Design and Development of Molecular Beacon-Based Real-Time PCR Assays to Identify Clostridioides difficile Types of Main Evolutionary Clades
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design Overview
2.2. Selection of C. difficile Genomes and Genomes Mapping
2.3. C. difficile Strains Selection, Culture, and Genomic DNA Extraction
2.4. cdtR Gene Amplification
2.5. Analysis of the cdtR Sequences and Design of Real-Time PCR Primers and Probes
2.6. MB-Based Real-Time PCR Assays
3. Results
3.1. Characteristics of C. difficile Strains and Origin of the cdtR Sequences
3.2. cdtR Sequences Alignment and Analysis
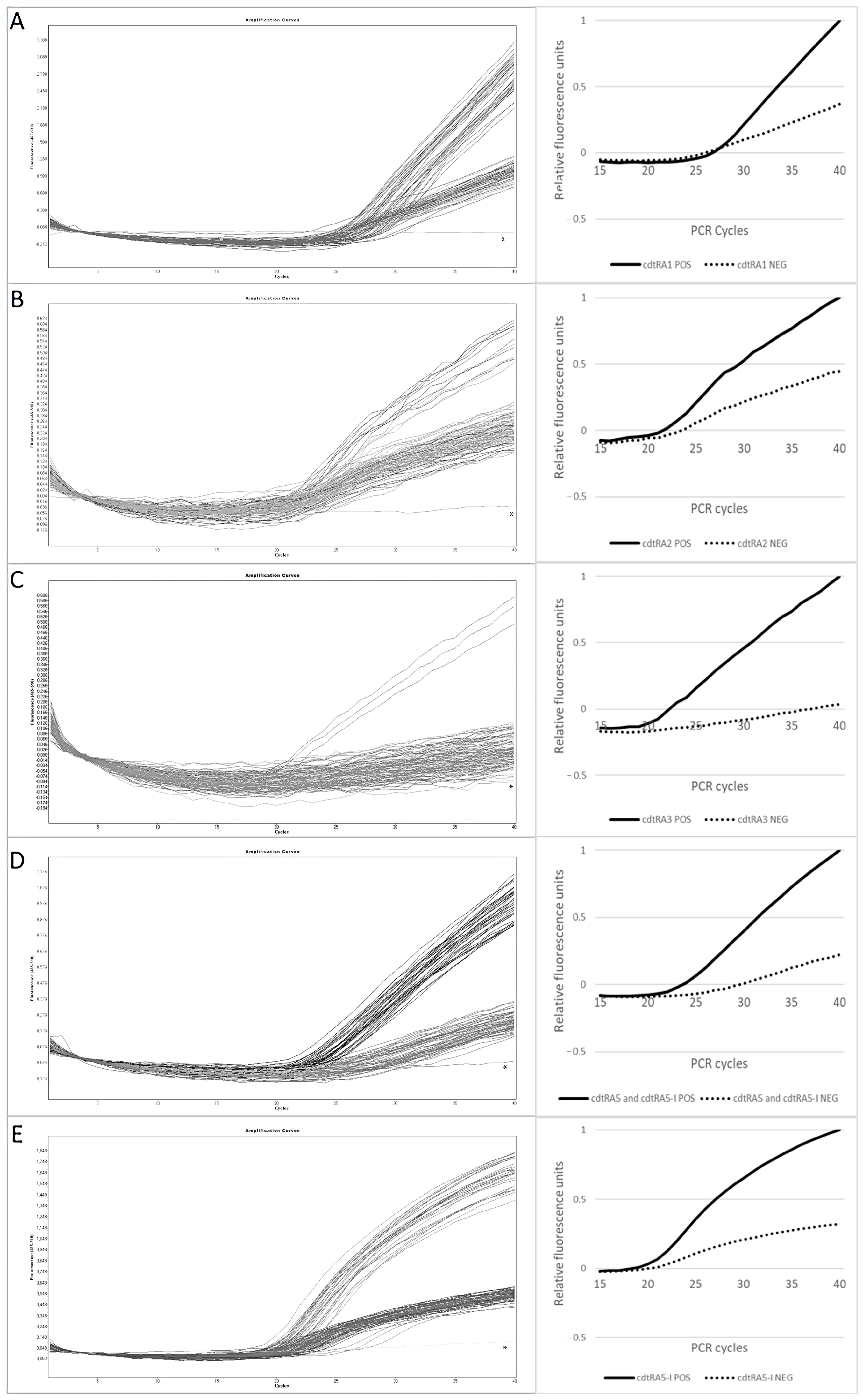
3.3. Optimization of the MB-Based Real-Time PCR Assays
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kuijper, E.J.; Coignard, B.; Tüll, P.; Poxton, I.; Brazier, J.; Duerden, B.; Delmée, M.; Mastrantonio, P.; Gastmeier, P.; Barbut, F.; et al. Emergence of Clostridium difficile-Associated Disease in North America and Europe. Clin. Microbiol. Infect. 2006, 12, 2–18. [Google Scholar] [CrossRef]
- Loo, V.G.; Poirier, L.; Miller, M.A.; Oughton, M.; Libman, M.D.; Michaud, S.; Bourgault, A.-M.; Nguyen, T.; Frenette, C.; Kelly, M.; et al. A Predominantly Clonal Multi-Institutional Outbreak of Clostridium difficile–Associated Diarrhea with High Morbidity and Mortality. N. Engl. J. Med. 2005, 353, 2442–2449. [Google Scholar] [CrossRef]
- Elliott, B.; Androga, G.O.; Knight, D.R.; Riley, T.V. Clostridium difficile Infection: Evolution, Phylogeny and Molecular Epidemiology. Infect. Genet. Evol. 2017, 49, 1–11. [Google Scholar] [CrossRef]
- Lim, S.C.; Knight, D.R.; Riley, T.V. Clostridium difficile and One Health. Clin. Microbiol. Infect. 2020, 26, 857–863. [Google Scholar] [CrossRef]
- Hensgens, M.P.M.; Keessen, E.C.; Squire, M.M.; Riley, T.V.; Koene, M.G.J.; De Boer, E.; Lipman, L.J.A.; Kuijper, E.J. Clostridium difficile Infection in the Community: A Zoonotic Disease? Clin. Microbiol. Infect. 2012, 18, 635–645. [Google Scholar] [CrossRef]
- Fatima, R.; Aziz, M. The Hypervirulent Strain of Clostridium difficile: NAP1/B1/027—A Brief Overview. Cureus 2019, 11, e3977. [Google Scholar] [CrossRef]
- He, M.; Miyajima, F.; Roberts, P.; Ellison, L.; Pickard, D.J.; Martin, M.J.; Connor, T.R.; Harris, S.R.; Fairley, D.; Bamford, K.B.; et al. Emergence and Global Spread of Epidemic Healthcare-Associated Clostridium difficile. Nat. Genet. 2013, 45, 109–113. [Google Scholar] [CrossRef]
- Spigaglia, P.; Mastrantonio, P.; Barbanti, F. Antibiotic Resistances of Clostridium difficile. Adv. Exp. Med. Biol. 2018, 1050, 137–159. [Google Scholar] [CrossRef]
- Keessen, E.C.; Gaastra, W.; Lipman, L.J.A. Clostridium difficile Infection in Humans and Animals, Differences and Similarities. Vet. Microbiol. 2011, 153, 205–217. [Google Scholar] [CrossRef]
- Loo, V.G.; Bourgault, A.-M.; Poirier, L.; Lamothe, F.; Michaud, S.; Turgeon, N.; Toye, B.; Beaudoin, A.; Frost, E.H.; Gilca, R.; et al. Host and Pathogen Factors for Clostridium difficile Infection and Colonization. N. Engl. J. Med. 2011, 365, 1693–1703. [Google Scholar] [CrossRef]
- Liu, C.; Monaghan, T.; Yadegar, A.; Louie, T.; Kao, D. Insights into the Evolving Epidemiology of Clostridioides difficile Infection and Treatment: A Global Perspective. Antibiotics 2023, 12, 1141. [Google Scholar] [CrossRef]
- Shaw, H.A.; Preston, M.D.; Vendrik, K.E.W.; Cairns, M.D.; Browne, H.P.; Stabler, R.A.; Crobach, M.J.T.; Corver, J.; Pituch, H.; Ingebretsen, A.; et al. The Recent Emergence of a Highly Related Virulent Clostridium difficile Clade with Unique Characteristics. Clin. Microbiol. Infect. 2020, 26, 492–498. [Google Scholar] [CrossRef]
- De Roo, A.C.; Regenbogen, S.E.; De Roo, A.C.; Regenbogen, S.E. Clostridium difficile Infection: An Epidemiology Update. Clin. Colon Rectal Surg. 2020, 33, 49–57. [Google Scholar] [CrossRef]
- Goorhuis, A.; Bakker, D.; Corver, J.; Debast, S.B.; Harmanus, C.; Notermans, D.W.; Bergwerff, A.A.; Dekker, F.W.; Kuijper, E.J. Emergence of Clostridium difficile Infection Due to a New Hypervirulent Strain, Polymerase Chain Reaction Ribotype 078. Clin. Infect. Dis. 2008, 47, 1162–1170. [Google Scholar] [CrossRef]
- Kachrimanidou, M.; Tzika, E.; Filioussis, G. Clostridioides (Clostridium) difficile in Food-Producing Animals, Horses and Household Pets: A Comprehensive Review. Microorganisms 2019, 7, 667. [Google Scholar] [CrossRef]
- Weese, J.S. Clostridium (Clostridioides) difficile in Animals. J. Vet. Diagn. Investig. 2020, 32, 213–221. [Google Scholar] [CrossRef]
- Mitchell, M.; Nguyen, S.V.; MacOri, G.; Bolton, D.; McMullan, G.; Drudy, D.; Fanning, S. Clostridioides difficile as a Potential Pathogen of Importance to One Health: A Review. Foodborne Pathog. Dis. 2022, 19, 806–816. [Google Scholar] [CrossRef]
- Spigaglia, P.; Barbanti, F.; Faccini, S.; Vescovi, M.; Criscuolo, E.M.; Ceruti, R.; Gaspano, C.; Rosignoli, C. Clostridioides difficile in Pigs and Dairy Cattle in Northern Italy: Prevalence, Characterization and Comparison between Animal and Human Strains. Microorganisms 2023, 11, 1738. [Google Scholar] [CrossRef]
- Hain-Saunders, N.M.R.; Knight, D.R.; Bruce, M.; Riley, T.V. Clostridioides difficile Infection and One Health: An Equine Perspective. Environ. Microbiol. 2022, 24, 985–997. [Google Scholar] [CrossRef]
- Uzal, F.A.; Navarro, M.A.; Asin, J.; Boix, O.; Ballarà-Rodriguez, I.; Gibert, X. Clostridial Diarrheas in Piglets: A Review. Vet. Microbiol. 2023, 280, 109691. [Google Scholar] [CrossRef]
- Songer, J.G.; Uzal, F.A. Clostridial Enteric Infections in Pigs. J. Vet. Diagn. Investig. 2005, 17, 528–536. [Google Scholar] [CrossRef]
- Goyal, M.; Hauben, L.; Pouseele, H.; Jaillard, M.; De Bruyne, K.; van Belkum, A.; Goering, R. Retrospective Definition of Clostridioides difficile PCR Ribotypes on the Basis of Whole Genome Polymorphisms: A Proof of Principle Study. Diagnostics 2020, 10, 1078. [Google Scholar] [CrossRef]
- Blau, K.; Berger, F.K.; Mellmann, A.; Gallert, C. Clostridioides difficile from Fecally Contaminated Environmental Sources: Resistance and Genetic Relatedness from a Molecular Epidemiological Perspective. Microorganisms 2023, 11, 2497. [Google Scholar] [CrossRef]
- Knight, D.R.; Riley, T.V. Genomic Delineation of Zoonotic Origins of Clostridium difficile. Front. Public Health 2019, 7, 164. [Google Scholar] [CrossRef]
- Knight, D.R.; Squire, M.M.; Collins, D.A.; Riley, T.V. Genome Analysis of Clostridium difficile PCR Ribotype 014 Lineage in Australian Pigs and Humans Reveals a Diverse Genetic Repertoire and Signatures of Long-Range Interspecies Transmission. Front. Microbiol. 2017, 7, 2138. [Google Scholar] [CrossRef]
- Knetsch, C.W.; Connor, T.R.; Mutreja, A.; van Dorp, S.M.; Sanders, I.M.; Browne, H.P.; Harris, D.; Lipman, L.; Keessen, E.C.; Corver, J.; et al. Whole Genome Sequencing Reveals Potential Spread of Clostridium difficile between Humans and Farm Animals in the Netherlands, 2002 to 2011. Eurosurveillance 2014, 19, 20954. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.S.; Hung, Y.P.; Lee, J.C.; Syue, L.S.; Hsueh, P.R.; Ko, W.C. Clostridioides difficile Infection: An Emerging Zoonosis? Expert Rev. Anti. Infect. Ther. 2021, 19, 1543–1552. [Google Scholar] [CrossRef]
- Knight, D.R.; Kullin, B.; Androga, G.O.; Barbut, F.; Eckert, C.; Johnson, S.; Spigaglia, P.; Tateda, K.; Tsai, P.J.; Riley, T.V. Evolutionary and Genomic Insights into Clostridioides difficile Sequence Type 11: A Diverse Zoonotic and Antimicrobial-Resistant Lineage of Global One Health Importance. mBio 2019, 10, 1–17. [Google Scholar] [CrossRef]
- Kurka, H.; Ehrenreich, A.; Ludwig, W.; Monot, M.; Rupnik, M.; Barbut, F.; Indra, A.; Dupuy, B.; Liebl, W. Sequence Similarity of Clostridium difficile Strains by Analysis of Conserved Genes and Genome Content Is Reflected by Their Ribotype Affiliation. PLoS ONE 2014, 9, e86535. [Google Scholar] [CrossRef]
- Knetsch, C.W.; Terveer, E.M.; Lauber, C.; Gorbalenya, A.E.; Harmanus, C.; Kuijper, E.J.; Corver, J.; van Leeuwen, H.C. Comparative Analysis of an Expanded Clostridium difficile Reference Strain Collection Reveals Genetic Diversity and Evolution through Six Lineages. Infect. Genet. Evol. 2012, 12, 1577–1585. [Google Scholar] [CrossRef] [PubMed]
- Knight, D.R.; Imwattana, K.; Kullin, B.; Guerrero-Araya, E.; Paredes-Sabja, D.; Didelot, X.; Dingle, K.E.; Eyre, D.W.; Rodríguez, C.; Riley, T.V. Major Genetic Discontinuity and Novel Toxigenic Species in Clostridioides difficile Taxonomy. eLife 2021, 10, e64325. [Google Scholar] [CrossRef]
- Imwattana, K.; Knight, D.R.; Kullin, B.; Collins, D.A.; Putsathit, P.; Kiratisin, P.; Riley, T.V. Clostridium difficile Ribotype 017–Characterization, Evolution and Epidemiology of the Dominant Strain in Asia. Emerging Microbes and Infections. Emerg. Microbes Infect. 2019, 8, 796–807. [Google Scholar] [CrossRef]
- Knight, D.R.; Elliott, B.; Chang, B.J.; Perkins, T.T.; Riley, T.V. Diversity and Evolution in the Genome of Clostridium difficile. Clin. Microbiol. Rev. 2015, 28, 721–741. [Google Scholar] [CrossRef]
- Knetsch, C.W.; Lawley, T.D.; Hensgens, M.P.; Corver, J.; Wilcox, M.W.; Kuijper, E.J. Current Application and Future Perspectives of Molecular Typing Methods to Study Clostridium difficile Infections. Eurosurveillance 2013, 18, 20381. [Google Scholar] [CrossRef]
- Rodríguez, A.; Rodríguez, M.; Córdoba, J.J.; Andrade, M.J. Design of Primers and Probes for Quantitative Real-Time PCR Methods. Methods Mol. Biol. 2015, 1275, 31–56. [Google Scholar] [CrossRef] [PubMed]
- Prajapati, G.K.; Kumar, A.; Wany, A.; Pandey, D.M. Molecular Beacon Probe (MBP)-Based Real-Time PCR. Methods Mol. Biol. 2023, 2638, 273–287. [Google Scholar] [CrossRef]
- Arya, M.; Shergill, I.S.; Williamson, M.; Gommersall, L.; Arya, N.; Patel, H.R.H. Basic Principles of Real-Time Quantitative PCR. Expert Rev. Mol. Diagn. 2005, 5, 209–219. [Google Scholar] [CrossRef]
- Edwards, K.J.; Logan, J.M.J. Mutation Detection by Real-Time PCR. In Real-Time PCR: Current Technology and Application; Caister Academic Press: Poole, UK, 2009; pp. 185–210. [Google Scholar]
- Wang, C.; Yang, C.J. Application of Molecular Beacons in Real-Time PCR. In Molecular Beacons; Nature Publishing Group: London, UK, 2014; pp. 45–59. [Google Scholar] [CrossRef]
- Lager, M.; Mernelius, S.; Löfgren, S.; Söderman, J. Real-Time PCR Typing of Escherichia coli Based on Multiple Single Nucleotide Polymorphisms-a Convenient and Rapid Method. Clin. Lab. 2016, 62, 349–355. [Google Scholar] [CrossRef]
- Greig, D.R.; Hickey, T.J.; Boxall, M.D.; Begum, H.; Gentle, A.; Jenkins, C.; Chattaway, M.A. A Real-Time Multiplex PCR for the Identification and Typing of Vibrio cholerae. Diagn. Microbiol. Infect. Dis. 2018, 90, 171–176. [Google Scholar] [CrossRef]
- Birdsell, D.N.; Vogler, A.J.; Buchhagen, J.; Clare, A.; Kaufman, E.; Naumann, A.; Driebe, E.; Wagner, D.M.; Keim, P.S. TaqMan Real-Time PCR Assays for Single-Nucleotide Polymorphisms Which Identify Francisella tularensis and Its Subspecies and Subpopulations. PLoS ONE 2014, 9, 223. [Google Scholar] [CrossRef]
- Ben Shabat, M.; Mikula, I.; Gerchman, I.; Lysnyansky, I. Development and Evaluation of a Novel Single-Nucleotide-Polymorphism Real-Time PCR Assay for Rapid Detection of Fluoroquinolone-Resistant Mycoplasma bovis. J. Clin. Microbiol. 2010, 48, 2909–2915. [Google Scholar] [CrossRef][Green Version]
- Dikdan, R.J.; Marras, S.A.E.; Field, A.P.; Brownlee, A.; Cironi, A.; Hill, D.A.; Tyagi, S. Multiplex PCR Assays for Identifying All Major Severe Acute Respiratory Syndrome Coronavirus 2 Variants. J. Mol. Diagn. 2022, 24, 309–319. [Google Scholar] [CrossRef]
- Täpp, I.; Malmberg, L.; Rennel, E.; Wik, M.; Syvänen, A.C. Homogeneous Scoring of Single-Nucleotide Polymorphisms: Comparison of the 5′-Nuclease TaqMan® Assay and Molecular Beacon Probes. Biotechniques 2000, 28, 732–738. [Google Scholar] [CrossRef]
- Wattiau, P.; Fretin, D. Real-Time PCR Typing of Single Nucleotide Polymorphism in DNA Containing Inverted Repeats. Biotechniques 2006, 41, 544–546. [Google Scholar] [CrossRef]
- Carter, G.P.; Lyras, D.; Allen, D.L.; Mackin, K.E.; Howarth, P.M.; O’Connor, J.R.; Rood, J.I. Binary Toxin Production in Clostridium difficile Is Regulated by CdtR, a LytTR Family Response Regulator. J. Bacteriol. 2007, 189, 7290–7301. [Google Scholar] [CrossRef] [PubMed]
- Bouvet, P.J.M.; Popoff, M.R. Genetic Relatedness of Clostridium difficile Isolates from Various Origins Determined by Triple-Locus Sequence Analysis Based on Toxin Regulatory Genes tcdC, tcdR, and cdtR. J. Clin. Microbiol. 2008, 46, 3703–3713. [Google Scholar] [CrossRef][Green Version]
- Lyon, S.A.; Hutton, M.L.; Rood, J.I.; Cheung, J.K.; Lyras, D. CdtR Regulates TcdA and TcdB Production in Clostridium difficile. PLoS Pathog. 2016, 12, e1005758. [Google Scholar] [CrossRef]
- Monot, M.; Eckert, C.; Lemire, A.; Hamiot, A.; Dubois, T.; Tessier, C.; Dumoulard, B.; Hamel, B.; Petit, A.; Lalande, V.; et al. Clostridium difficile: New Insights into the Evolution of the Pathogenicity Locus. Sci. Rep. 2015, 5, 15023. [Google Scholar] [CrossRef]
- Janezic, S.; Dingle, K.; Alvin, J.; Accetto, T.; Didelot, X.; Crook, D.W.; Borden Lacy, D.; Rupnik, M. Comparative Genomics of Clostridioides difficile Toxinotypes Identifies Module-Based Toxin Gene Evolution. Microb. Genom. 2020, 6, mgen000449. [Google Scholar] [CrossRef]
- Marras, S.A.E.; Kramer, F.R.; Tyagi, S. Genotyping SNPs with Molecular Beacons. Methods Mol. Biol. 2003, 212, 111–128. [Google Scholar]
- Vet, J.A.M.; Marras, S.A.E. Design and Optimization of Molecular Beacon Real-Time Polymerase Chain Reaction Assays. Methods Mol. Biol. 2005, 5, 15023. [Google Scholar] [CrossRef]
- Kolpashchikov, D.M. An Elegant Biosensor Molecular Beacon Probe: Challenges and Recent Solutions. Scientifica 2012, 2012, 928783. [Google Scholar] [CrossRef]
- Sebaihia, M.; Wren, B.W.; Mullany, P.; Fairweather, N.F.; Minton, N.; Stabler, R.; Thomson, N.R.; Roberts, A.P.; Cerdeño-Tárraga, A.M.; Wang, H.; et al. The Multidrug-Resistant Human Pathogen Clostridium difficile Has a Highly Mobile, Mosaic Genome. Nat. Genet. 2006, 38, 779–786. [Google Scholar] [CrossRef]
- Marras, S.A.E. Selection of Fluorophore and Quencher Pairs for Fluorescent Nucleic Acid Hybridization Probes. Methods Mol. Biol. 2006, 335, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Knetsch, C.W.; Kumar, N.; Forster, S.C.; Connor, T.R.; Browne, H.P.; Harmanus, C.; Sanders, I.M.; Harris, S.R.; Turner, L.; Morris, T.; et al. Zoonotic Transfer of Clostridium difficile Harboring Antimicrobial Resistance between Farm Animals and Humans. J. Clin. Microbiol. 2018, 56, e01384-17. [Google Scholar] [CrossRef] [PubMed]
- Imwattana, K.; Rodríguez, C.; Riley, T.V.; Knight, D.R. A Species-Wide Genetic Atlas of Antimicrobial Resistance in Clostridioides difficile. Microb. Genom. 2021, 7, 000696. [Google Scholar] [CrossRef]
- Dingle, K.E.; Elliott, B.; Robinson, E.; Griffiths, D.; Eyre, D.W.; Stoesser, N.; Vaughan, A.; Golubchik, T.; Fawley, W.N.; Wilcox, M.H.; et al. Evolutionary History of the Clostridium difficile Pathogenicity Locus. Genome Biol. Evol. 2014, 6, 36–52. [Google Scholar] [CrossRef]
- Markovska, R.; Dimitrov, G.; Gergova, R.; Boyanova, L. Clostridioides difficile, a New “Superbug”. Microorganisms 2023, 11, 845. [Google Scholar] [CrossRef] [PubMed]
- Abad-Fau, A.; Sevilla, E.; Martín-Burriel, I.; Moreno, B.; Bolea, R. Update on Commonly Used Molecular Typing Methods for Clostridioides difficile. Microorganisms 2023, 11, 1752. [Google Scholar] [CrossRef]
- Tümmler, B. Molecular Epidemiology in Current Times. Environ. Microbiol. 2020, 22, 4909–4918. [Google Scholar] [CrossRef]
- Knight, D.R.; Squire, M.M.; Riley, T.V. Nationwide Surveillance Study of Clostridium difficile in Australian Neonatal Pigs Shows High Prevalence and Heterogeneity of PCR Ribotypes. Appl Environ. Microbiol. 2015, 81, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Chrysostomou, A.C.; Aristokleous, A.; Rodosthenous, J.H.; Christodoulou, C.; Stathi, G.; Kostrikis, L.G. Detection of Circulating SARS-CoV-2 Variants of Concern (VOCs) Using a Multiallelic Spectral Genotyping Assay. Life 2023, 13, 304. [Google Scholar] [CrossRef] [PubMed]
- Artika, I.M.; Dewi, Y.P.; Nainggolan, I.M.; Siregar, J.E.; Antonjaya, U. Real-Time Polymerase Chain Reaction: Current Techniques, Applications, and Role in COVID-19 Diagnosis. Genes 2022, 13, 2387. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Wang, Y.; Zhang, W.; Li, W.; Bai, L.; Lu, J.; Ma, C.; Wu, Y. A Rapid Multiplex Real-Time PCR Detection of Toxigenic Clostridioides difficile Directly from Fecal Samples. 3 Biotech 2023, 13, 54. [Google Scholar] [CrossRef] [PubMed]
- Kilic, A.; Alam, M.J.; Tisdel, N.L.; Shah, D.N.; Yapar, M.; Lasco, T.M.; Garey, K.W. Multiplex Real-Time PCR Method for Simultaneous Identification and Toxigenic Type Characterization of Clostridium difficile from Stool Samples. Ann. Lab. Med. 2015, 35, 306–313. [Google Scholar] [CrossRef]
- Kouhsari, E.; Douraghi, M.; Barati, M.; Yaseri, H.F.; Talebi, M.; Abbasian, S.; Moqarabzadeh, V.; Amirmozafari, N. Rapid Simultaneous Molecular Stool-Based Detection of Toxigenic Clostridioides difficile by Quantitative TaqMan Real-Time PCR Assay. Clin. Lab. 2019, 65, 461–469. [Google Scholar] [CrossRef]
- Jayaratne, P.A.; Monkman, L.; Broukhanski, G.; Pillai, D.R.; Lee, C. Real-Time Polymerase Chain Reaction Method for Detection of Toxigenic Clostridium difficile from Stools and Presumptive Identification of NAP1 Clone. Diagn. Microbiol. Infect. Dis. 2013, 75, 121–123. [Google Scholar] [CrossRef]
- Bélanger, S.D.; Boissinot, M.; Clairoux, N.; Picard, F.J.; Bergeron, M.G. Rapid Detection of Clostridium difficile in Feces by Real-Time PCR. J. Clin. Microbiol. 2003, 41, 730–734. [Google Scholar] [CrossRef]
- Metcalf, D.S.; Scott Weese, J. Binary Toxin Locus Analysis in Clostridium difficile. J. Med. Microbiol. 2011, 60, 1137–1145. [Google Scholar] [CrossRef]
- Bilverstone, T.W.; Minton, N.P.; Kuehne, S.A. Phosphorylation and Functionality of CdtR in Clostridium difficile. Anaerobe 2019, 58, 103–109. [Google Scholar] [CrossRef]
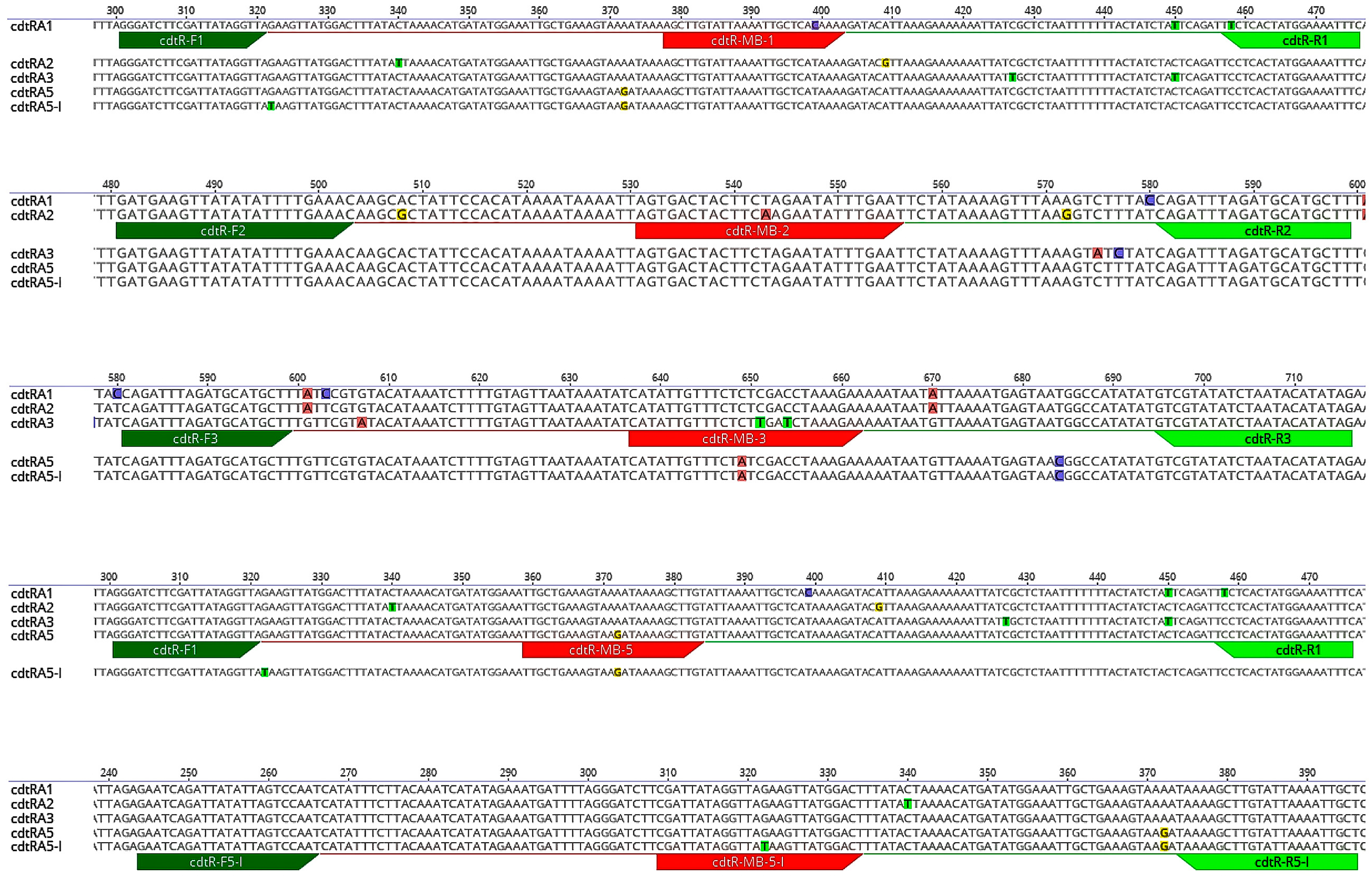

| Parameter | PCR Product | Primers | MB Probes | MB Stem | MB Loop | References |
|---|---|---|---|---|---|---|
| GC content | optimal 21% | optimal 23% | 70–100% | [35,37,38,44,52,53] | ||
| Tm 1 | Optimal 48 °C. Tm of primers should not differ > 2 °C | 3–7 °C > Ta 2 PCR | >Ta PCR | >Tm stem | [35,37,38,44,52,53] | |
| Length | 100–200 bp. Primers and probe distance: 20–50 bp | 15–30 bp (optimal 25 bp) | 40 bp (linear) | 7 bp | 26 bp | [35,37,38,44,52,53] |
| Poly-G/C | Avoided | [35,38,44,52,53,54] | ||||
| Self- dimers | Avoided | MBs have a secondary structure | [35,38,44,52,53,54] | |||
| Primer- dimers | Avoided | Probe or primer dimers should be avoided | [35,38,44,52,53,54] | |||
| G at 5′-end | Avoided | [35,38,44,52,53,54] |
| Clade | cdtR Allele | RT 1 | Toxin A 2 | Toxin B 2 | Binary Toxin CDT 2 | cdtR Gene Sequences from | Total cdtR Sequences (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genomes | ISS Strains | |||||||||||
| Human | Animal | Food/Environment | Unknown | Human | Animal | |||||||
| C1 | cdtRA1 | 001 | + | + | − | 2 | 2 | 1 | 5 (0.9) | |||
| 002 | + | + | − | 1 | 1 (0.2) | |||||||
| 012 | + | + | − | 1 | 5 | 1 | 7 (1.3) | |||||
| 014 | + | + | − | 1 | 8 | 5 | 14 (2.5) | |||||
| 018 | + | + | − | 1 | 10 | 11 (2) | ||||||
| 020 | + | + | − | 1 | 6 | 5 | 12 (2.2) | |||||
| 056 | + | + | − | 1 | 1 (0.2) | |||||||
| 087 | + | + | − | 1 | 1 (0.2) | |||||||
| 106 | + | + | − | 1 | 6 | 3 | 10 (1.8) | |||||
| 607 | + | + | − | 8 | 8 (1.5) | |||||||
| C2 | cdtRA2 | 019 | + | + | + | 1 | 1 | 2 (0.4) | ||||
| 027 | + | + | + | 14 | 1 | 2 | 8 | 25 (4.5) | ||||
| 036 | + | + | + | 1 | 1 (0.2) | |||||||
| 153 | + | + | + | 1 | 1 | 2 (0.4) | ||||||
| 176 | + | + | + | 1 | 1 (0.2) | |||||||
| 181 | + | + | + | 2 | 2 (0.4) | |||||||
| C3 | cdtRA3 | 023 | + | + | + | 2 | 3 | 2 | 7 (1.3) | |||
| 063 | + | + | + | 1 | 1 (0.2) | |||||||
| 212 | + | + | + | 1 | 1 (0.2) | |||||||
| C5 | cdtRA5 | 033 | − | − | + | 19 | 5 | 5 | 4 | 8 | 41 (7.5) | |
| 045 | + | + | + | 2 | 2 | 1 | 5 (0.9) | |||||
| 127 | + | + | + | 33 | 10 | 1 | 44 (8) | |||||
| 288 | − | − | + | 1 | 1 | 2 (0.4) | ||||||
| 126 | + | + | + | 6 | 6 | 12 (2.2) | ||||||
| cdtRA5-I | 126 | + | + | + | 66 | 3 | 12 | 7 | 88 (16) | |||
| 078 | + | + | + | 165 | 41 | 5 | 14 | 17 | 242 (44) | |||
| 193 | + | + | + | 1 | 1 (0.2) | |||||||
| 620 | + | + | + | 1 | 1 | 1 | 3 (0.5) | |||||
| 322 | 68 | 15 | 3 | 93 | 49 | 550 | ||||||
| cdtR Alleles | Primers and Probes | Sequence | Name | SNPs | Amplicon Size |
|---|---|---|---|---|---|
| cdtRA1 | Forward | 5′-GGGATCTTCGATTATAGGTTA-3′ | cdtR-F1 | 399C | 176 bp |
| Reverse | 5′-GAAATTTTCCATAGTGAGGA-3′ | cdtR-R1 | |||
| Probe 1 | 5′-FAM-CGCGATCAGCTTGTATTAAAATTGCTCACAAAAGATCGCG-BHQ1-3′ | cdtR-MB-1 | |||
| cdtRA2 | Forward | 5′-GATGAAGTTATATATTTTGAAAC-3′ | cdtR-F2 | 543A | 119 bp |
| Reverse | 5′-AAGCATGCATCTAAATCTG-3′ | cdtR-R2 | |||
| Probe | 5′-FAM-CGCGATCAGTGACTACTTCAAGAATATTTGAATGATCGCG-BHQ1-3′ | cdtR-MB-2 | |||
| cdtRA3 | Forward | 5′-CAGATTTAGATGCATGCTT-3′ | cdtR-F3 | 651T and 654T | 136 bp |
| Reverse | 5′-CTATATGTATTAGATATACGAC-3′ | cdtR-R3 | |||
| Probe | 5′-FAM-CGCGATCCATATTGTTTCTCTTGATCTAAAGAAGATCGCG-BHQ1-3′ | cdtR-MB-3 | |||
| cdtRA5 and cdtRA5-I | Forward | 5′-GGGATCTTCGATTATAGGTTA-3′ | cdtR-F1 | 372G | 176 bp |
| Reverse | 5′-GAAATTTTCCATAGTGAGGA-3′ | cdtR-R1 | |||
| Probe | 5′-FAM-CGCGATCTTGCTGAAAGTAAGATAAAAGCTTGTGATCGCG-BHQ1-3′ | cdtR-MB-5 | |||
| cdtRA5-I | Forward | 5′-GAATCAGATTATATTAGTCCAAT-3′ | cdtR-F5-I | 322T | 153 bp |
| Reverse | 5′-AGCAATTTTAATACAAGCTTTTA-3′ | cdtR-R5-I | |||
| Probe | 5′-FAM-CGCGATCCGATTATAGGTTATAAGTTATGGACTGATCGCG-BHQ1-3′ | cdtR-MB-5-I |
| Program | Cycles | Analysis Mode | Target (°C) | Acquisition Mode | Hold (hh:mm:ss) | Ramp Rate (°C/s) | Acquisition (per °C) |
|---|---|---|---|---|---|---|---|
| pre-incubation | 1 | None | 95 | None | 00:05:00 | 4.4 | |
| amplification | 40 | Quantification | 95 | None | 00:00:10 | 4.4 | |
| 48 | Single | 00:00:15 | 1.5 | 5 | |||
| 72 | None | 00:00:01 | 4.4 | ||||
| cooling | 1 | None | 40 | None | 00:00:10 | 1.5 |
| cdtR Allele Assay | Range of F 1 (40th Cycle) | Mean F ± STD 2 (40th Cycle) | RFU 3 | |||
|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | Positive | Negative | |
| cdtRA1 | 2.22–3.35 | 0.79–1.54 | 2.75 ± 0.35 | 1 ± 0.05 | 1 | 0.3 |
| cdtRA2 | 0.47–0.61 | 0.15–0.32 | 0.55 ± 0.05 | 0.24 ± 0.05 | 1 | 0.4 |
| cdtRA3 | 0.49–0.57 | 0–0.18 | 0.56 ± 0.04 | 0.02 ± 0.05 | 1 | 0 |
| cdtRA5 | 0.84–1.16 | 0.14–0.36 | 0.98 ± 0.09 | 0.22 ± 0.08 | 1 | 0.25 |
| cdtRA5-I | 1.39–1.79 | 0.11–0.55 | 1.59 ± 0.12 | 0.51 ± 0.10 | 1 | 0.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Criscuolo, E.M.; Barbanti, F.; Spigaglia, P. Design and Development of Molecular Beacon-Based Real-Time PCR Assays to Identify Clostridioides difficile Types of Main Evolutionary Clades. Microbiol. Res. 2024, 15, 354-370. https://doi.org/10.3390/microbiolres15010024
Criscuolo EM, Barbanti F, Spigaglia P. Design and Development of Molecular Beacon-Based Real-Time PCR Assays to Identify Clostridioides difficile Types of Main Evolutionary Clades. Microbiology Research. 2024; 15(1):354-370. https://doi.org/10.3390/microbiolres15010024
Chicago/Turabian StyleCriscuolo, Enrico Maria, Fabrizio Barbanti, and Patrizia Spigaglia. 2024. "Design and Development of Molecular Beacon-Based Real-Time PCR Assays to Identify Clostridioides difficile Types of Main Evolutionary Clades" Microbiology Research 15, no. 1: 354-370. https://doi.org/10.3390/microbiolres15010024
APA StyleCriscuolo, E. M., Barbanti, F., & Spigaglia, P. (2024). Design and Development of Molecular Beacon-Based Real-Time PCR Assays to Identify Clostridioides difficile Types of Main Evolutionary Clades. Microbiology Research, 15(1), 354-370. https://doi.org/10.3390/microbiolres15010024






