cyanide Removal | |
|---|---|---|
| (mg/L) | (%) | |
| B1 | 348.6 ± 14.59 | 23.24 ± 0.97 |
| B4 | 200.7 ± 13.78 | 13.38 ± 0.92 |
| B8 | 173 ± 11.81 | 11.53 ± 0.79 |
| B11 | 504.5 ± 19.81 | 33.63 ± 1.41 |
| B12 | 205.5 ± 12.40 | 13.7 ± 0.83 |
| Biomass-free control | 28.7 ± 5.55 | 1.91 ± 0.1 |
| Colony Morphology | Cell Morphology | Spore-Forming Bacteria | ||
|---|---|---|---|---|
| Shape: | Irregular | Shape: | Short rod-shaped | Positive |
| Color: | Beige | Gram: | Positive | |
| Edge: | Entire | |||
| Surface appearance: | Rough and matte | |||
| Elevation: | Plane convex | |||
| Opacity: | Opaque | |||
| Texture: | Dry | |||
| Acetate Concentration (g/L) | Maximum Increase of Cell Mass (g/L) | Cyanide Biodegradation Efficiency (%) | ||
|---|---|---|---|---|
| 400 mg of Cyanide/L | 1500 mg of Cyanide/L | 400 mg of Cyanide/L | 1500 mg of Cyanide/L | |
| 1 | 0.345 ± 0.035 | 0.55 ± 0.028 | 75 ± 0.81 | 44 ± 1.08 |
| 2 | 0.655 ± 0.035 | 0.99 ± 0.014 | 79 ± 0.05 | 47 ± 0.14 |
| 4 | 0.71 ± 0.014 | 1.38 ± 0.056 | 85 ± 0.28 | 50 ± 0.61 |
| 6 | 0.785 ± 0.063 | 1.41 ± 0.007 | 87 ± 0.43 | 53 ± 0.44 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uribe-Ramírez, D.; Cristiani-Urbina, E.; Morales-Barrera, L. Biodegradation of Free Cyanide by a New Isolated Alkaliphilic Bacillus licheniformis Strain. Microbiol. Res. 2024, 15, 33-49. https://doi.org/10.3390/microbiolres15010003
Uribe-Ramírez D, Cristiani-Urbina E, Morales-Barrera L. Biodegradation of Free Cyanide by a New Isolated Alkaliphilic Bacillus licheniformis Strain. Microbiology Research. 2024; 15(1):33-49. https://doi.org/10.3390/microbiolres15010003
Chicago/Turabian StyleUribe-Ramírez, Daniel, Eliseo Cristiani-Urbina, and Liliana Morales-Barrera. 2024. "Biodegradation of Free Cyanide by a New Isolated Alkaliphilic Bacillus licheniformis Strain" Microbiology Research 15, no. 1: 33-49. https://doi.org/10.3390/microbiolres15010003
APA StyleUribe-Ramírez, D., Cristiani-Urbina, E., & Morales-Barrera, L. (2024). Biodegradation of Free Cyanide by a New Isolated Alkaliphilic Bacillus licheniformis Strain. Microbiology Research, 15(1), 33-49. https://doi.org/10.3390/microbiolres15010003








