The Detection of Circulating Cell-Free DNA for the Diagnosis of Schistosoma in Immigrants from African Countries in Italy
Abstract
:1. Introduction
2. Materials and Methods
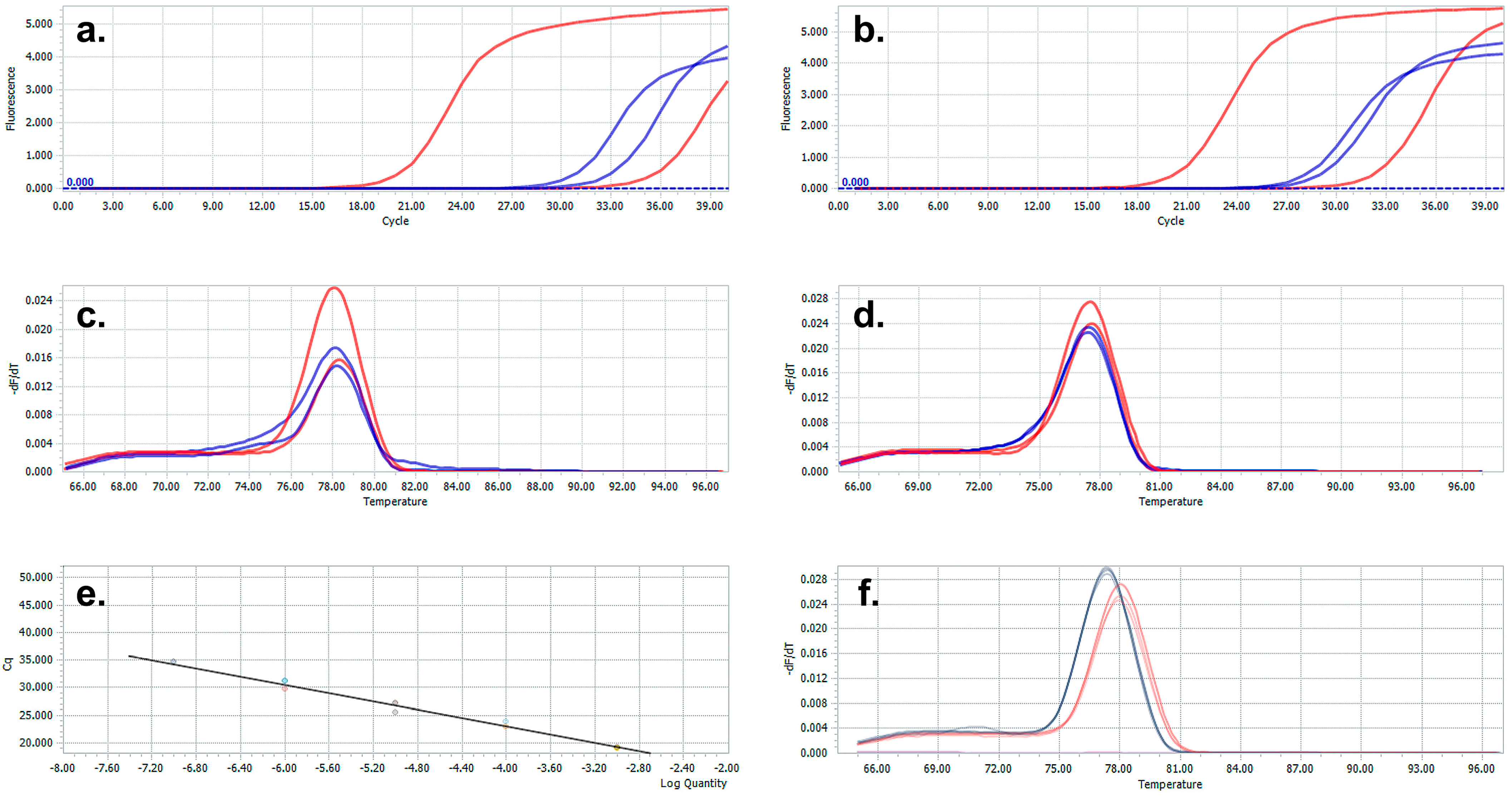
3. Results and Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rey, O.; Webster, B.L.; Huyse, T.; Rollinson, D.; Van den Broeck, F.; Kincaid-Smith, J.; Onyekwere, A.; Boissier, J. Population genetics of African Schistosoma species. Infect. Genet. Evol. 2021, 89, 104727. [Google Scholar] [CrossRef]
- Colley, D.G.; Bustinduy, A.L.; Secor, W.E.; King, C.H. Human schistosomiasis. Lancet 2014, 383, 2253–2264. [Google Scholar] [CrossRef]
- Engels, D.; Sinzinkayo, E.; Gryseels, B. Day-to-day egg count fluctuation in Schistosoma mansoni infection and its operational implications. Am. J. Trop. Med. Hyg. 1996, 54, 319–324. [Google Scholar] [CrossRef]
- Wickmann, D.; Panning, M.; Quack, T.; Kramme, S.; Burchard, G.; Grevelding, C.; Drosten, C. Diagnosing schistosomiasis by detection of cell-free parasite DNA in human plasma. PLoS Negl. Trop. Dis. 2009, 3, e422. [Google Scholar] [CrossRef] [PubMed]
- INMP; ISS; SIMM. I Controlli Alla Frontiera. La Frontiera dei Controlli. In Border Checks Kept in Check. Health Checks and Protection Pathways for Migrants upon Arrival and While Hosted in Reception Centers; Ministry of Health: Rome, Italy, 2017. Available online: http://www.salute.gov.it/imgs/C_17_pubblicazioni_2624_allegato.pdf (accessed on 1 December 2022).
- Hinz, R.; Norbert, G.; Schwarz, C.; Hahn, A.; Frickmann, H. Serological approaches for the diagnosis of schistosomiasis—A review. Mol. Cell. Probes 2017, 31, 2–21. [Google Scholar] [CrossRef]
- Fuss, A.; Mazigo, H.D.; Tappe, D.; Kasang, C.; Mueller, A. Comparison of sensitivity and specificity of three diagnostic test to detect Schistosoma mansoni infections in school children in Mwanza region, Tanzania. PLoS ONE 2018, 13, e020249. [Google Scholar] [CrossRef]
- Utzinger, J.; Becker, S.; Van Lieshout, L.; Van Dam, J.; Knopp, S. New diagnostic tools in schistosomiasis. Clin. Microbiol. Infect. 2015, 21, 529–542. [Google Scholar]
- Aryeetey, Y.A.; Essien-Baidoo, S.; Larbi, I.A.; Ahmed, K.; Amoah, A.S.; Obeng, B.B.; Van Lieshout, L.; Yazdanbakhsh, M.; Boakye, D.A.; Verweij, J.J. Molecular diagnosis of Schistosoma infections in urine samples of school children in Ghana. Am. J. Trop. Med. Hyg. 2013, 88, 1028–1031. [Google Scholar] [CrossRef]
- Pontes, L.A.; Dias-Neto, E.; Rabello, A. Detection by polymerase chain reaction of Schistosoma mansoni DNA in human serum and feces. Am. J. Trop. Med. Hyg. 2002, 66, 157–162. [Google Scholar] [CrossRef]
- Wichmann, D.; Poppert, S.; Von Thien, H.; Clerinx, J.; Dieckmann, S.; Mogens, J.; Parola, P.; Richter, J.; Shunk, M.; Zanger, A.; et al. Prospective European-wide multicentre study on a blood based real-time PCR for the diagnosis of acute schistosomiasis. BMC Infect. Dis. 2013, 13, 55. [Google Scholar] [CrossRef]
- Berriman, M.; Haas, B.J.; LoVerde, P.T.; Wilson, R.A.; Dillon, G.P.; Cerqueira, G.C.; Mashiyama, S.T.; Al-Lazikani, B.; Andrade, L.F.; Ashton, P.D.; et al. The genome of the blood fluke Schistosoma mansoni. Nature 2009, 460, 352–358. [Google Scholar] [CrossRef]
- Weerakoon, K.G.; McManus, D.P. Cf-DNA as a Diagnostic Tool for Human Parasitic Infections. Trends Parasitol. 2016, 32, 378–391. [Google Scholar] [CrossRef]
- Weerakoon, K.G.; Gordon, A.C.; Williams, G.M.; Cai, P.; Gobert, G.N.; Olveda, R.M.; Ross, A.G.; Olveda, D.U.; McManus, D.P. Droplet digital PCR diagnosis of human schistosomiasis: Parasite cf-DNA detection in diverse clinical samples. J. Infect. Dis. 2015, 216, 1611–1622. [Google Scholar] [CrossRef]
- Weerakoon, K.G.; Gordon, A.C.; Cai, P.; Gobert, G.N.; Duke, M.; Williams, G.M.; McManus, D.P. A novel duplex ddPCR assay for the diagnosis of schistosomiasis japonica: Proof of concept in an experimental mouse model. Parasitology 2017, 144, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.M.; Chiu, R.W. Prenatal diagnosis: Progress through plasma nucleic acids. Nat. Rev. Genet. 2016, 8, 71–77. [Google Scholar]
- Goessl, C. Diagnostic potential of circulating nucleic acids for oncology. Expert Rev. Mol. Diagn. 2003, 3, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Kato-Hayashi, N.; Yasuda, M.; Yuasa, J.; Isaka, S.; Haruki, K.; Ohmae, H.; Osada, Y.; Kanazawa, Y. Use of cell-free circulating schistosome DNA in serum, urine, semen, and saliva to monitor a case of refractory imported schistosomiasis hematobia. J. Clin. Microbiol. 2013, 51, 3435–3438. [Google Scholar] [CrossRef]
- Ullah, H.; Qadeer, A.; Giri, B.R. Detection of circulating cf-DNA to diagnose Schistosoma japonicum infection. Acta Trop. 2020, 211, 105604. [Google Scholar] [CrossRef] [PubMed]
- Ullah, H.; Arbab, S.; Inayat Ullah Khan, M.; Li, K.; Muhammad, N.; Qadeer, A.; Dan, W.; Otake Sato, M. Circulating cell-free mitochondrial DNA fragment: A possible marker for early detection of Schistosoma japonicum. Infect. Genet. Evol. 2021, 88, 104683. [Google Scholar] [CrossRef] [PubMed]
- Real-Time PCR Guide: Design, Validation, Analysis, and Troubleshooting. Integrated DNA Technologies. September 2020. Available online: https://go.idtdna.com/202009qPCRCampaignConsolidatedGuideParts1-qpcrguide-part-3.htm (accessed on 1 December 2022).
- Hamburger, J.; Xu, X.; Xin, Y.; Ramzy, R.; Jourdane, A.; Ruppel, A. A polymerase chain reaction assay for detecting snails infected with bilharzia parasites (Schistosoma mansoni) from very early prepatency. Am. J. Trop. Med. Hyg. 1998, 59, 872–876. [Google Scholar] [CrossRef]
- Guegan, H.; Fillaux, J.; Charpentier, E.; Robert-Gangneux, F.; Chauvin, P.; Guemas, E. Real-time PCR for diagnosis of imported schistosomiasis. PLoS Negl. Trop. Dis. 2019, 13, e0007711. [Google Scholar] [CrossRef] [PubMed]
| Serology + | Serology − | Total Number | |
|---|---|---|---|
| Cell-fee DNA PCR + | 29 | 3 | 32 |
| Cell-free DNA PCR − | 23 | 47 | 70 |
| Total | 52 | 50 | 102 |
| Specificity % | 94 (47/50) | ||
| Concordance rate % | 74.51 ((29 + 47)/102) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dimartino, V.; Scopelliti, F.; Cattani, C.; Nicolella, G.; Cavani, A. The Detection of Circulating Cell-Free DNA for the Diagnosis of Schistosoma in Immigrants from African Countries in Italy. Microbiol. Res. 2023, 14, 2034-2040. https://doi.org/10.3390/microbiolres14040137
Dimartino V, Scopelliti F, Cattani C, Nicolella G, Cavani A. The Detection of Circulating Cell-Free DNA for the Diagnosis of Schistosoma in Immigrants from African Countries in Italy. Microbiology Research. 2023; 14(4):2034-2040. https://doi.org/10.3390/microbiolres14040137
Chicago/Turabian StyleDimartino, Valentina, Fernanda Scopelliti, Caterina Cattani, Gianluca Nicolella, and Andrea Cavani. 2023. "The Detection of Circulating Cell-Free DNA for the Diagnosis of Schistosoma in Immigrants from African Countries in Italy" Microbiology Research 14, no. 4: 2034-2040. https://doi.org/10.3390/microbiolres14040137
APA StyleDimartino, V., Scopelliti, F., Cattani, C., Nicolella, G., & Cavani, A. (2023). The Detection of Circulating Cell-Free DNA for the Diagnosis of Schistosoma in Immigrants from African Countries in Italy. Microbiology Research, 14(4), 2034-2040. https://doi.org/10.3390/microbiolres14040137







