Abstract
The production of biosurfactants from organic wastes has received significant attention due to its potential cost savings. This study involved the isolation of biosurfactant-producing microorganisms from waste sources. The surfactant properties of the 37 studied isolates were assessed by reducing surface tension and their emulsifying properties, determined by the emulsification index E24. We assessed the ability of these isolated strains to produce biosurfactants using various waste substrates, namely potato peelings, waste cooking oil and sunflower cake. Our results showed that sunflower cake exhibited better growth and biosurfactant production for most of the strains studied. This highlights that sunflower cake is a potentially effective and economical substrate for the production of biosurfactants. The most effective strains allowing to achieve an emulsification index above 50% and reduce surface tension below 40 mN m−1 were Enterobacter sp. 2pp, strain 2wfo, Peribacillus sp. 1mo, Sphingomonas sp. 2mo, Ochrobactrum sp. 5mo, Shouchella sp. 6mo, Bacillus sp. 1os, Bacillus sp. 2os. Among these strains, both previously known strains as biosurfactant producers and previously unknown strains were found. Thus, we found that among representatives of the genus Sphingomonas there are effective producers of biosurfactants. The highest yield of biosurfactant on a medium with glycerol and glucose was shown by the Bacillus sp. 2os strain of 0.501 and 0.636 g L−1, respectively.
1. Introduction
Rising environmental concerns have propelled a quest for novel, eco-friendly methodologies across diverse domains. One such area of interest involves the investigation of substituting synthetic surfactants with biologically derived alternatives [1,2]. These biologically derived surfactants, termed biosurfactants, are currently under exploration for their potential applications in agriculture, the bioremediation of oil-contaminated soils, and enhanced oil recovery.
However, despite the burgeoning interest in biosurfactants, their practical applications remain restricted due to the high production costs associated with them. Potential resolutions to this challenge encompass the development and scaling of technological processes and reduction in the expenses tied to raw materials utilized. An emerging strategy involves the use of organic waste as a primary raw material, presenting a promising avenue to significantly diminish the cost of biosurfactants while simultaneously fostering the integration of waste materials into recycling cycles [3,4,5].
Organic wastes from food and agricultural industries, industrial byproducts such as wastewater, raw glycerol, and waste generated from meat production, as well as oil-contaminated soil, are all considered viable substrates for biosurfactant production. These waste materials are rich in nutrients, encompassing a wide array of sources such as sugarcane molasses, crop cakes, banana, orange, and potato peels, waste frying oils, coconut oils, rapeseed oils, moringa and cassava residues, distillery waste, effluents from food and vegetable production, coffee wastewater, among others [6]
The selection of the substrate for biosurfactant production is pivotal as various carbon sources exert differing effects on product yield. For instance, Aparna et al. (2012) discovered that Pseudomonas sp. 2B yielded varying amounts of rhamnolipid, producing 4.14, 4.38, 3.24, and 4.09 g L−1 when cultivated on glycerol, coconut oil cake, orange peel, and whey, respectively [7]. Similarly, Moussa (2014) observed that the rhamnolipid yield of Pseudomonas aeruginosa TMN was 2.9 ± 0.02 g L−1 on a glucose substrate and 1.35 ± 0.01 g L−1 on glycerol, while sucrose resulted in a notably lower yield of 0.91 g L−1 [8].
The metabolic pathways and resultant homologues of biosurfactants display significant variability contingent upon the strain used, as well as the substrate employed [9,10,11,12]. Ndlovu et al. (2017) studied surfactin analogues obtained from Bacillus amyloliquefasciens ST34, noting the dominance of surfactins C13–C15 across samples. Similarly, P. aeruginosa ST5 generated six rhamnolipid congeners, with Rha–C10–C10 being the most abundant [13]. Mouafo et al. (2017) highlighted the ability of three Lactobacillus strains to produce biosurfactants with higher lipid content on a glycerol substrate compared to sugarcane molasses [12]. This difference was attributed to the mechanism of glycerol consumption primarily directed toward the lipolytic pathway and gluconeogenesis, allowing for the production of fatty acids and sugars [14].
Moreover, the properties of resulting surfactants are contingent upon the substrate used. Surfactant biomolecules, classified into surfactants and emulsifiers, exhibit distinct roles: surfactants reduce surface tension while emulsifiers partake in the formation and stabilization of emulsions [15]. Some biomolecules, however, possess both surfactant and emulsifying properties. Distinguishing these properties necessitates the application of diverse evaluation methodologies [16]. Biosurfactants are identified through methods assessing the reduction in surface and interfacial tension, while bioemulsifiers form stable emulsions without significant alterations in surface/interfacial tension across phases. For instance, Stoimenova (2014) reported the ability of an indigenous strain of industrial wastewater Pseudomonas fluorescens to produce glycolipid biosurfactants in a medium containing hexadecane, mineral oil, vegetable oil, and glycerol, with the highest emulsifying capability observed in the vegetable oil medium [17].
The influence of substrate on biosurfactant biosynthesis highlights the need for careful substrate selection. The use of organic wastes as substrates for the synthesis of biosurfactants has not been sufficiently studied, since only a small number of wastes have been studied as substrates. In particular, the study has not previously been carried out on the isolation of microorganisms and the build-up of biosurfactants on different types of organic municipal and agricultural wastes. In the present study, isolates were obtained and tested for their ability to synthesize biosurfactants on various organic wastes. Subsequently, the most effective waste-microorganism pair was selected, giving the highest biosurfactant productivity.
2. Materials and Methods
2.1. Waste Sampling
To isolate microorganisms, the following organic wastes were selected: waste from grease traps and oil traps of a water utility (Chelnyvodokanal LLC, Naberezhnye Chelny, Russia), potato peelings and used frying oil (catering restaurants, Kazan, Russia), rapeseed and sunflower cake (JSC Kazan Oil Extraction Plant, Kazan, Russia), oil-contaminated soil; soil contaminated with motor oil (car service areas, Kazan, Russia).
2.2. Isolation of Microorganisms from Wastes
Isolates of microorganisms from wastes were cultivated using a minimal medium (g L−1): NaNO3 2.0, KH2PO4 0.5, K2HPO4 1.0, MgSO4 7H2O 0.5, KCl 0.1, FeSO4 7H2O 0.01. The pH was adjusted to 7.0 with 1 N HCl/NaOH. The investigated wastes were added as the sole carbon source to the autoclaved medium. Various carbon sources were separately introduced into the medium. The initial waste materials, such as potato peelings, rapeseed and sunflower cake, oil-contaminated soil, soil contaminated with motor oils, and waste from water utility grease traps and oily sludge, as well as waste frying oil, were included in the mineral medium at a volume of 2% (v/v) [18].
Bacterial strains were inoculated in 250 mL flasks containing 100 mL of medium and incubated at 120 rpm at 28 °C for 72 h. Following incubation, the strains were subjected to the limiting dilution method and subsequently sown on a solid MPA medium. Individual isolates of microorganisms were obtained by evaluating the morphological characteristics of colonies on a solid medium in a Petri dish and through microscopic analysis utilizing an Axio Lab A1 light microscope (Carl Zeiss, Jena, Germany).
2.3. Assessment of Biosurfactant Production Ability
The assessment of emulsifying ability was conducted using the E24 method [19]. All measurements were carried out in triplicate. This procedure involved combining an equal volume of cell-free strain culture supernatant with crude oil. The cell-free supernatant was obtained via centrifugation of the liquid cell culture (10 min, 5000 rpm). The resulting mixture was vortexed and allowed to stand for 24 h at room temperature. Subsequently, the height of the emulsified column was measured, and E24 was calculated using the formula:
The cell-free strain culture supernatant was analyzed for the reduction in water surface tension (ST) using the Du Nouy ring method with a K20 tensiometer (KRUSS, Hamburg, Germany) at room temperature [20].
2.4. Determination of Strain Species
The total genomic DNA of the isolates was extracted utilizing a K-Sorb reagent kit for DNA extraction on microcolumns (SINTHOL Company, Moscow, Russia). Subsequently, the nucleotide sequence of the samples was determined through the Sanger sequencing method employing an ABI 3730 DNA Analyzer (Life Technologies, Carlsbad, CA, USA). Genomic libraries were constructed using general bacterial primers 27f-1492r [21]. The sequences obtained for each strain were matched with the sequences from the database using the BLAST NCBI. The sequences were aligned using MEGA 10.0 software.
2.5. Cultivation of Isolates on Different Waste Types
The chosen isolates were cultured on four distinct substrates: pure glycerol, potato peelings, waste frying oil, and sunflower cake. For the preparation of potato peelings and sunflower cake, both were dried at 55 °C for 4 days, crushed into fine powder, and subsequently, a 10% (w/v) solution of the powder in distilled water was autoclaved. The solution was filtered through gauze to obtain a clear filtrate, which was then added sterilely at a 4% (v/v) concentration to the previously described mineral medium. Waste frying oil and glycerol were directly added to the medium at a 2% (v/v) concentration before autoclaving. Biosurfactant producers were grown in 500 mL flasks containing 200 mL of medium at 120 rpm at 28 °C for 72 h.
2.6. Extraction of Biosurfactants
The cell culture was centrifuged at 5000 rpm for 10 min at room temperature to obtain cell-free supernatant. Extraction of biosurfactants was carried out using the acid precipitation method—bringing the cell-free supernatant to pH = 2 using 5 N HCl. The acidified supernatant was kept at 4 °C overnight and then centrifuged at 4 °C for 40 min at 3700 rpm. The biosurfactant precipitate was dissolved in a mixture of methanol and chloroform in a ratio of 2:1 and filtered using a 0.22 µm filter (Sartorius, Gottingen, Germany). The biosurfactant was obtained by evaporating the solvent on a rotary evaporator (IKA, Staufen im Breisgau, Germany) and evaluated gravimetrically.
2.7. Statistical Analysis
The error bars depicted in the figures indicate the standard error of means derived from the replicates. To evaluate the data within groups of values (characterizing each substrate type and individual strain), weights were assigned, and the weighted arithmetic means were computed. The statistical analysis was carried out using Statistica 10.0 software (StatSoft Inc., Tulsa, OK, USA). Graphs were generated using Microsoft Excel 2016 MSO (Microsoft, Redmond, WA, USA).
3. Results and Discussion
3.1. Isolation of Biosurfactant-Producing Microorganisms from Waste
Microorganisms capable of biosurfactant synthesis were isolated from various categories of wastes, including agricultural sources (potato peelings, rapeseed, and sunflower cake), industrial sources (oil-contaminated soil and soil contaminated with motor oils), and municipal wastes (waste from grease traps and water utility oily sludge, waste frying oil).
A total of 37 strains were isolated from these waste materials, and their genera were identified through Sanger sequencing (Table 1).

Table 1.
Microorganisms isolated from waste.
The investigation revealed a significant variance in the number of strains isolated from different waste sources. The largest number of strains was retrieved from oil-contaminated soils, while the smallest count originated from rapeseed cake. Among the total isolated strains, 33 were identified at the genus level, and four strains remained unidentified.
The dominant genus observed among these isolates was Sphingomonas sp., with ten strains, followed by six strains attributed to the genus Bacillus sp. Notably, five strains were isolated from potato peelings, predominantly affiliated with the genus Enterobacter sp., a common genus found in organic wastes [22]. The prevalence of Sphingomonas sp. in natural environments, such as soils, is noteworthy as they demonstrate the ability to degrade hydrocarbons [23].
The majority of the strains from oil-contaminated soils (10 strains) were divided between Sphingomonas and Bacillus, with each genus accounting for four strains. Conversely, the minimum number of strains emerged from rapeseed cake, predominantly featuring Pantoea sp. 1rc. The Pantoea genus encompasses a diverse array of bacteria isolated from various environments and is commonly found in the rhizosphere of rapeseed [24].
3.2. Evaluation of Isolates for Their Surface Tension Reduction Abilities
The isolates were assessed for their emulsifying and water surface tension reduction abilities, primarily on a conventional medium containing glycerol (Table 2). Research indicates that a crucial criterion for substantial surface-active properties is the reduction of surface tension to around 40 mN m−1 [25]. Consequently, among the isolates tested, six strains demonstrated notable surface tension reduction capabilities. Notable strains encompassed 3sc, Sphingomonas sp. 5sc (derived from sunflower cake), Peribacillus sp. 1mo, Sphingomonas sp. 2mo, Ochrobactrum sp. 5mo (isolated from soil contaminated with motor oil), as well as Bacillus sp. 5os (from oil-contaminated soil). Among these, there are well-recognized biosurfactant producers, such as representatives of the Peribacillus genus (previously known as Brevibacterium) [26], reducing surface activity to levels of 25.9–27.6 mN m−1 [27,28]. Additionally, the Ochrobactrum genus, isolated from motor oil-contaminated soil, managed to reduce the surface tension of the growth medium from 70 to 30.8 mN m−1 [29]. An intriguing aspect of this study is that the three members of the genus Sphingomonas we isolated exhibited significant emulsifying activity. In particular, strain 2mo, identified as Sphingomonas sp., showed a remarkable surface activity of 39.7 mN m−1. Whereas previously, other studies noted that most Sphingomonas strains do not have the ability to produce biosurfactants [30].

Table 2.
Surface tension on culture media with different carbon substrates.
The subsequent phase involved selecting more cost-effective substrates to augment biosurfactant production. To this end, three substrate types—potato peelings, used frying oil, and sunflower cake—were chosen. The efficacy of all isolated strains on these selected waste materials was evaluated by measuring the degree of surface tension reduction (Table 2). These particular wastes were identified as the most high-volume waste sources in the city of Kazan, Republic of Tatarstan, Russia.
An analysis of these waste sources for cultivating biosurfactant producers revealed certain trends (Table 2). Notably, sunflower cake emerged as the most effective substrate, facilitating significant biosurfactant production with noteworthy surface activity in nine strains spanning various genera: 2wfo, Sphingomonas sp. 2mo, Nocardiopsis sp. 3mo, Ochrobactrum sp. 5mo, Shouchella sp. 6mo, Bacillus sp. 1os, Bacillus sp. 2os, Bacillus sp. 3os, Bacillus sp. 5os.
Conversely, used frying oil was the least effective substrate for biosurfactant production, leading to an effective reduction in surface tension in only four strains of genus—Sphingomonas sp. 2mo, Ochrobactrum sp. 5mo, Bacillus sp. 2os, Bacillus sp. 3os.
Among representatives of the genus Bacillus, for example, the species Bacillus toyonensis is known, which was previously isolated from oil-contaminated areas and demonstrated the ability to reduce surface tension to 47 mN m−1 [31]. Similarly, Chaurasia et al. (2020) isolated Bacillus tequilensis LK5.4 from soybean, displaying the capacity to reduce the surface tension of the culture medium by up to 40% [32]. Nocardiopsis sp. B4, isolated from seawater, exhibited a surface tension decrease to 29 mN m−1 during cultivation, with an E24 emulsification index of 80% [3].
3.3. Evaluation of Emulsifying Properties of Isolates
In the subsequent stage, all isolates were examined for the presence of emulsifying properties in their produced metabolites. Emulsification is considered significant when the emulsification index exceeds 50% [33,34] (Table 3).

Table 3.
E24 values in culture media with different carbon substrates.
On traditional glycerol-based sources, eight strains—identified as genus Enterobacter sp. 2pp, 4pp (from potato peelings), Pantoea sp. 1rc (from rapeseed cake), 3sc (from sunflower cake), Peribacillus sp. 1mo, Sphingomonas sp. 2mo, Ochrobactrum sp. 5mo (from soil contaminated with motor oil), and Bacillus sp. 1os (from oil-contaminated soil)—demonstrated substantial emulsifying activity (over 50%).
Several of these genus have been previously identified for their emulsification capabilities. For instance, Rabiei (2013) noted that a consortium of Enterobacter sp. genus representatives could produce biosurfactants with an emulsification index exceeding 70%, thereby reducing the surface tension of the nutrient medium from 72 to 31 mN m−1 [35]. Additionally, Essghaier et al. (2023) demonstrated that endophytes of Pantoea alhagi species are capable of producing biosurfactants with an emulsifying activity of 82% [36]. The species representative of B. tequilensis LK5.4 exhibited a maximum emulsification index of 52% [32].
Upon assessing new waste substrates, it was observed that the selected waste materials yielded substances with comparatively lower emulsifying activity than glycerol. Specifically, while glycerol facilitated the synthesis of bioemulsifiers in 8 strains, potato peelings and cake showed 4, and used frying oil demonstrated this trait in 3 strains.
These results indicated that metabolites from strains exhibiting high surface activity may not always exhibit high emulsifying activity. However, metabolites of such strains as 4pp, Peribacillus sp. 1mo, Sphingomonas sp. 2mo, and Ochrobactrum sp. 5mo have both of these activities. It is known from the literature that many bacterial strains produce a diverse mixture of analogues and congeners of biosurfactants under the influence of a single carbon source present in the nutrient medium [37]. Moreover, studies show that surface tension values and emulsification indices may not be uniformly correlated for the same strains [16]. Microorganisms are capable of synthesizing various mixtures of heteropolysaccharides, lipopolysaccharides, lipoproteins and proteins, potentially indicating varying degrees of biosurfactant and bioemulsifying properties. It is known that substances with low molecular weight have mainly surface activity, while substances with high molecular weight are effective as emulsion stabilizers [38]. The combination of polysaccharides, fatty acids and protein components in bioemulsifiers allows achieving greater emulsifying potential [16]. Some studies note the influence of the substrate C/N ratio on surface and emulsifying activity, which is not detected at the same C/N values [39]. Notably, other studies also note the ability of microorganisms of the genera Peribacillus sp., Sphingomonas sp. and Ochrobactrum sp. to produce both biosurfactants and bioemulsifiers, while representatives of the genera Enterobacter sp. and Pantoea sp. produce substances that are mainly biosurfactants.
It’s evident that different strains exhibited varied performance across different substrates. Notably, strains such as Bacillus sp. 2os, Bacillus sp. 3os и Sphingomonas sp. 2mo displayed activity across multiple substrates, whereas Peribacillus sp. 1mo solely showed effectiveness in the glycerol substrate. This variability might be attributed to the individual metabolic capabilities of each specific strain.
The process of identifying the most effective substrate involved the assignment of weight to each parameter (Supplement Table S1). Weight scores were designated to the lowest surface tension (ST) values and the highest E24 values, which were then used to calculate the weighted arithmetic mean (Supplement Table S2). Based on these assessments, it was determined that sunflower cake served as the most effective medium for cultivating microorganisms with surface-active properties. In contrast, glycerol displayed greater suitability for emulsifying properties. Sunflower cake not only supported the growth of microorganisms with emulsifying properties but also secured the second-highest position in the ranking. Considering our objective to select a cost-effective substrate for further investigations, sunflower cake was selected, while potato peelings were identified as the least effective substrate. The efficacy of sunflower cake as a substrate for biosurfactant production has been corroborated by numerous studies [40,41]. For instance, Ciurko et al. (2022) reported that sunflower cake enabled the enhancement of Bacillus subtilis surfactin at a concentration of 1.19 ± 0.03 g L−1 [39].
During the evaluation of the most effective strains, the individual weights for each value and their corresponding weighted arithmetic mean were analyzed. Remarkably, the most effective strains, those with weight scores surpassing 20, included eight specific strains: Enterobacter sp. 2pp, strain 2wfo, Peribacillus sp. 1mo, Sphingomonas sp. 2mo, Ochrobactrum sp. 5mo, Shouchella sp. 6mo, Bacillus sp. 1os, Bacillus sp. 2os.
3.4. Biosurfactants Yield
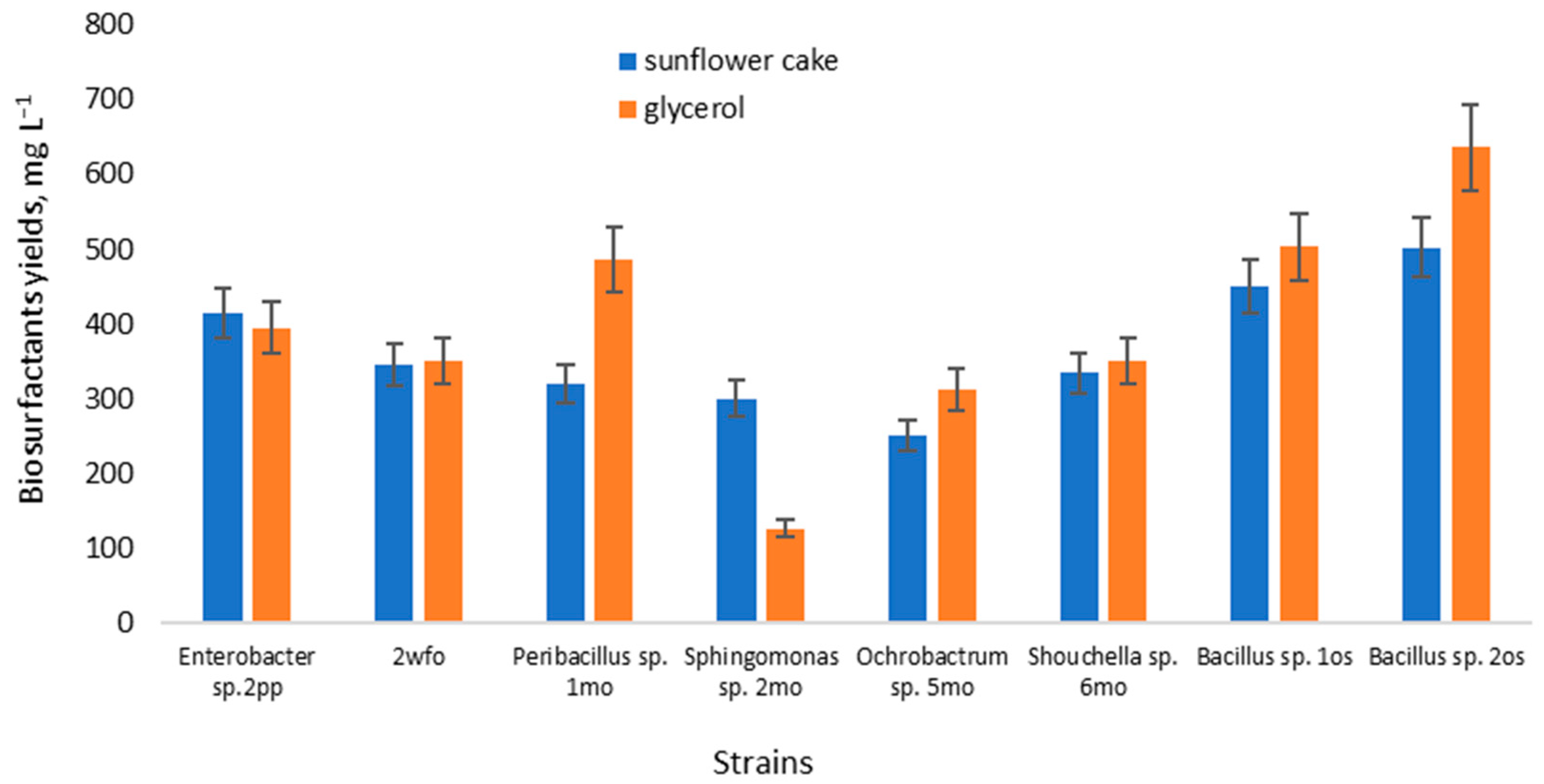
The yield of biosurfactants for proficient strains in a medium based on sunflower cake was evaluated (Figure 1). Additionally, the yield of biosurfactants in a conventional glycerol-containing medium was also assessed for comparison.
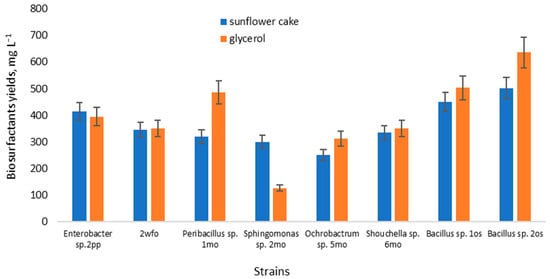
Figure 1.
Biosurfactant yield from glycerol and sunflower cake medium.
In the glycerol-based medium, the biosurfactant yield ranged from 126–636 mg L−1, while in the medium with sunflower cake, it was between 250–502 mg L−1. Notably, the strain of Sphingomonas sp. 2mo exhibited the lowest yield in the glycerol-based medium, while a strain of the genus Ochrobactrum sp. 5mo showed the lowest yield in the sunflower cake medium. These yields align with existing literature data [8,42]. Rane et al. (2017) reported biosurfactant production in minimal media, noting 0.207 g L−1 when glucose served as the sole carbon source, and 0.241 g L−1 with molasses [18]. However, biosurfactant production wasn’t feasible using whey as the carbon source. Additionally, Rane found that agro-wastes, bagasse, and orange peel extracts yielded 0.127 and 0.089 g L−1 of biosurfactant, respectively. Das (2018) discovered that Pseudomonas azotoformans AJ15 yielded rhamnolipids in the range of 0.6–0.76 g L−1 while cultivating on potato peelings ranging from 5–15 g L−1 [43]. Similar concentrations of sugarcane waste resulted in yields of 0.6–0.97 g L−1.
Notably, the strain Bacillus sp. 2os achieved the highest yield in both glycerol and sunflower cake mediums. Other studies confirm the potential of this genus to attain biosurfactant yields within the range of 1–2.5 g L−1 [30]. However, these results indicate the need for further optimization of the nutrient medium. Despite the slightly superior yield observed based on glycerol, sunflower cake exhibited comparable results and proved to be more economically advantageous.
The manifestation of surface-active and emulsifying properties in biosurfactants of the same microorganisms on different substrates can be explained by the different hydrophilic-lipophilic balances of these substrates. The different hydrophilic-lipophilic balance of sunflower cake and glycerol ensures the synthesis of biosurfactants of different molecular weights with different properties [44].
4. Conclusions
The findings of this study revealed that strains isolated from various sources, including potato peelings, rapeseed and sunflower cake, oil-contaminated soil, soil contaminated with motor oils, waste from water utility grease traps and oily sludge, as well as used frying oil, possess the capacity for biosurfactant production. From these isolates, 8 microorganisms were identified as displaying the greatest potential in biosurfactant synthesis with surfactant and emulsifying properties (Enterobacter sp. 2pp, strain 2wfo, Peribacillus sp. 1mo, Sphingomonas sp. 2mo, Ochrobactrum sp. 5mo, Shouchella sp. 6mo, Bacillus sp. 1os, Bacillus sp. 2os), which will be utilized in further investigations. In this study, for the first time, representatives of the genus Sphingomonas were isolated that have the ability to synthesize biosurfactants with significant emulsifying and surfactant properties. Based on the biosurfactant yields, the Bacillus sp. 2os strain demonstrated the highest biosurfactant production capability—0.501 and 0.636 g L−1 for glycerol and glucose, respectively. The study investigated the feasibility of utilizing three types of waste as carbon sources: potato peelings, rapeseed and sunflower cake, and waste frying oil. It was determined that among all substrates, sunflower cake exhibited the most promising potential for biosurfactant production. This substrate led to the most substantial reduction in surface tension and the highest emulsification index among the majority of the strains tested.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microbiolres15010010/s1, Table S1: Weight scores; Table S2: Weighted arithmetic means.
Author Contributions
Conceptualization, S.S.; methodology, L.B.; validation, A.G.; formal analysis, P.K.; investigation, L.B. and A.K.; data curation, S.S.; writing—original draft preparation, A.G. and L.B.; writing—review and editing, P.K.; visualization, A.K.; supervision, S.S.; funding acquisition, P.K. All authors have read and agreed to the published version of the manuscript.
Funding
The work has been performed with the financial support of the Russian Science Foundation, grant No. 23-24-00611.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Mohanty, S.S.; Koul, Y.; Varjani, S.; Pandey, A.; Ngo, H.H.; Chang, J.S.; Wong, J.W.C.; Bui, X.T. A Critical Review on Various Feedstocks as Sustainable Substrates for Biosurfactants Production: A Way towards Cleaner Production. Microb. Cell Fact. 2021, 20, 120. [Google Scholar] [CrossRef] [PubMed]
- Galitskaya, P.; Karamova, K.; Biktasheva, L.; Galieva, G.; Gordeev, A.; Selivanovskaya, S. Lipopeptides Produced by Bacillus Mojavensis P1709 as an Efficient Tool to Maintain Postharvest Cherry Tomato Quality and Quantity. Agriculture 2022, 12, 609. [Google Scholar] [CrossRef]
- Khopade, A.; Biao, R.; Liu, X.; Mahadik, K.; Zhang, L.; Kokare, C. Production and Stability Studies of the Biosurfactant Isolated from Marine Nocardiopsis sp. B4. Desalination 2012, 285, 198–204. [Google Scholar] [CrossRef]
- Carolin, C.F.; Senthil Kumar, P.; Mohanakrishna, G.; Hemavathy, R.V.; Rangasamy, G.; M Aminabhavi, T. Sustainable Production of Biosurfactants via Valorisation of Industrial Wastes as Alternate Feedstocks. Chemosphere 2023, 312, 137326. [Google Scholar] [CrossRef] [PubMed]
- Gaur, V.K.; Sharma, P.; Sirohi, R.; Varjani, S.; Taherzadeh, M.J.; Chang, J.-S.; Yong Ng, H.; Wong, J.W.C.; Kim, S.-H. Production of Biosurfactants from Agro-Industrial Waste and Waste Cooking Oil in a Circular Bioeconomy: An Overview. Bioresour. Technol. 2022, 343, 126059. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Patil, Y.; Rale, V. Biosurfactant Production: Emerging Trends and Promising Strategies. J. Appl. Microbiol. 2019, 126, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Aparna, A.; Srinikethan, G.; Smitha, H. Production and Characterization of Biosurfactant Produced by a Novel Pseudomonas sp. 2B. Colloids Surf. B Biointerfaces 2012, 95, 23–29. [Google Scholar] [CrossRef]
- Moussa, T.A.A.; Mohamed, M.S.; Samak, N. Production and Characterization of Di-Rhamnolipid Produced by Pseudomonas Aeruginosa TMN. Braz. J. Chem. Eng. 2014, 31, 867–880. [Google Scholar] [CrossRef]
- Bonmatin, J.-M.; Laprevote, O.; Peypoux, F. Diversity Among Microbial Cyclic Lipopeptides: Iturins and Surfactins. Activity-Structure Relationships to Design New Bioactive Agents. Comb. Chem. High Throughput Screen. 2012, 6, 541–556. [Google Scholar] [CrossRef]
- Das, P.; Mukherjee, S.; Sen, R. Substrate Dependent Production of Extracellular Biosurfactant by a Marine Bacterium. Bioresour. Technol. 2009, 100, 1015–1019. [Google Scholar] [CrossRef]
- Singh, A.K.; Rautela, R.; Cameotra, S.S. Substrate Dependent in Vitro Antifungal Activity of Bacillus Sp Strain AR2. Microb. Cell Fact. 2014, 13, 67. [Google Scholar] [CrossRef] [PubMed]
- Mouafo, T.H.; Mbawala, A.; Ndjouenkeu, R. Effect of Different Carbon Sources on Biosurfactants’ Production by Three Strains of Lactobacillus spp. Biomed Res. Int. 2018, 2018, 5034783. [Google Scholar] [CrossRef] [PubMed]
- Ndlovu, T.; Rautenbach, M.; Khan, S.; Khan, W. Variants of Lipopeptides and Glycolipids Produced by Bacillus Amyloliquefaciens and Pseudomonas Aeruginosa Cultured in Different Carbon Substrates. AMB Express 2017, 7, 109. [Google Scholar] [CrossRef]
- Santos, D.; Rufino, R.; Luna, J.; Santos, V.; Sarubbo, L. Biosurfactants: Multifunctional Biomolecules of the 21st Century. Int. J. Mol. Sci. 2016, 17, 401. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh-Sani, M.; Hamishehkar, H.; Khezerlou, A.; Azizi-Lalabadi, M.; Azadi, Y.; Nattagh-Eshtivani, E.; Fasihi, M.; Ghavami, A.; Aynehchi, A.; Ehsani, A. Bioemulsifiers Derived from Microorganisms: Applications in the Drug and Food Industry. Adv. Pharm. Bull. 2018, 8, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Uzoigwe, C.; Burgess, J.G.; Ennis, C.J.; Rahman, P.K.S.M. Bioemulsifiers Are Not Biosurfactants and Require Different Screening Approaches. Front. Microbiol. 2015, 6, 245. [Google Scholar] [CrossRef] [PubMed]
- Stoimenova, E.; Vasileva-Tonkova, E.; Sotirova, A.; Galabova, D.; Lalchev, Z. Evaluation of Different Carbon Sources for Growth and Biosurfactant Production by Pseudomonas Fluorescens Isolated from Wastewaters. Z. Fur Naturforschung Sect. C J. Biosci. 2009, 64, 96–102. [Google Scholar] [CrossRef]
- Rane, A.N.; Baikar, V.V.; Ravi Kumar, D.V.; Deopurkar, R.L. Agro-Industrial Wastes for Production of Biosurfactant by Bacillus Subtilis ANR 88 and Its Application in Synthesis of Silver and Gold Nanoparticles. Front. Microbiol. 2017, 8, 492. [Google Scholar] [CrossRef]
- Cooper, D.G.; Goldenberg, B.G. Surface-Active Agents from Two Bacillus Species. Appl. Environ. Microbiol. 1987, 53, 224–229. [Google Scholar] [CrossRef]
- Yin, H.; Qiang, J.; Jia, Y.; Ye, J.; Peng, H.; Qin, H.; Zhang, N.; He, B. Characteristics of Biosurfactant Produced by Pseudomonas Aeruginosa S6 Isolated from Oil-Containing Wastewater. Process Biochem. 2009, 44, 302–308. [Google Scholar] [CrossRef]
- Ludwig, W. Nucleic Acid Techniques in Bacterial Systematics and Identification. Int. J. Food Microbiol. 2007, 120, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Waghmare, P.R.; Patil, S.M.; Jadhav, S.L.; Jeon, B.H.; Govindwar, S.P. Utilization of Agricultural Waste Biomass by Cellulolytic Isolate Enterobacter sp. SUK-Bio. Agric. Nat. Resour. 2018, 52, 399–406. [Google Scholar] [CrossRef]
- Pei, X.H.; Zhan, X.H.; Wang, S.M.; Lin, Y.S.; Zhou, L.X. Effects of a Biosurfactant and a Synthetic Surfactant on Phenanthrene Degradation by a Sphingomonas Strain. Pedosphere 2010, 20, 771–779. [Google Scholar] [CrossRef]
- Berg, G. Rhizobacteria of Oilseed Rape Antagonistic to Verticillium Dahliae Var. Longisporum STARK. Z Pflanzenkr Pflanzenschutz 1996, 103, 20–30. [Google Scholar]
- Kalvandi, S.; Garousin, H.; Pourbabaee, A.A.; Farahbakhsh, M. The Release of Petroleum Hydrocarbons from a Saline-Sodic Soil by the New Biosurfactant-Producing Strain of Bacillus sp. Sci. Rep. 2022, 12, 19770. [Google Scholar] [CrossRef] [PubMed]
- Montecillo, J.A.V.; Bae, H. Reclassification of Brevibacterium Frigoritolerans as Peribacillus Frigoritolerans Comb. Nov. Based on Phylogenomics and Multiple Molecular Synapomorphies. Int. J. Syst. Evol. Microbiol. 2022, 72, 005389. [Google Scholar] [CrossRef]
- Khondee, N.; Ruamyat, N.; Luepromchai, E.; Sikhao, K.; Hawangchu, Y. Bioconversion of Lignocellulosic Wastes to Zwitterionic Biosurfactants by an Alkaliphilic Bacterium: Process Development and Product Characterization. Biomass Bioenergy 2022, 165, 106568. [Google Scholar] [CrossRef]
- Manetsberger, J.; Caballero Gómez, N.; Soria-Rodríguez, C.; Benomar, N.; Abriouel, H. Simply Versatile: The Use of Peribacillus Simplex in Sustainable Agriculture. Microorganisms 2023, 11, 2540. [Google Scholar] [CrossRef]
- Ibrahim, H.M.M. Characterization of Biosurfactants Produced by Novel Strains of Ochrobactrum Anthropi HM-1 and Citrobacter Freundii HM-2 from Used Engine Oil-Contaminated Soil. Egypt. J. Pet. 2018, 27, 21–29. [Google Scholar] [CrossRef]
- Renard, P.; Canet, I.; Sancelme, M.; Wirgot, N.; Deguillaume, L.; Delort, A.M. Screening of Cloud Microorganisms Isolated at the Puy de Dôme (France) Station for the Production of Biosurfactants. Atmo. Chem. Phys. 2016, 16, 12347–12358. [Google Scholar] [CrossRef]
- Darwesh, O.M.; Mahmoud, M.S.; Barakat, K.M.; Abuellil, A.; Ahmad, M.E. Improving the Bioremediation Technology of Contaminated Wastewater Using Biosurfactants Produced by Novel Bacillus Isolates. Heliyon 2021, 7, e08616. [Google Scholar] [CrossRef] [PubMed]
- Chaurasia, L.K.; Tamang, B.; Tirwa, R.K.; Lepcha, P.L. Influence of Biosurfactant Producing Bacillus Tequilensis LK5.4 Isolate of Kinema, a Fermented Soybean, on Seed Germination and Growth of Maize (Zea mays L.). 3 Biotech 2020, 10, 297. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, E.; Rubinovitz, C.; Gottlieb, A.; Rosenhak, S.; Ron, E.Z. Production of Biodispersan by Acinetobacter Calcoaceticus A2. Appl. Environ. Microbiol. 1988, 54, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Mendes Lopes, E. Emulsification Properties of Bioemulsifiers Produced by Wild-Type and Mutant Bradyrhizobium Elkanii Strains. J. Bioremediat. Biodegrad. 2014, 5, 1000245. [Google Scholar] [CrossRef]
- Rabiei, A.; Sharifinik, M.; Niazi, A.; Hashemi, A.; Ayatollahi, S. Core Flooding Tests to Investigate the Effects of IFT Reduction and Wettability Alteration on Oil Recovery during MEOR Process in an Iranian Oil Reservoir. Appl. Microbiol. Biotechnol. 2013, 97, 5979–5991. [Google Scholar] [CrossRef] [PubMed]
- Essghaier, B.; Mallat, N.; Khwaldia, K.; Mottola, F.; Rocco, L.; Hannachi, H. Production and Characterization of New Biosurfactants/Bioemulsifiers from Pantoea Alhagi and Their Antioxidant, Antimicrobial and Anti-Biofilm Potentiality Evaluations. Molecules 2023, 28, 1912. [Google Scholar] [CrossRef] [PubMed]
- Sen, R. Response Surface Optimization of the Critical Media Components for the Production of Surfactin. J. Chem. Technol. Biotechnol. 1997, 68, 263–270. [Google Scholar] [CrossRef]
- Jiménez-Peñalver, P.; Koh, A.; Gross, R.; Gea, T.; Font, X. Biosurfactants from Waste: Structures and Interfacial Properties of Sophorolipids Produced from a Residual Oil Cake. J. Surfactants Deterg. 2020, 23, 481–486. [Google Scholar] [CrossRef]
- Ciurko, D.; Czyżnikowska, Ż.; Kancelista, A.; Łaba, W.; Janek, T. Sustainable Production of Biosurfactant from Agro-Industrial Oil Wastes by Bacillus Subtilis and Its Potential Application as Antioxidant and ACE Inhibitor. Int. J. Mol. Sci. 2022, 23, 10824. [Google Scholar] [CrossRef]
- Umar, A.A.; Saaid, I.B.M.; Sulaimon, A.A.; Pilus, R.B.M. A Review of Petroleum Emulsions and Recent Progress on Water-in-Crude Oil Emulsions Stabilized by Natural Surfactants and Solids. J. Pet. Sci. Eng. 2018, 165, 673–690. [Google Scholar] [CrossRef]
- Saimmai, A.; Rukadee, O.; Onlamool, T.; Sobhon, V.; Maneerat, S. Isolation and Functional Characterization of a Biosurfactant Produced by a New and Promising Strain of Oleomonas Sagaranensis AT18. World J. Microbiol. Biotechnol. 2012, 28, 2973–2986. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.Y.; Yeh, K.L.; Lu, W.B.; Lin, C.L.; Chang, J.S. Rhamnolipid Production with Indigenous Pseudomonas Aeruginosa EM1 Isolated from Oil-Contaminated Site. Bioresour. Technol. 2008, 99, 1157–1164. [Google Scholar] [CrossRef] [PubMed]
- Das, A.J.; Kumar, R. Utilization of Agro-Industrial Waste for Biosurfactant Production under Submerged Fermentation and Its Application in Oil Recovery from Sand Matrix. Bioresour. Technol. 2018, 260, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Liepins, J.; Balina, K.; Soloha, R.; Berzina, I.; Lukasa, L.K.; Dace, E. Glycolipid Biosurfactant Production from Waste Cooking Oils by Yeast: Review of Substrates, Producers and Products. Fermentation 2021, 7, 136. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).