Abstract
Background: The detection of neutralizing anti-SARS-CoV-2 antibodies is important since they represent the subset of antibodies able to prevent the virus to invade human cells. The aim of this study is to evaluate the clinical performances of an in-house pseudovirus neutralization test (pVNT) versus a commercial surrogate neutralization test (sVNT). Material and Methods: A total of 114 RT-PCR positives samples from 75 COVID-19 patients were analyzed using a pVNT and an sVNT technique. Fifty-six pre-pandemic samples were also analyzed to assess the specificity of the two techniques. An analysis of the repeatability and the reproducibility of the pVNT was also performed. Results: A coefficient of variation (CV) of 10.27% for the repeatability of the pVNT was computed. For the reproducibility test, CVs ranged from 16.12% for low NAbs titer to 6.40% for high NAbs titer. Regarding the clinical sensitivity, 90 RT-PCR positive samples out of 114 were positive with the pVNT (78.94%), and 97 were positive with the sVNT (84.21%). About the clinical specificity, all 56 pre-pandemic samples were negative in both techniques. When comparing the sVNT to the pVNT, the specificity and sensibility were 66.67% (95%CI: 47.81–85.53%) and 98.88% (95%CI: 96.72–99.99%), respectively. Conclusions: The results obtained with the automated sVNT technique are consistent with those obtained with the pVNT technique developed in-house. The results of the various repeatability and reproducibility tests demonstrate the good robustness of the fully manual pVNT technique.
1. Introduction
The gold standard method for detecting SARS-CoV-2 infection is the real-time reverse transcription polymerase chain reaction (RT-PCR) [1,2,3]. The detection of antibodies directed against the spike protein (S protein), the receptor binding domain (RBD) or the nucleocapsid (N) in the serum or plasma of convalescent patients allows for the monitoring of the development of adaptive immunity after SARS-CoV-2 infection or vaccination [2,3,4,5]. Following the infection or the vaccination, different antibodies are produced targeting distinctive epitopes of the virus, including the spike protein. Nevertheless, not all of these antibodies are able to efficiently neutralize the entry of the virus into the host cell (i.e., binding a non-essential epitope for the entrance mechanism). Therefore, neutralizing antibodies (NAbs) are of particular importance because they can prevent the binding of the protein S RBD to the angiotensin-converting enzyme 2 (ACE2) receptor present at the surface of human cells, preventing the entry of the virus [6,7,8].
Nowadays, the gold standard method for the detection of NAbs is the plaque reduction neutralization test (PRNT) [9]. Based on the measurement of cell lysis due to viral infection, this test is able to detect antibodies that are able to neutralize the entry of the virus in the host cell [10,11]. In the case of SARS-CoV-2, the technique requires the use of live pathogens and must therefore be performed in a biosafety level 3 (BL3) laboratory. In addition, it requires expensive installations and skillful and meticulous staff resulting in a high workload preventing the wide implementation of this technique, even in research laboratories [11,12]. Similar neutralization techniques based on pseudoviral particles (also called pseudovirus neutralization tests, pVNTs) have been developed and present the advantage of being able to be used in BL2 laboratories and allow for a higher throughput [13,14,15]. Moreover, Cantoni D. et al. reported in their three-level meta-analysis a high level of correlation between the assay based on a pseudovirus compared to those based on a live virus [16].
Besides these “live” tests which mimic human cell infection, the use of a SARS-CoV-2 surrogate virus neutralization test (sVNT) has been presented as an alternative. Such tests are based on the antibody-mediated blockage of the interaction between the RBD (arbored on magnetic beads for example) and the ACE-2 receptor labeled with a signaling molecule (i.e., chemiluminescence). The advantage of these automated techniques is their ease of use, their high capacity of implementation in routine laboratories, as well as their shortened turn-around time. Moreover, compared to “live” tests, the sVNT is less expensive. The performance of these sVNT methods have been claimed to be suboptimal in some reports, although this may differ depending on test parameters and the selected antibodies [5,17,18,19]. This study aims at evaluating the clinical performance of our in-house developed pVNT technique and to compare the results with an automated sVNT.
2. Materials and Methods
2.1. Patient Samples
One hundred and fourteen samples from 75 patients with a confirmed SARS-CoV-2 RT-PCR were retrospectively included from 26 March 2020 to 6 January 2021. Among them, 39 were females (median age = 45; min–max: 24–95 years) and 36 were males (median age = 62; min–max: 24–88 years). Multiple sequential sera were available from 41 patients. Seventeen patients required hospitalization and were categorized as severe patients according to the WHO criteria [20]. Information on the days since the onset of symptoms was collected from medical records and was available for 63 patients. When data about the symptoms were not available (n = 12), the day of diagnosis (i.e., RT-PCR result) was used. Among these samples, 10 samples with high NAb titers and 10 with low titers were used to perform the repeatability and reproducibility analysis. Furthermore, 56 sera collected before the start of the pandemic in Belgium (March 2020) were analyzed to assess the specificity of both antibody methods. The population has already been used for other reports [21].
2.2. Sample Collection
Blood samples were collected into serum gel tubes (BD SST II Advance®, Becton Dickinson, NJ, USA) and centrifuged for 10 min at 1740× g on a Sigma 3-16KL centrifuge. Sera were stored in the laboratory serum biobank at −20 °C from the collection date. Frozen samples were thawed for 1 h at room temperature on the day of the analysis. Re-thawed samples were vortexed before the analysis. All samples were collected at the Clinique Saint-Luc (Bouge, Namur, Belgium). The study protocol was in accordance with the Declaration of Helsinki (approval number: 2020-006149-2).
2.3. Analytical Procedures
2.3.1. RT-PCR
The RT-PCR for SARS-CoV-2 determination in the nasopharyngeal swab sample was performed on the LightCycler® 480 instrument II (Roche Diagnostics®, Bâle, Switzerland) using the LightMix® Modular SARS-CoV E-gene set according to the recommendations from the manufacturer.
2.3.2. Pseudovirus Neutralization Test
A pseudovirus neutralization test was used to assess the neutralization capacity of infected patients’ sera. Pseudoviruses were gathered from E-enzyme (catalog number: SCV2-PsV-614G). SARS-CoV-2 pseudoviral particles are replication-deficient Moloney murine leukemia virus (MLV or MuLV) pseudotyped with the SARS-CoV-2 spike protein carrying the wild type D614G genotype. They also contain the open reading frame for firefly luciferase as a reporter. Briefly, HEK293T hACE2 cells were seeded at the density of 8500 cells/well in a white 384-well cell culture plate. The sera used were heat-inactivated by a water bath at 54 °C for 30 min and then serially diluted in a culture medium (Dulbecco’s modified Eagle medium) supplemented with 10% of fetal bovine serum. Thereafter, samples were mixed in a 1:4 ratio with pseudovirus and incubated for 2 h at 37 °C. This mixture was added to the cells and incubated for 48 h at 37 °C. The reading was performed by adding firefly luciferase reagent to measure the activity of luciferase which is proportional to the cells infected by the pseudovirus. Raw data obtained in relative luminescence units were normalized to the positive control where cells were incubated with pseudovirus in the absence of serum. The NAbs titer was determined as the dilution of serum at which 50% of the infectivity is inhibited (IC50) as determined by a nonlinear sigmoid regression model. A sample with a pVNT titer dilution−1 below 20 was considered negative. The detailed protocol has already been described elsewhere [22].
2.3.3. Surrogate Virus Neutralization Test
The iFlash-2019-nCoV NAbs assay is a one-step competitive paramagnetic particle chemiluminescent immunoassay (CLIA) for the quantitative determination of 2019-nCoV NAbs in human serum and plasma. The assay detects NAbs that block the binding of RBD and ACE2. Firstly, NAbs (if present) react with the RBD antigen coated on paramagnetic microparticles to form a complex. Secondly, the acridinium ester-labeled ACE2 conjugate is added to competitively bind to the RBD-coated particles, which have not been neutralized by the NAbs (if present) from the sample and form another reaction mixture. Under magnetic field, magnetic particles are adsorbed to the wall of the reaction tube, and unbound materials are washed away by the wash buffer. The resulting chemiluminescent reaction is measured as relative light units (RLUs), with an inverse relationship between the amount of NAbs and the RLUs detected. A result <10.0 AU/mL is considered as negative and a result ≥10.0 AU/mL is considered as positive (manufacturer’s information). The sVNTs were performed on an iFlash1800 automated MCLIA analyzer from Shenzhen YHLO Biotech Co., Ltd (Shenzhen, China).
2.3.4. Statistical Analysis
Descriptive statistics were used to interpret the results. Means, a 95% confidence interval (95% CI) and the standard deviation (SD) were computed. Sensitivity was defined as the proportion of correctly identified COVID-19 positive patients initially positive by RT-PCR SARS-CoV-2 determination in nasopharyngeal swab samples. Specificity was defined as the proportion of pre-pandemic samples classified as negative. A nonlinear regression model was performed to assess the long-term kinetics of NAbs in COVID-19 patients. Pearson regressions were computed to assess the correlation between pVNT and sVNT results and between NAbs titers and days post positive RT-PCR. To challenge the sVNT manufacturer’s cut-off, a ROC curve analysis was performed. Repeatability was assessed by analyzing the same sample 10 times in a row by the same operator; a sample considered with a low NAbs titer and another with a medium-high NAbs titer were analyzed. Reproducibility was assessed by analyzing the same sample 10 times on 10 different plates. A coefficient of variation (CV) was then calculated. Data analysis was performed using GraphPad Prism® software (version 9.1.0, San Diego, CA, USA). A p-value < 0.05 was used as the significant level.
3. Results
3.1. Repeatability and Reproducibility
Regarding the repeatability, NAb titers obtained with pVNT ranged from 228.0 to 326.5 pVNT titer dilution−1, with a mean of 267.10 titer dilution−1 (95% CI: 245.70–286.30, SD: 27.41) for the sample with a medium-high NAbs titer. The CV was 10.27%. For the sample with the low NAbs titer, NAb titers obtained ranged from 20.5 to 35.4 pVNT titer dilution−1, with a mean of 25.5 titer dilution−1 (95% CI: 24.4–30.7, SD: 4.44). The CV was 16.12%. Regarding the reproducibility assessment, the NAb titers ranged from 20.53 to 35.45 titer dilution−1, with a mean of 27.54 pVNT titer dilution−1 (95% CI: 22.42–30.40, SD: 4.44). The coefficient of variation was 16.12%. For the top dilution range sample, NAb titers ranged from 155.50 to 194.10 titer dilution−1. The mean was 170.60 titer dilution−1 (95% CI: 162.80–178.40, SD: 10.93), and the coefficient of variation was 6.40%.
3.2. Clinical Sensitivity
Among the 114 COVID-19 RT-PCR positive samples analyzed in the pVNT, 90 (70.94%) were positive for NAbs. Among the 24 samples considered as negative (i.e., pVNT titer dilution−1 < 20), 45.83% of samples (n = 11/24) were collected between 0 and 10 days following RT-PCR positivity, 24% (n = 6/24) between 10 and 20 days and 28% were obtained beyond 20 days (n = 7/24). Among the seven negative samples after 20 days, two had NAb titers close to the positivity threshold (i.e., 19.54 for the sample at day 44, and 18.51 for the sample at day 109). These two samples were positive for their previous sampling. Regarding the same samples analyzed with the sVNT technique, 97 (84.21%) were considered positive. Among the 17 samples considered as negative (sVNT titer < 10 AU/mL), 70.58% (n = 12/17) were between 0 and 10 days post positive RT-PCR, and 29.41% (n = 5/17) were between 10 and 20 days (Figure 1). The tracking of individual NAb titers is shown in Supplementary Figure S1.
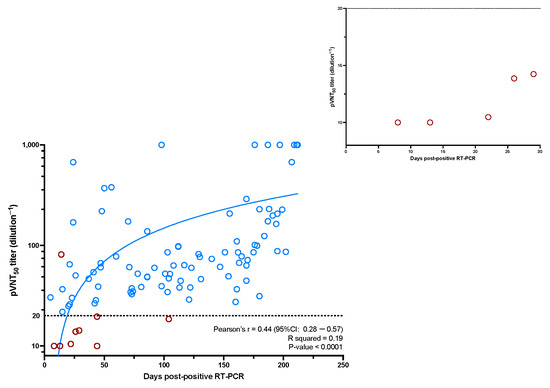
Figure 1.
The evolution of the NAbs titer as a function of days post positive RT-PCR. A zoom on the negative results obtained before 30 days post positive RT-PCR is represented. Dots circled in red are those for which there is a discordant result compared to the sVNT technique. Dots circled in blue are those for which there is a concordant result compared to the sVNT technique.
3.3. Clinical Specificity
Among the 56 pre-pandemic samples analyzed using the pVNT, all samples were negatives (100% specificity). The same specificity of 100% was also found using the sVNT.
3.4. sVNT vs. pVNT
Using the pVNT as the reference method, the sensitivity of the sVNT was 66.67% (95% CI: 47.81–85.53%) and the specificity was 98.88% (95% CI: 96.72–99.99%) (Table 1).

Table 1.
Contingency table of sVNT versus pVNT results. Sensitivity and specificity of sVNT are also presented.
The eight sVNT-positive and pVNT-negative samples had sVNT titers that ranged from 10.50 AU/mL to 17.21 AU/mL except for one sample with a titer of 37.98 AU/mL. This sample had a borderline pVNT titer dilution−1 of 19.54. The only pVNT-positive and sVNT-negative sample had a pVNT titer dilution−1 of 81.25. This sample had an sVNT titer of 8.40 AU/mL. Discordant results are highlighted in Figure 1. The sVNT adapted cut-off was 9.85 AU/mL, and none of the nine discordant results became concordant using this latter cut-off. As observed in Figure 2, the correlation between the pVNT and the sVNT showed a Pearson’s r of 0.67 (95%CI: 0.56–0.76) (p-value < 0.05). The R squared for the best fit curve was 0.47.
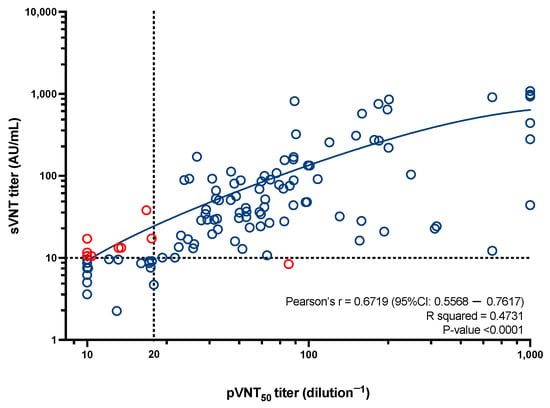
Figure 2.
A representation of the results obtained with the pVNT in relation to the results obtained with the sVNT. Dotted lines represented the cut-off of each technique, i.e., a pVNT titer dilution−1 below 20 for the pVNT, and a titer below 10 AU/mL for the sVNT. Red dots represent discordant results. Pearson’s R, the R squared of the best fit curve and the p-value are presented.
4. Discussion
Currently, the plate reduction neutralization test is considered the gold standard method for the measurement of NAbs but suffers from several limitations due to the requirement of a BL3 working environment, as well as rigorous and laborious manipulations. The use of the pVNT presents several advantages including the requirement of only a BL2 security level laboratory [23,24]. In addition, pseudoviruses are easier to produce and quantify and have been reported to generate similar results as the PRNT, permitting us to consider this technique as a valuable alternative to the gold standard method [13,25].
The comparison of the sVNT technique to the pVNT technique showed a sensitivity of 98.88% (95% CI: 96.72–99.99%) with only one positive result with the pVNT but negative with the sVNT (but close to the positivity threshold). The specificity between the two techniques was 66.67% (95% CI: 47.81–85.53%) which represents eight samples negative in the pVNT and positive in the sVNT. Studies have already compared the results obtained with sVNT techniques versus the PRNT, and the results are consistent with those observed in our cohort [26,27,28,29]. Mariën et al. reported a strong correlation in a cohort of 316 samples (r of 0.85) between the sVNT and the PRNT [30]. Tan et al. also reported an R2 of 0.84 using a cohort of 60 samples between the sVNT and the pVNT [31]. Using seven sVNT assays with the PRNT as the reference, Graninger et al. reported a sensitivity above 95.00% for all the sVNTs tested. The repeatability test shows a relatively low coefficient of variation (10.27%). For the reproducibility test, the coefficient of variation is higher in the bottom of the dilution range (16.12%) than in the top of the dilution range (6.40%). These data allow for the method to be considered reliable and robust.
Khoury et al. were the first to demonstrate that NAbs correlated well with protection against SARS-CoV-2 infections. They also highlight that the efficacy of COVID-19 vaccines is correlated with the ability of these vaccines to induce NAbs production [8,32]. Favresse et al. also showed an increase in NAb titers along with disease severity. They also demonstrated a higher level of NAbs in the vaccinated populationit.
It is interesting to note that only 79.94% of RT-PCR positive samples were pVNT-positive. Results considered as negative results in the pVNT were mostly located shortly after positive RT-PCR. This is probably due to the maturation process of the neutralizing antibodies. Indeed, during a viral infection such as COVID-19, antigen-presenting cells (APCs) will expose different fragments of the antigen to T-Helper which will then stimulate B-cells to produce antibodies against this epitope of the antigen. Depending on which epitope the antibody is directed against, the antibody–antigen interaction will not be the same. The effectiveness of the antibodies in neutralizing key fragments of the virus’s anchoring proteins that allow it to interact with human cells will also differ [33]. In the case of COVID-19, antibodies are said to be neutralizing when they prevent the interaction between protein S and the ACE2 receptor; antibodies directed against protein S but not preventing this interaction are therefore not neutralizing [34]. The fact that some samples remain negative beyond 30 days may be the result of a lack of NAbs production, although some of the negative samples are borderline in terms of the results, i.e., 19.54 for the sample at day 44, and 18.51 for the sample at day 104. Jeewandara et al. also pointed out a relationship between the development of NAbs and the time of exposure to the virus. In their study, patients with prolonged exposure to SARS-CoV-2 had a higher NAbs titer than patients who cleared the virus earlier [35]. This link between NAbs and the time of the exposure to the virus is not well developed as in most hospitals, patients are sent home when they become negative via RT-PCR, which does not allow for long-term follow-up.
5. Conclusions
In conclusion, this study provides a comprehensive analysis of the clinical performance of the pVNT and the sVNT in detecting NAbs against SARS-CoV-2. Despite the gold standard status of the PRNT, requiring high biosafety level laboratories and extensive resources, the findings highlight the efficacy of the pVNT and the sVNT as viable alternatives for evaluating adaptive immunity post-infection or vaccination. This study showcases that the pVNT, requiring only biosafety level 2 facilities, offers comparable results to the PRNT, underscoring its potential as a practical and accessible method for widespread implementation.
Moreover, this study reveals a high degree of clinical sensitivity and specificity in both the pVNT and the sVNT, with the sVNT demonstrating ease of use and cost-effectiveness for routine laboratory implementation. The correlation between the pVNT and the sVNT results further validates the reliability of the sVNT in detecting NAbs, despite some discrepancies that call for cautious interpretation in certain scenarios.
Overall, this study underlines the importance of developing and validating accessible, reliable neutralization assays for SARS-CoV-2 to enhance our understanding of immune responses and to facilitate the global effort in monitoring and combating the COVID-19 pandemic. Future research should aim to refine these assays further and explore their application in diverse populations to ensure the comprehensive surveillance and management of SARS-CoV-2 immunity worldwide.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microbiolres15010028/s1, Figure S1: Kinetics of NAbs since the symptom onset in non-hospitalized and hospitalized patients. Blue plain lines represent non-hospitalized patients and red plain lines represent hospitalized patients. Samples obtained at different times for the same patient are connected by dotted lines.
Author Contributions
Conceptualization, C.G., J.D. and J.F.; methodology, J.D. and J.F.; software, C.G. and J.D.; validation, J.F. and J.D.; formal analysis, J.D. and J.F.; investigation, C.G. and J.F.; resources, J.D.; data curation, J.F.; writing—original draft preparation, C.G. and J.D.; writing—review and editing, C.D., J.F., J.-M.D. and V.M.; supervision, J.D.; project administration, J.D. and J.F.; funding acquisition, J.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of CHU UCL-Namur (protocol code 2020-006149-2).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data is not available online, but can be shared by the authors if requested.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- van Kasteren, P.B.; van Der Veer, B.; van Den Brink, S.; Wijsman, L.; de Jonge, J.; van Den Brandt, A.; Molenkamp, R.; Reusken, C.B.; Meijer, A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020, 128, 104412. [Google Scholar] [CrossRef] [PubMed]
- Chau, C.H.; Strope, J.D.; Figg, W.D. COVID-19 Clinical Diagnostics and Testing Technology. Pharmacotherapy 2020, 40, 857–868. [Google Scholar] [CrossRef]
- Ward, S.; Lindsley, A.; Courter, J.; Assa’ad, A. Clinical testing for COVID-19. J. Allergy Clin. Immunol. 2020, 146, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Ding, C.; Li, J.; Wang, Y.; Guo, H.; Lu, Z.; Wang, J.; Zheng, C.; Jin, T.; Gao, Y.; et al. Characteristics of patients with coronavirus disease (COVID-19) confirmed using an IgM-IgG antibody test. J. Med. Virol. 2020, 92, 2004–2010. [Google Scholar] [CrossRef] [PubMed]
- Simon, G.; Favresse, J.; Gillot, C.; Closset, M.; Catry, E.; Dogne, J.M.; Douxfils, J.; Wieers, G.; Bayart, J.L. Kinetics and ability of binding antibody and surrogate virus neutralization tests to predict neutralizing antibodies against the SARS-CoV-2 Omicron variant following BNT162b2 booster administration. Clin. Chem. Lab. Med. 2023, 61, 1875–1885. [Google Scholar] [CrossRef]
- Chen, P.; Nirula, A.; Heller, B.; Gottlieb, R.L.; Boscia, J.; Morris, J.; Huhn, G.; Cardona, J.; Mocherla, B.; Stosor, V.; et al. SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with COVID-19. N. Engl. J. Med. 2021, 384, 229–237. [Google Scholar] [CrossRef]
- Shi, R.; Shan, C.; Duan, X.; Chen, Z.; Liu, P.; Song, J.; Song, T.; Bi, X.; Han, C.; Wu, L.; et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 2020, 584, 120–124. [Google Scholar] [CrossRef]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef]
- Perera, R.A.; Mok, C.K.; Tsang, O.T.; Lv, H.; Ko, R.L.; Wu, N.C.; Yuan, M.; Leung, W.S.; Chan, J.M.; Chik, T.S.; et al. Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), March 2020. Eurosurveillance 2020, 25, 2000421. [Google Scholar] [CrossRef] [PubMed]
- Lau, E.H.Y.; Tsang, O.T.Y.; Hui, D.S.C.; Kwan, M.Y.W.; Chan, W.H.; Chiu, S.S.; Ko, R.L.W.; Chan, K.H.; Cheng, S.M.S.; Perera, R.; et al. Neutralizing antibody titres in SARS-CoV-2 infections. Nat. Commun. 2021, 12, 63. [Google Scholar] [CrossRef] [PubMed]
- Muruato, A.E.; Fontes-Garfias, C.R.; Ren, P.; Garcia-Blanco, M.A.; Menachery, V.D.; Xie, X.; Shi, P.Y. A high-throughput neutralizing antibody assay for COVID-19 diagnosis and vaccine evaluation. Nat. Commun. 2020, 11, 4059. [Google Scholar] [CrossRef]
- Lee, W.T.; Girardin, R.C.; Dupuis, A.P.; Kulas, K.E.; Payne, A.F.; Wong, S.J.; Arinsburg, S.; Nguyen, F.T.; Mendu, D.R.; Firpo-Betancourt, A.; et al. Neutralizing Antibody Responses in COVID-19 Convalescent Sera. J. Infect. Dis. 2021, 223, 47–55. [Google Scholar] [CrossRef]
- Nie, J.; Li, Q.; Wu, J.; Zhao, C.; Hao, H.; Liu, H.; Zhang, L.; Nie, L.; Qin, H.; Wang, M.; et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020, 9, 680–686. [Google Scholar] [CrossRef]
- Case, J.B.; Rothlauf, P.W.; Chen, R.E.; Liu, Z.; Zhao, H.; Kim, A.S.; Bloyet, L.M.; Zeng, Q.; Tahan, S.; Droit, L.; et al. Neutralizing Antibody and Soluble ACE2 Inhibition of a Replication-Competent VSV-SARS-CoV-2 and a Clinical Isolate of SARS-CoV-2. Cell Host Microbe 2020, 28, 475–485.e5. [Google Scholar] [CrossRef]
- Vogel, A.B.; Kanevsky, I.; Che, Y.; Swanson, K.A.; Muik, A.; Vormehr, M.; Kranz, L.M.; Walzer, K.C.; Hein, S.; Güler, A.; et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature 2021, 592, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Cantoni, D.; Wilkie, C.; Bentley, E.M.; Mayora-Neto, M.; Wright, E.; Scott, S.; Ray, S.; Castillo-Olivares, J.; Heeney, J.L.; Mattiuzzo, G.; et al. Correlation between pseudotyped virus and authentic virus neutralisation assays, a systematic review and meta-analysis of the literature. Front. Immunol. 2023, 14, 1184362. [Google Scholar] [CrossRef] [PubMed]
- Douxfils, J.; Gillot, C.; De Gottal, E.; Vandervinne, S.; Bayart, J.L.; Dogne, J.M.; Favresse, J. Efficient Maternal to Neonate Transfer of Neutralizing Antibodies after SARS-CoV-2 Vaccination with BNT162b2: A Case-Report and Discussion of the Literature. Vaccines 2021, 9, 907. [Google Scholar] [CrossRef] [PubMed]
- Douxfils, J.; Gillot, C.; Mullier, F.; Favresse, J. Post-SARS-CoV-2 vaccination specific antibody decrease—Thresholds for determining seroprevalence and seroneutralization differ. J. Infect. 2021, 83, e4–e5. [Google Scholar] [CrossRef] [PubMed]
- Favresse, J.; Gillot, C.; Di Chiaro, L.; Eucher, C.; Elsen, M.; Van Eeckhoudt, S.; David, C.; Morimont, L.; Dogné, J.M.; Douxfils, J. Neutralizing Antibodies in COVID-19 Patients and Vaccine Recipients after Two Doses of BNT162b2. Viruses 2021, 13, 1364. [Google Scholar] [CrossRef] [PubMed]
- Marshall, J.C.; Murthy, S.; Diaz, J.; Adhikari, N.K.; Angus, D.C.; Arabi, Y.M.; Baillie, K.; Bauer, M.; Berry, S.; Blackwood, B.; et al. A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect. Dis. 2020, 20, e192–e197. [Google Scholar] [CrossRef] [PubMed]
- Gillot, C.; Favresse, J.; Maloteau, V.; Dogne, J.M.; Douxfils, J. Dynamics of Neutralizing Antibody Responses Following Natural SARS-CoV-2 Infection and Correlation with Commercial Serologic Tests. A Reappraisal and Indirect Comparison with Vaccinated Subjects. Viruses 2021, 13, 2329. [Google Scholar] [CrossRef]
- Gillot, C.; Favresse, J.; Maloteau, V.; Dogné, J.M.; Douxfils, J. Identification of SARS-CoV-2 Neutralizing Antibody with Pseudotyped Virus-based Test on HEK-293T hACE2 Cells. Bio Protoc. 2022, 12, e4377. [Google Scholar] [CrossRef]
- Xiong, H.L.; Wu, Y.T.; Cao, J.L.; Yang, R.; Liu, Y.X.; Ma, J.; Qiao, X.Y.; Yao, X.Y.; Zhang, B.H.; Zhang, Y.L.; et al. Robust neutralization assay based on SARS-CoV-2 S-protein-bearing vesicular stomatitis virus (VSV) pseudovirus and ACE2-overexpressing BHK21 cells. Emerg. Microbes Infect. 2020, 9, 2105–2113. [Google Scholar] [CrossRef]
- Donofrio, G.; Franceschi, V.; Macchi, F.; Russo, L.; Rocci, A.; Marchica, V.; Costa, F.; Giuliani, N.; Ferrari, C.; Missale, G. A Simplified SARS-CoV-2 Pseudovirus Neutralization Assay. Vaccines 2021, 9, 389. [Google Scholar] [CrossRef]
- VanBlargan, L.A.; Goo, L.; Pierson, T.C. Deconstructing the Antiviral Neutralizing-Antibody Response: Implications for Vaccine Development and Immunity. Microbiol. Mol. Biol. Rev. 2016, 80, 989–1010. [Google Scholar] [CrossRef]
- Perera, R.; Ko, R.; Tsang, O.T.Y.; Hui, D.S.C.; Kwan, M.Y.M.; Brackman, C.J.; To, E.M.W.; Yen, H.L.; Leung, K.; Cheng, S.M.S.; et al. Evaluation of a SARS-CoV-2 Surrogate Virus Neutralization Test for Detection of Antibody in Human, Canine, Cat, and Hamster Sera. J. Clin. Microbiol. 2021, 59. [Google Scholar] [CrossRef]
- Gauger, P.C.; Vincent, A.L. Serum virus neutralization assay for detection and quantitation of serum-neutralizing antibodies to influenza A virus in swine. Methods Mol. Biol. 2014, 1161, 313–324. [Google Scholar] [CrossRef]
- Valcourt, E.J.; Manguiat, K.; Robinson, A.; Chen, J.C.; Dimitrova, K.; Philipson, C.; Lamoureux, L.; McLachlan, E.; Schiffman, Z.; Drebot, M.A.; et al. Evaluation of a commercially-available surrogate virus neutralization test for severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). Diagn. Microbiol. Infect. Dis. 2021, 99, 115294. [Google Scholar] [CrossRef]
- Graninger, M.; Jani, C.M.; Reuberger, E.; Prüger, K.; Gaspar, P.; Springer, D.N.; Borsodi, C.; Weidner, L.; Rabady, S.; Puchhammer-Stöckl, E.; et al. Comprehensive Comparison of Seven SARS-CoV-2-Specific Surrogate Virus Neutralization and Anti-Spike IgG Antibody Assays Using a Live-Virus Neutralization Assay as a Reference. Microbiol. Spectr. 2023, 11, e0231422. [Google Scholar] [CrossRef]
- Marien, J.; Michiels, J.; Heyndrickx, L.; Nkuba-Ndaye, A.; Ceulemans, A.; Bartholomeeusen, K.; Madinga, J.; Mbala-Kingebeni, P.; Vanlerberghe, V.; Ahuka-Mundeke, S.; et al. Evaluation of a surrogate virus neutralization test for high-throughput serosurveillance of SARS-CoV-2. J. Virol. Methods 2021, 297, 114228. [Google Scholar] [CrossRef]
- Tan, C.W.; Chia, W.N.; Qin, X.; Liu, P.; Chen, M.I.; Tiu, C.; Hu, Z.; Chen, V.C.; Young, B.E.; Sia, W.R.; et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 2020, 38, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Pishko, A.M.; Bussel, J.B.; Cines, D.B. COVID-19 vaccination and immune thrombocytopenia. Nat. Med. 2021, 27, 1145–1146. [Google Scholar] [CrossRef] [PubMed]
- Rhorer, J.; Ambrose, C.S.; Dickinson, S.; Hamilton, H.; Oleka, N.A.; Malinoski, F.J.; Wittes, J. Efficacy of live attenuated influenza vaccine in children: A meta-analysis of nine randomized clinical trials. Vaccine 2009, 27, 1101–1110. [Google Scholar] [CrossRef] [PubMed]
- Rodda, L.B.; Netland, J.; Shehata, L.; Pruner, K.B.; Morawski, P.A.; Thouvenel, C.D.; Takehara, K.K.; Eggenberger, J.; Hemann, E.A.; Waterman, H.R.; et al. Functional SARS-CoV-2-Specific Immune Memory Persists after Mild COVID-19. Cell 2021, 184, 169–183.e17. [Google Scholar] [CrossRef]
- Jeewandara, C.; Jayathilaka, D.; Gomes, L.; Wijewickrama, A.; Narangoda, E.; Idampitiya, D.; Guruge, D.; Wijayamuni, R.; Manilgama, S.; Ogg, G.S.; et al. SARS-CoV-2 neutralizing antibodies in patients with varying severity of acute COVID-19 illness. Sci. Rep. 2021, 11, 2062. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).