Abstract
Among the oldest marine species on the planet, the genus Salinispora is often encountered inhabiting sediments and other marine creatures in tropical and subtropical marine settings. This bacterial genus produces a plethora of natural products. The purpose of this study was to examine the potential for salinispora-based natural products (NPs) to combat the SARS-CoV-2 virus. The RCSB PDB was used to obtain the crystal structures of proteins 3CLpro and PLpro. All 125 NPs were obtained from online databases. Using Autodock Vina software v1.2.0 the molecular docking process was carried out after the proteins and ligands were prepared. Assessments of binding affinities and interacting amino acids were rigorously examined prior to MD simulations. The docking experiments revealed 35 NPs in total for both 3CLpro and PLpro, with high docking scores ranging from −8.0 kcal/mol to −9.0 kcal/mol. However, a thorough binding residue analyses of all docked complexes filtered nine NPs showing strong interactions with HIS: 41 and CYS: 145 of 3CLpro. Whereas, for PLpro, merely six NPs presented good interactions with residues CYS: 111, HIS: 272, and ASP: 286. Further research was conducted on residue–residue and ligand–residue interactions in both the filtered docked complexes and the Apo-protein structures using the Protein Contacts Atlas website. All complexes were found to be stable in CABS-flex 2.0 MD simulations conducted at various time frames (50, 125, 500, and 1000 cycles). In conclusion, salinaphthoquinone B appears to be the most promising metabolite, based on favorable amino acid interactions forming stable confirmations towards 3CLpro and PLpro enzymes, acting as a dual inhibitor.
Keywords:
Salinispora; natural products; marine drugs; repurposing; antiviral; Mpro; 3CLpro; PLpro; SARS-CoV-2; S. arenicola; S. pacifica; S. tropica 1. Introduction
A worldwide health crisis has been sparked by SARS-CoV-2, which causes severe acute respiratory syndrome [1,2]. SARS-CoV-2 was discovered for the first time in Wuhan City in China in December 2019, and it traveled rapidly around the world [3,4,5]. The pandemic has now been ongoing for over three years, and the majority of people have either succeeded in getting a SARS-CoV-2 vaccine or have already contracted the virus. Roughly 7.0 million people have died as a consequence of the estimated 771.5 million COVID-19 infections as of October 2023 all around the globe, as reported by the WHO coronavirus dashboard. The vulnerability of the public health and medical care systems in numerous countries has indeed been unmasked by the COVID-19 pandemic [6]. The SARS-CoV-2 3C-like protease (3CLpro/Mpro/nsp5) and the papain-like protease (nsp3) play an essential role in viral gene replication, transcription, and expression [7,8,9,10]. The viral polyproteins are processed synergistically by both PLpro and Mpro enzymes. Amongst SARS-CoV and SARS-CoV-2, Mpro and PLpro demonstrated strong sequence similarities of 86% and 96% [11,12], indicating their significance as cogent drug targets. This explains the rising trend of researchers choosing to develop targeted therapies against coronaviruses using the 3C-like and papain-like proteases as primary targets.
Discovering effective antiCOVID-19 treatments is continuously necessary due to the risk presented by the emergence of additional variants featuring higher virulence as well as the increased incidence of viral resistance to approved treatments [13]. Ever since the COVID-19 pandemic began, numerous computational and clinical experimentations have been carried out to reposition various drugs to restrict 3CLpro and PLpro activities as possible therapeutic options [14,15]. Among the repositioned pharmaceutical medicines, remdesivir, danoprevir, darunavir, oseltamivir, lopinavir, umifenovir, ritonavir, favipiravir [16,17], and riamilovir [18] have already been encouraged for therapeutic applications for COVID-19 treatments. The Food and Drug Administration (FDA, USA) recently, in 2022, officially authorized the use of a ritonavir and nirmatrelvir combination (Paxlovid) in case of emergency as a SARS-CoV-2 medication [19,20]. Moreover, several antiviral compounds, recombinant soluble angiotensin-converting enzyme 2 (ACE2), monoclonal antibodies, vaccinations, corticosteroids, interferon therapies, and herbal remedies are among the several possible therapeutic agents to combat COVID-19. Furthermore, numerous vaccine candidates are being developed, some of which are presently undergoing various stages of clinical trials, thanks to our growing understanding of the structure and infection processes of SARS-CoV-2 [21]. Vaccines are just a preventive measure and even today there is no definite cure for COVID-19. In addition, there may be negative side effects to these aforementioned therapeutic strategies. Consequently, it is imperative to continue searching for relatively safer therapeutic molecules to treat COVID-19.
In our work, we focus on the discovery of natural compounds with required antiviral activities as they could be less sensitive to virus variant mutations. Natural products (NPs) from plants and of marine origin have long been thought to be a major source of therapeutic substances. Plant-based natural products have proven effective in inhibiting a variety of viral proteins and, in fact, exhibit strong antiviral properties [22,23]. Secondary metabolites from marine organisms are well known as crucial natural product sources that can be used in the fight against COVID-19 [24,25]. The marine ecosystem is a reservoir of naturally occurring substances called marine natural products (MNPs) that are highly distinct and physiologically potent [26,27]. Evaluations on the antimicrobial activities of MNPs against numerous diseases have already been explored [28,29]. Examples include the significant in vitro actions against the 3C-like protease that natural products of marine origin demonstrated [30,31]. Recently, chetomin was identified as a potential inhibitor of 3CLpro in an in silico study [32]. However, a thorough investigation of MNPs’ prospects as inhibitors of SARS-CoV-2 main protease has not yet been fully accomplished.
The goal of this study was to evaluate whether natural products from the genus Salinispora might be used as 3CLpro and PLpro inhibitors by conducting an in silico assessment (see Figure 1). Pre-screening against the 3CLpro and PLpro proteins using a built library made up of NPs from various marine sources revealed Salinispora metabolites that showed strong affinities and advantageous residue interactions. Additionally, the low molecular weight and established antimicrobial properties [33,34,35,36,37,38] of Salinispora metabolites were the main reasons for their selection to conduct this work. Small molecules in general are perceived as potentially effective protease inhibitors, especially in the instance of 3CLpro protein [39]. Out of 125 NPs, this study identified a select few NPs by placing more emphasis on the amino acid interactions rather than just the docking scores. This is because there is no assurance that high docking scores equate to therapeutic effectiveness [40,41].
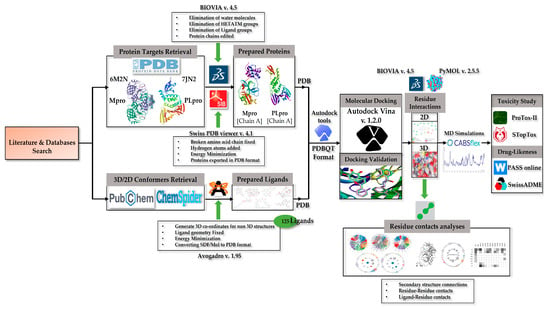
Figure 1.
Schematic of the work flow used in this study.
This paper, probably for the first time, has proposed an initiative for Salinispora NPs to be conceived as repurposed prospective SARS-CoV-2 protease inhibitor candidates. In the end, an evaluation of the toxicity and pharmacokinetic properties of filtered NPs was performed. Though it is early to discuss marine bacterial genus Salinispora-based medications for the current treatment module of COVID-19, overall, this study may provide a solid basis for further experimental study in this area.
2. Materials and Methods
2.1. Protein Retrieval
The most well-known and extensively researched COVID-19 proteins were investigated and learned about through published works. The crystal structures of the SARS-CoV-2 proteases, as characterized by X-ray diffraction, were located using the RCSB PDB database [42]. For this work, the 3D structure of the main protease (Mpro/3CLpro), also referred to as 3C-like protease (PDB ID: 6M2N), resolved at 2.20 Å was used [43]. Similarly, the 3D structure of the enzyme known as PLpro, or papain-like protease, resolved at 1.93 Å was considered (PDB ID: 7JN2) [44].
2.2. Ligand Retrieval
Through a comprehensive literature study, knowledge of NPs from the genus Salinospora was explored. Salinispora, a genus of marine actinomycete, has three species, including S. arenicola, S. pacifica, and S. tropica, according to a literature search [45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103]. The species S. arenicola consists of 84 natural products, whereas S. pacifica consists of 36 natural products. On the other hand, S. tropica consists of 34 natural products after excluding the common ones (refer to Table S1). The genus Salinispora consists of around 125 unique natural products to date [104]. A thorough database search was conducted to discover all of the ligands’ 2D and/or 3D conformers. First, SDF chemically formatted 3D conformers of ligands were obtained from the PubChem database [105]. In the scenario where 3D structures were unattainable, 2D conformers were retrieved. The ligands that did not appear in the PubChem database were acquired from a secondary source, the ChemSpider database. The ligands were retrieved in the Mol chemical format from the ChemSpider database [106]. We succeeded in obtaining all 125 NPs.
2.3. Visualization Software
The protein structures of 3CLpro and PLpro were viewed and explored using BIOVIA Discovery Studio Visualizer 4.5 and PyMOL 2.5.5 [107,108]. Our knowledge of the number of HETATM groups and amino acid chains was obtained from these tools. As opposed to that, the ligands were visualized using Avogadro software v. 1.95 [109,110].
2.4. Docking Preparations
It was essential to make the proteins and ligands ready for the docking procedure. Using the information gathered during the visualization process, BIOVIA Discovery Studio Visualizer was used to produce both target proteins. In contrast to the PLpro protein, which only had one amino acid chain A, the 3CLpro protein contained amino acid chains A, B, C, and D. Both proteins demonstrated the existence of ligand groups, HETATM, and water molecules. Thus, undesired molecules and amino acid chains other than the ligand groups were eliminated from 3D structures. In this investigation, only chain A from both proteins was considered. In addition, hydrogen atoms were inserted, energy reduction was carried out, and Swiss-pdb viewer application [111] was applied to possibly repair the disrupted amino acid chain. After the energy minimization operation, the ligand groups were knocked out to preserve the active site from collapsing. Additionally, these structures were exported in the Swiss-pdb viewer’s protein data bank format before being converted using Autodock tools, a feature of MGL tools, to the needful PDBQT format [112].
The Avogadro software (version 1.95) was employed for the preparation of 2D and 3D conformers of all molecules. Avogadro was initially employed to access the 2D structures and then to produce the 3D coordinates necessary to convert the ligands to 3D. All 2D structures were thus transformed into 3D conformers and saved. When all 125 ligands were in 3D format, geometry optimization and energy minimization were carried out on each of them individually. All molecules obtained in PDB chemical format were changed to PDBQT using auto dock tools by adding Gasteiger charges required for molecular docking.
2.5. Docking Validation
Both 3CLpro and PLpro crystallographic structures included their respective inhibitors attached by default. 5, 6, and 7-trihydroxy-2-phenyl-4H-chromen-4-one (ID—3WL), a novel inhibitor, was bound to the active site of 3CLpro. Similar to 3CLpro, the active site of PLpro had 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl) ethyl] benzamide (ID—Y41) attached. For the reliability of any docking outcomes, it is important to validate the docking procedure. For the validation process, the default inhibitors attached to the crystals were re-docked in the same pockets (active sites) of the respective proteins. The original crystal structures were then superimposed with the re-docked structures to perform this verification step (refer to Figure 2). The binding affinities of inhibitors and the RMSD values for superimposed complexes were accessed using the BIOVIA Discovery studio visualizer program.
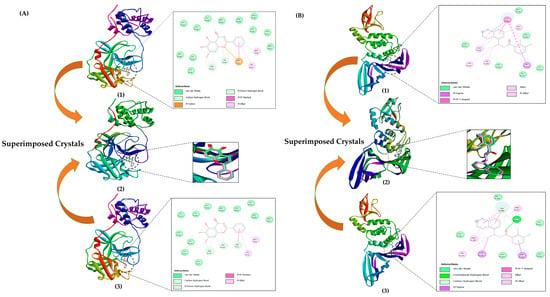
Figure 2.
Docking validation: (A) (1) 3CLpro crystal with a re-docked inhibitor in the catalytic site, (2) superimposed 3CLpro crystals with re-docked and attached inhibitor, (3) original 3CLpro crystal with an attached inhibitor in the catalytic site (PDB ID: 6M2N). (B) (1) Original PLpro crystal (PDB ID: 7JN2) with an attached inhibitor in the active site, (2) superimposed PLpro crystals with re-docked and attached inhibitor, (3) PLpro crystal with a re-docked inhibitor in the active site. Re-docking experiments for the proteins 3CLpro and PLpro indicate the level of accuracy of the docking protocol used in the study.
2.6. Molecular Docking
The molecular docking process was carried out by using a recent Autodock Vina 1.2.0 release [113,114]. The grid box was constructed and placed using knowledge of the essential amino acids for 3CLpro and PLpro, as well as the known functional site. One hundred and twenty-five ligands were individually docked once the program first opened the 3CLpro protein file in PDBQT format. The 3CLpro cavity’s volume was 1386 Å3, whereas the PLpro protein’s active site volume was 1280 Å3. For the 3CLpro protein, the grid box was placed on the active site at X = −34 Å, Y = −67 Å, Z = 41 Å, and for the PLpro protein, it was fixed at X = 47 Å, Y = 32 Å, Z = 2 Å. All 125 ligands were also finally docked with PLpro. All molecules were kept flexible, while both enzymes were maintained rigid. The docking protocol was carried out three times in order to assure accuracy. The docking process provided us with upto 30 binding conformations and the docking scores for each of the 125 ligands with the 3CLpro and PLpro proteins.
2.6.1. Binding Affinity and Residue Analyses
We made sure to convey the docking calculation results in this paper in an understandable manner. On the basis of the determined values for binding affinity, three main categories—low, moderate, and high—were used to group the data that are shown in Section 3.1.2 and Section 3.2.2. The moderate category was further split into lower-moderate and upper-moderate divisions. The resulting 250 protein–ligand complexes were explored for binding and non-binding residue analyses. BIOVIA Discovery Studio Visualizer 4.5 and PyMOL 2.5.5 assisted in revealing all the residues involved by providing us with the necessary 2D and 3D interaction maps.
2.6.2. Selection of Best Interaction Complexes, 3CLpro and PLpro
Determining whether the ligands are effective at suppressing the activity of target proteins requires a thorough understanding of the interacting residues. The inhibitory potential of ligands must not be solely correlated with high binding affinities.
The complexes were filtered based on the following selection criteria:
- The ligand interacted with critical catalytic amino acids;
- The ligand made one or more hydrogen bond (RMSD Å ≤ 3.00);
- The ligand interacted with at least one of the catalytic amino acids to generate electrostatic connections;
- The predetermined threshold RMSD cutoff distance (Å ≤ 5.00) for electrostatic bonding was not exceeded.
The complexes were rejected based on the criteria stated below:
- No interactions with crucial catalytic residues;
- No hydrogen bonds associated with key residues;
- Only hydrophobic interactions (VdW forces) occurred;
- Exceeded the maximum predetermined RMSD cutoff distance threshold for electrostatic associations (Å ≥ 5.00).
2.6.3. Residue–Residue and Ligand–Residue Contact Analyses
Utilizing the Protein Contact Atlas website [115], further investigation of residue interactions was carried out for both the Apo-protein structures (energy minimized) and the candidate docked protein complexes. The interactions formed between the ligands and the catalytic amino acids of the proteins in the case of docked complexes were evident through the asteroid plots, and ligand residue matrices provided insights into the number of contacts formed. Contrarily, for the Apo structures, chord plots emphasized relationships at the secondary structure level, while asteroid plots revealed the inter-residue relationships among the critical amino acids present in the catalytic sites of both proteins.
2.7. Molecular Dynamics Simulation
For the Apo-proteins (no ligands attached) and protein–ligand complexes of 3CLpro and PLpro, molecular dynamics simulations were carried out using the CABS-flex 2.0 website [116]. The CABS-flex 2.0 tool illustrates excellent agreement with NMR spectroscopic data on protein flexibility and is very useful for fast simulations of protein residue flexibility evaluations. PDB files for the shortlisted complexes were provided to the webserver. The simulations for the complexes that made the short list were run at different simulation periods, such as 50, 125, 500, and 1000 cycles, with the default values for simulation temperatures and random number generator seed offered by the server. The RMSF charts for the indicated three distinct run periods were essential for assessing the stability of the docked complexes overtime.
2.8. Toxicity Profile Evaluation
ProTox II and StopTox web servers were used to determine the probable toxicity of leading ligands [117,118]. By promptly estimating the hazardous potential of ligands in an internet-based setting, ProTox-II reduces the need for in vivo testing involving animals. This server offered information on the lethal dose concentrations (mg/kg body weight), toxicity level classification, and toxicity endpoints. A simple-to-operate online application called StopTox provided quick and accurate estimates of the likelihood that ligands may or may not cause acute toxicity.
2.9. Pharmacokinetics Evaluation
On the Swiss-ADME system [119], the drug-like characteristics of our chosen NPs were investigated using their respective canonical SMILES (see Table S2). Applying the Lipinski framework (Ro5 criteria), proposed by Christopher A. Lipinski, on our candidate NPs was the first phase in the process. The molecular weight, the consensus logP-value, the hydrogen bond donors, and the acceptors were among these criteria. To enhance further the precision of our prediction, the TPSA and rotatable bond parameter were applied to determine the likelihood that the ligand would be orally potent. The bioavailability, solubility, and likelihood of GI absorption were noted in the second phase. A basic and straightforward mathematical formula was applied to calculate an absorption percentage value in order to correctly predict GI absorption [120,121,122].
The ability of NPs to penetrate cells and the blood–brain barrier was also examined using the boiled egg diagram provided to us by the Swiss-ADME analysis. Additionally, having an average precision of approximately 95%, the PASSonline program allowed us to predict the bioactivity spectra for our chosen NPs based on their respective structural formulae. On this platform, the predictions are made on the basis of the information stored for over 300,000 organic compounds [123].
3. Results and Discussion
The surface morphology of the ligand binding pocket and the positioning of the residues in the said active site need to be specifically examined when evaluating the bioactivities of target proteins for a rational structure-based drug design.
3.1. SARS-CoV-2 Main Protease Protein (3CLpro/Mpro)
3.1.1. 3CLpro Enzyme Structure and Residue–Residue Interactions in Active Site
The ORF1a-encodes 3-chymotrypsin-like protease (3CLpro/Nsp5) assists in the cleavage of pp1ab through eleven conserved splicing sites. 3CLpro is a prime pharmacological target because it has three times as many proteolytic cleavage sites compared with PLpro [124]. About 306 residues make up the 3CLpro protein. One polypeptide is all that is present in the asymmetric unit. Nevertheless, a dimer is created upon the association of protomers A and B. Three domains make up each protomer. The first domain (amino acids 8–101) and the second domain (amino acids 102–184) have antiparallel beta-barrel structures. Five alpha-helices are organized into a globular cluster in the third domain (amino acids 201–303), which is linked to the second domain by an elongated loop region (amino acids 185–200) [125,126].
The Apo structure of the 3CLpro chain A was selected for molecular docking and simulation experiments. The RMSF values for HIS: 41 (Å = 1.09) and CYS: 145 (Å = 1.43) were reported by the simulation results for the Apo structure in Figure 3A.
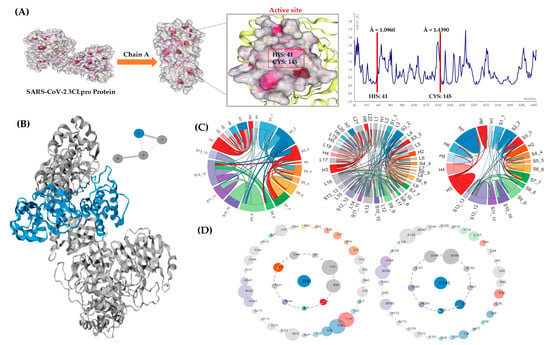
Figure 3.
In-depth 3CLpro analyses. (A) Three-dimensional Apo structure of 3CLpro with the active site and CABS-flex RMSF plot. Protein contact atlas output (B) shows chain A selected for analysis. (C) Chord plots depicting loops and secondary structure interactions for 3CLpro enzyme. (D) Asteroid plots depicting residue contacts with HIS: 41 and CYS: 145. The atomic scale interaction number percentage is represented by the circumference of the sphere. Residues are colored differently according to their respective secondary structures (from cyan blue to magenta high).
The tertiary structure, and consequently the function of the 3CLpro protein, are most likely determined by the orientation of interactions among the various secondary structural components. In Figure 3C, all non-covalent connections at the secondary structure levels are presented in the form of chord plots. Residue HIS: 41, which forms 52 atomic interactions with its immediate neighbors, is located in region S3_3 (colored red), presented as the outer edge of the chord plot. Amino acid CYS: 145, which is situated at the S11_11 region on the chord plot’s edge, is presented in a bright purple color. A total of 49 atomic connections are made by CYS: 145 with its immediate neighbors. These connections among the secondary structures are depicted as chords and each chord’s thickness depends on the total number of atomic connections among the residues. The plot depicting loops shows regions L5 and L13 that comprise HIS: 41 and CYS: 145 residues, correspondingly. The associations among the secondary structures indicate a strong correlation.
The inter-residue interactions within chain A (see Figure 3B) of protein are indicated by the asteroid plots made for the active site residues of the 3CLpro Apo structure (see Figure 3D). A direct non-covalent relationship (the inner shell) between residue HIS: 41 and residues L27, P39, R40, V42, I43, C44, Y54, C145, and H164 is shown in the first plot. Similarly, in the case of CYS: 145, the direct connections are made with nine residues such as L27, N28, H41, G143, S144, G146, S147, H163, and H164. The outer shell of these plots represents indirect non-covalent contacts. Residues L27 and H164 share a direct link with both H41 and C145. Therefore, alterations to L27 and H164 may have a direct impact on ligand binding, whereas mutations to outer-shell residues may have an indirect impact on ligand binding.
3.1.2. 3CLpro Docking
Presented below in Table 1 are all the 125 NPs docked with the 3CLpro protein and categorized by their docking scores.

Table 1.
Depicting the range of binding affinities obtained for the target 3CLpro.
Category 1 of Table 1 indicates that 31 NPs showed low docking scores in the range of −4.0 and −5.9 kcal/mol (refer to Table S3). In this category, 2-methyl-N-(2′-phenylethyl) butyramide, rifamycin B, sal-GBL1, and salinipostin K showed the highest docking values of −5.9 kcal/mol. On the other hand, salinilactone B, D, and F showed the lowest scores, ranging from −4.7 to −4.9 kcal/mol. The category 2 section comprises 33 ligands that showed lower-moderate docking, ranging from −6 to −6.9 kcal/mol (see Table S4). Ligands such as arenamide C, arenicolide B, arenimycin B, emericellamide A, and isopimara-8, 15-diene showed the highest affinity scores. However, lomaiviticin E and salinipostin E indicated the lowest docking score at −6.0 kcal/mol. The 39 NPs in the upper-moderate class had docking scores between −7.0 and −7.9 kcal/mol (refer to Table S5). The highest affinity scores were demonstrated by neolymphostin B and rifamycin S at −7.9 kcal/mol. Saliniketal A, salinosporamide D, and sporolide A, on the other hand, displayed docking values of −7.0 kcal/mol. Last but not least, the 22 NPs in category 3 (see Table S6) had strong affinity values that exceeded −8.0 kcal/mol towards the 3CLpro enzyme. The lowest binding energies for ikarugamycin and arenimycin D were −9.0 and −8.7 kcal/mol, respectively. The remaining 20 NPs, however, had docking scores that varied between −8.0 and −8.5 kcal/mol. The binding affinities in the re-docking experiment of 3CLpro obtained −8.0 kcal/mol for ligand 3WL, with the RMSD value 0.86 Å for superimposed structures (see Figure 2A).
3.1.3. Best Amino Acid Interaction Complexes, 3CLpro
Residue HIS: 41 is crucial for residue CYS: 145 to deprotonate in order for it to target the glutamine backbone at the P1 location of the peptide substrate. The involvement of this catalytic dyad residue in the protease activity of 3CLpro was found to be highly significant when researchers carried out alanine substitutions to eliminate the side chains of HIS: 41 and CYS: 145 [127]. Based on this knowledge, we used the aforementioned screening criteria (in Section 2.6.2) to select docked complexes that showed favorable amino acid interactions with the challenging catalytic dyad. In Figure 2A, docking validation revealed residue interactions that were very comparable to those in the original protein crystal structure (PDB ID 6M2N). This indicates that a high degree of ligand binding mode similarity was obtained in the re-docking experiment, demonstrating the accuracy of the docking procedure. Independent of their binding affinities, 9 out of 125 docked complexes were ultimately chosen as the optimum complexes for 3CLpro (See Figure 4 and Table 2).
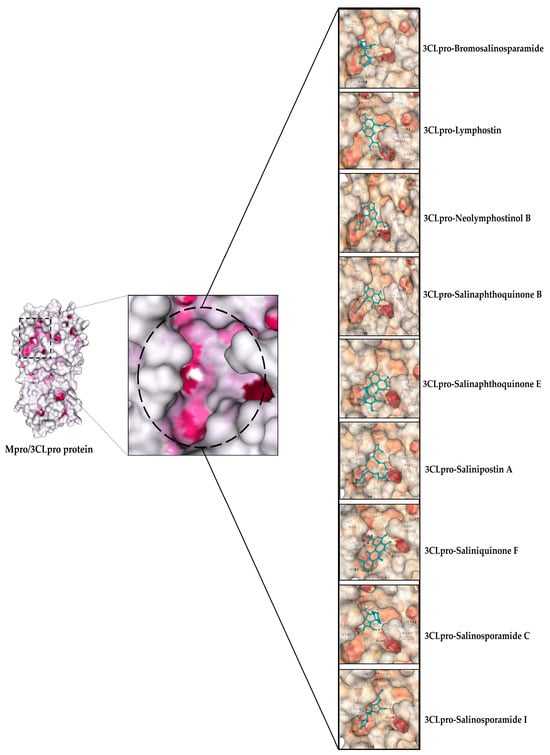
Figure 4.
Depicting ligands docked in the active site pocket of 3CLpro. The active site of the protein colored by B-factor and ligands are shown in blue.

Table 2.
NPs showing desired residue interactions for 3CLpro protein.
The interactions between amino acids that supposedly occurred during the formation of the complex with the 3CLpro enzyme are shown in the table below.
Bromosalinosporamide formed an H bond (2.84 Å) and a pi-alkyl bond (4.05 Å) with HIS: 41 and CYS: 145, respectively. Lymphostin formed three pi-cation and one hydrogen bond with HIS: 41 (2.46 Å), and with CYS: 145 it formed a pi-alkyl bond (4.89 Å). Neolymphostinol B formed two pi-cation bonds (2.23 Å and 2.24 Å) with HIS: 41, whereas for CYS: 145 it formed two bonds: a pi-pi stacked bond (4.89 Å) and a hydrogen bond (2.46 Å). Salinaphthoquinone B formed two hydrogen (2.53 Å and 2.93 Å) and one alkyl bond (4.92 Å) with CYS: 145, and with HIS: 41 it formed a single pi-alkyl bond (4.23 Å), along with a carbon–hydrogen bond (3.51 Å). Similarly, salinaphthoquinone E formed a single pi-alkyl (4.36 Å) and carbon–hydrogen bond (3.59 Å) with HIS: 41, and in the case of CYS: 145, two hydrogen bonds (2.72 Å and 2.85 Å) and one alkyl bond (4.28 Å) were formed. In the case of salinipostin A, a pi-cation (4.11 Å) and a pi-alkyl bond (4.55 Å) were formed with HIS: 41, whereas a single hydrogen bond (2.71 Å) was made with CYS: 145. Saliniquinone F interacted with HIS: 41, making a pi-pi T stacked (4.64 Å) and a pi-sigma bond (3.92 Å), whereas with CYS: 145 a single hydrogen bond (3.06 Å) was formed. Salinosporamide C formed one hydrogen (2.18 Å) and two alkyl bonds (4.64 Å and 4.97 Å) with HIS: 41, whereas CYS: 145 interacted with Vander Waals forces. Salinosporamide I interacted with HIS: 41 and CYS: 145 by forming a single hydrogen (3.07 Å) and pi-alkyl bond (4.00 Å), respectively. Lymphostin, neolymphostinol B, salinaphthoquinone B, and E interacted weakly with HIS: 163 using Vander Waals forces (see Table 2 and Figure 5).
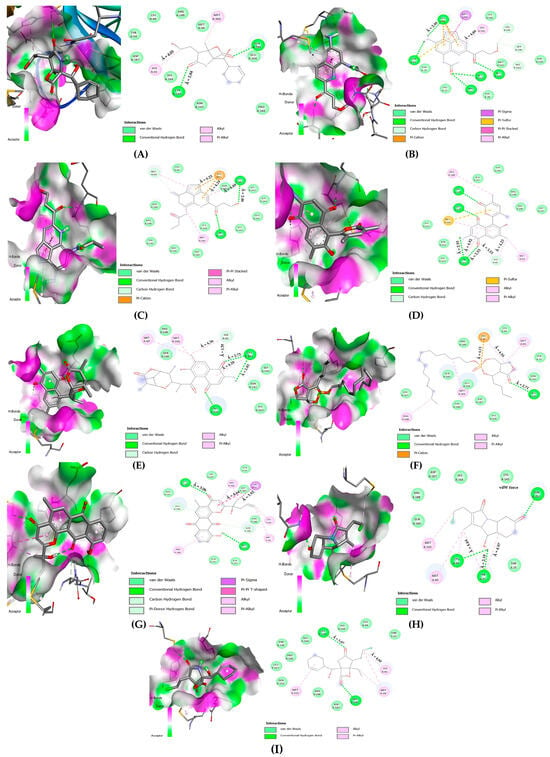
Figure 5.
Two-dimensional amino acid interaction maps. (A) 3CLpro-bromosalinosporamide, (B) 3CLpro-lymphostin, (C) 3CLpro-neolymphostinol B, (D) 3CLpro-salinaphthoquinone B, (E) 3CLpro-salinaphthoquinone E, (F) 3CLpro-saliniposin A, (G) 3CLpro-saliniquinone F, (H) 3CLpro-salinosporamide C, (I) 3CLpro-salinosporamide I.
3.1.4. Ligand–Residue Non-Covalent Interactions for 3CLpro Complexes
The nine docked complexes selected after passing the rigorous selection criteria were put through the Protein Contact Atlas webserver to gain an insight into the number of non-covalent atomic associations formed between the ligands and the catalytic dyad residues (refer to Figure 6).
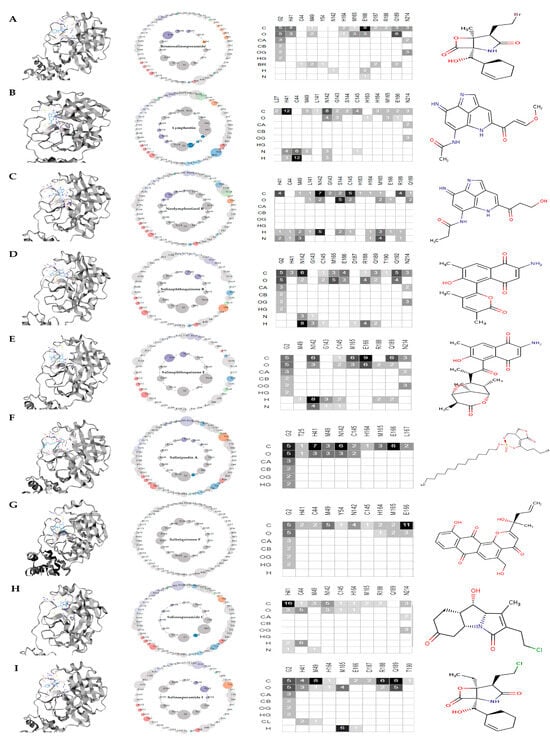
Figure 6.
Asteroid plots and ligand residue matrices. (A) 3CLpro-bromosalinosporamide, (B) 3CLpro-lymphostin, (C) 3CLpro-neolymphostinol B, (D) 3CLpro-salinaphthoquinone B, (E) 3CLpro-salinaphthoquinone E, (F) 3CLpro-saliniposin A, (G) 3CLpro-saliniquinone F, (H) 3CLpro-salinosporamide C, (I) 3CLpro-salinosporamide I. The atomic scale interaction number percentage is represented by the circumference of the sphere. Residues are colored according to their respective secondary structures. The atoms of the ligands are displayed as rows in the ligand residue matrices, and the residues that are in contact with the ligand are displayed as columns. In the matrices, the number of atomic associations has also been presented. The shades of grey (from light to dark) depend on the number of atomic contacts made.
In the first case of bromosalinosporamide, carbon and oxygen atoms made direct non-covalent associations with HIS: 41 by forming seven atomic connections. On the other hand, indirect associations were made with CYS: 145, as the residue lay on the outer shell of the asteroid plot (see Figure 6A).
In the second case, lymphostin associated in a direct manner with HIS: 41 residue by forming 12 carbon atom contacts, 6 atomic contacts via nitrogen, and about 12 hydrogen contacts. For CYS: 145 residue, the lymphostin made direct connections through carbon and nitrogen atoms by forming five non-covalent links in total (see Figure 6B).
In the third case, neolymphostinol B interacted with the amino acid HIS: 41 through direct non-covalent connections by forming four carbon, one hydrogen, and two nitrogen links. In the case of CYS: 145, the atomic connections were made by the carbon, oxygen, and hydrogen atoms of neolymphostinol B, forming a total of about eight contacts (see Figure 6C).
In the fourth case, salinaphthoquinone B attached non-covalently to HIS: 41 through the carbon atom, making three contacts, whereas the CYS: 145 was observed to be directly associated with not only the carbon atom but also the oxygen and hydrogen atoms, forming four atomic contacts in total (see Figure 6D).
In the fifth case, salinaphthoquinone E formed direct contacts with CYS: 145 residue via carbon, oxygen, hydrogen, and nitrogen atoms by forming six atomic connections (see Figure 6E).
In the sixth case, salinipostin A associated directly through non-covalent interactions with both HIS: 41 and CYS: 145 residues via carbon and oxygen atoms by forming 14 contacts (see Figure 6F).
In the seventh case, saliniquinone F interacted with residue HIS: 41 and CYS: 145 with carbon and hydrogen atoms by making three connections altogether (see Figure 6G).
In the eighth case, salinosporamide C strongly associated with amino acid HIS: 41 by forming 22 atomic connections made via carbon, oxygen, hydrogen, and nitrogen atoms. However, only a total of two atomic contacts were observed in the case of CYS: 145 residue, formed by the carbon and oxygen atoms of the salinosporamide C molecule (see Figure 6H).
Finally, in the ninth case, salinosporamide I was found in direct non-covalent association with HIS: 41 through carbon, oxygen, and chlorine atoms. A total of nine contacts were witnessed (see Figure 6I).
3.1.5. Simulations for 3CLpro-Ligand Complexes
The aforementioned complexes underwent molecular dynamics simulations on the CABS-flex 2.0 platform. This website may occasionally be utilized as an alternate simulation option if researchers encounter technological challenges or limitations.
For two key reasons, the simulations for all nine docked complexes were run at 50, 125, and 500 cycles. One reason was to monitor the degree of residue fluctuation as simulation cycles proceeded for longer. The second reason was to contrast them with the results of the RMSF plot generated by the 3CLpro Apo-protein simulation. The root mean square fluctuation values for residues HIS: 41 and CYS: 145 were 1.09 Å and 1.43 Å, respectively (see Figure 3A). When there was an overall reduction in the fluctuation scores of the residues in a complex, it may be argued that a rather stable protein–ligand conformation developed (see Figure 7)
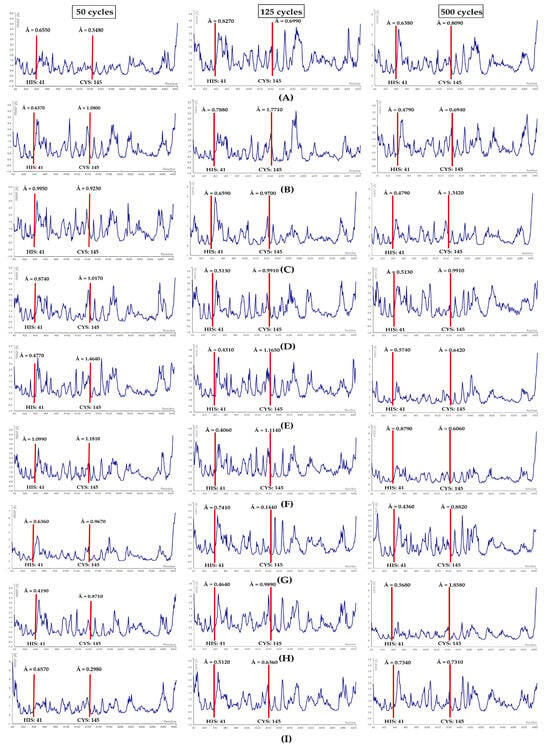
Figure 7.
RMSF plots. (A) 3CLpro-bromosalinosporamide, (B) 3CLpro-lymphostin, (C) 3CLpro-neolymphostinol B, (D) 3CLpro-salinaphthoquinone B, (E) 3CLpro-salinaphthoquinone E, (F) 3CLpro-salinipostin A, (G) 3CLpro-saliniquinone F, (H) 3CLpro-salinosporamide C, (I) 3CLpro-salinosporamide I. Red lines denote the location of the catalytic dyad residues.
In the first case, the 3CLpro-bromosalinosporamide complex at 50 cycles showed fluctuations of HIS: 41 and CYS: 145 to be 0.65 Å and 0.54 Å, correspondingly. At 125 cycles, a rise in the fluctuation was witnessed for both residues. However, the RMSF value for HIS: 41 dropped and for CYS: 145 slightly increased at 500 cycles (see Figure 7A).
In the second case, the 3CLpro-lymphostin complex when simulated at 50 cycles showed fluctuation scores of 0.63 Å for HIS: 41 and 1.08 Å for CYS: 145. Upon simulating at 125 cycles, an upward trend in fluctuations for both residues was noticed, as expected. Surprisingly, at 500 cycles there was a noteworthy decline in the RMSF values (see Figure 7B).
In the third case, 3CLpro-neolymphostinol B at 50 cycles displayed RMSF scores for HIS: 41 of 0.99Å and for CYS: 145 of 0.92 Å. Simulation at 125 and 500 cycles revealed that there was a drop in the fluctuation of HIS: 41, whereas a considerable increment in the RMSF score was evident in the case of CYS: 145 (see Figure 7C).
In the fourth case, 3CLpro-salinaphthoquinone B when simulated at 50 cycles demonstrated fluctuation values of 0.87 Å and 1.01 Å for HIS: 41 and CYS: 145, respectively. Upon performing simulations at 125 and 500 cycles, it was clearly evident that there were no fluctuations. In this study, it was truly the most stable conformation-forming complex in the case of 3CLpro (see Figure 7D).
In the fifth case, the 3CLpro-salinaphthoquinone E complex upon simulation obtained RMSF values of HIS: 41 and CYS: 145 of 0.47 Å and 1.46 Å, correspondingly. Overall, a drop in fluctuation was observed in further simulation runs at 125 and 500 cycles (see Figure 7E).
In the sixth case, 3CLpro-salinipostin A when subjected to simulation at a mere 50 cycles, the value obtained for HIS: 41 was 1.09 Å, which was similar to the Apo-protein simulation score. However, this was not the case for CYS: 145, as a fluctuation drop (1.18 Å) was noticeable. Subsequently, the RMSF values showed both upward and downward trends, but the reduction of overall fluctuation levels cannot be ignored (see Figure 7F).
In the seventh case, 3CLpro-saliniquinone F upon simulation at 50 cycles, the RMSF score for HIS: 41 was 0.63 Å and for CYS: 145 it was 0.96 Å. Simulation at 125 cycles showed that there was a slight increment in the fluctuation of HIS: 41. On the contrary, a sharp decline was evident for the residue CYS: 145, and vice versa at a 500 cycle simulation (see Figure 7G).
In the eighth case, the 3CLpro-salinosporamide C simulation depicted an RMSF value of HIS: 41 of 0.41 Å and of CYS: 145 of 0.87 Å. The RMSF scores indicated a continuous rise in fluctuations at 125 and 500 cycles (see Figure 7H).
Finally, for the ninth complex (3CLpro-salinosporamide I), the 50 cycle simulation provided an RMSF score for HIS: 41 of 0.65 Å and 0.29 Å. Similar to the salinosporamide C, we noticed a gradual upward trend in fluctuations as the simulation cycles were increased (see Figure 7I).
The RMSF values for the catalytic dyad residues remained within 0 to 3 Å for all nine docked complexes of 3CLpro. Also noteworthy is the fact that the RMSF values for the nine complexes showed an overall significant reduction when compared with the values obtained for the Apo structure of 3CLpro, potentially demonstrating the required stability for protein inhibition.
3.2. SARS-CoV-2 Papain-Like Protease Protein (PLpro)
3.2.1. PLpro Enzyme Structure and Residue–Residue Interactions in Active Site
In the natural immunological response to viral infection, PLpro plays a vital role [128,129,130]. It can eliminate the ubiquitin and ISG15 from cellular proteins to assist the virus in dodging the host’s natural immune response [131]. As a result, specific targeting of the catalytic triad (CYS: 111, HIS: 272, and ASP: 286 of PLpro) can inhibit its activity, preventing the viral replication and deregulation of biochemical cascades in infected cells. TRP: 93, TRP: 106, ASP: 108, and ASN: 109 are also known to contribute towards PLpro function [132]. The N-terminal ubiquitin-like region (Ubl, β1–3), the helical thumb domain (α2–7), the finger region (β4–7), and the palm region (β8–13) are the four sub-regions that constitute the SARS-CoV-2 PLpro protein. Four conserved cysteines create a zinc finger, a component of the “zinc ribbon” fold group 28, in the finger sub-region (C189 and C192 on the loop within β4–5, C224 and C226 on the loop within β6–7) [44]. Figure 8A depicts the location of the catalytic site and the simulation output for the PLpro Apo-protein structure. The PLpro enzyme has no chains (refer to Figure 8B). The plots show root mean square fluctuation values for CYS: 111 (Å = 1.50), HIS: 272 (Å = 1.66), and ASP: 286 (Å = 0.97), respectively.
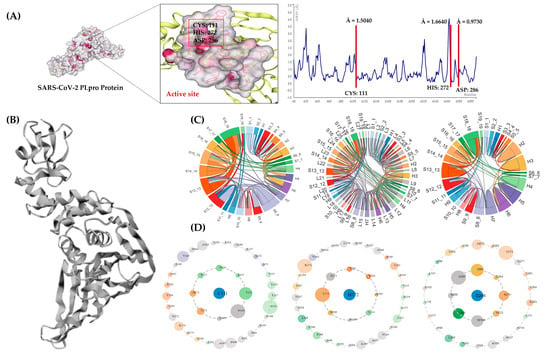
Figure 8.
In-depth PLpro analyses. (A) Three-dimensional Apo structure of PLpro with the active site and CABS-flex RMSF plot. Protein Contact Atlas output (B) shows PLpro crystal structure as presented on the webserver. (C) Chord plots depicting loops and secondary structure interactions for PLpro enzyme. (D) Asteroid plots depicting residue contacts with CYS: 111, HIS: 272, and ASP: 286. The atomic scale interaction number percentage is represented by the circumference of the sphere. Residues are colored according to their respective secondary structures (from cyan blue to magenta high).
The sequence of connections between the various secondary structural components most likely dictate the tertiary structure and, in turn, the function of the PLpro protein. In Figure 8C, the chord plots (including loops) show the location and the secondary structure relationship of the catalytic triad residues. The amino acid CYS: 111, with its immediate neighbors, formed 52 contacts. The position of CYS: 111 was in region H4 of the chord plot (colored as pastel green). The amino acid HIS: 272 situated at the S15_15 region on the chord plot is presented in an apricot orange color. A total of 50 atomic connections were made by HIS: 272 with its immediate neighbors. Finally, ASP: 286 formed 60 atomic associations with immediate neighbors, positioned in the S16_16 region on the chord plot and presented in a mustard yellow color. The associations among the secondary structures in PLpro appear to be dense, which means there is a strong correlation among them.
The asteroid plots in Figure 8D illustrate the non-covalent connections that exist between the three critical catalytic residues, CYS 111, HIS 272, and ASP 286. The first plot shows the direct relationships that the residue CYS: 111 formed with the amino acids N109, N110, Y112, L113, A114, T115, H272, and Y273. Similarly, in the second plot, the amino acid HIS: 272 is also shown to be directly connected to residues W106, C111, E263, Y264, T265, G271, Y273, and D286 via non-covalent linkages. Finally, direct correlations between the amino acids W106, A268, H272, K274, C284, I285, and L289 were seen in the plot generated for residue ASP: 286. There was no direct association found between CYS: 111 and ASP: 286. The amino acid HIS: 272 might be acting as a link between the two residues of concern. The residue Y273 formed direct non-covalent linkages with both C111 and H272 residues, whereas residue W106 was found to be commonly linked between amino acids H272 and D286. Thus, amino acids Y273 and W106 mutations may have a direct impact on the ligand binding. On the other hand, altering the outer-shell residues might have an impact on ligand binding, although indirectly.
3.2.2. PLpro Docking
Ligands docked with PLpro protein classified on the basis of affinity values are presented in Table 3.

Table 3.
Depicting the range of binding affinities obtained for the target PLpro.
In the instance of enzyme PLpro, category 1 had 46 NPs with low binding affinities that ranged from −4.0 to −5.9 kcal/mol (see Table S7). About 12 NPs, including arenimycin B, enterocin, salinilactone A, B, and G, salinipostin C, D, F, H, I, K, and salinosporamide F, demonstrated binding affinity with an energy of −5.9 kcal/mol. However, sporolide B exhibited the lowest binding affinity, measuring −4.4 kcal/mol. About 52 NPs with moderately low binding affinities ranging from −6.0 to −6.9 kcal/mol are listed in category 2 (refer to Table S8). A total of 8 NPs out of 52 revealed the highest binding affinity of −6.9 kcal/mol. These NPs were arenicolide C, desferrioxamine B, N-(2′-phenylethyl) isobutyramide, rifsaliniketal, salinaphthoquinone E, saliniquinone A, and saliniketal A and B. Further, the NPs with moderately high binding affinities starting from −7.0 kcal/mol up to −7.9 kcal/mol are presented in Table 3. In this group, 14 NPs, out of which 2 NPs, (namely cyclomarazine B and tirandalydigin) produced the lowest binding energies of −7.9 kcal/mol. The highest binding energies, −7.0 kcal/mol, were shown by salinipyrone A and saliniquinone C (see Table S9). Ultimately, Saliniquinone D and homoseongomycin, 2 of the 13 NPs in category 3, had the maximum binding affinities of −8.7 and −8.5 kcal/mol, respectively, whereas neolymphostinol A, C and saliniquinone B, E were shown to have a −8.0 kcal/mol affinity for PLpro (refer to Table S10). In the case of the docking validation experiment for PLpro, inhibitor Y41 achieved a score of −8.9 kcal/mol. The RMSD value for the superimposed structure was 1.03 Å. The RMSD value was lower than the threshold of 2.0 Å (see Figure 2B).
3.2.3. Best Residue Interaction Complexes, PLpro
Residues CYS: 111, HIS: 272, and ASP: 286 comprise the standard cysteine protease catalytic triad in the PLpro active site. With CYS: 111 functioning as a nucleophile in its commonly recognized thiolate form and ASP: 286 facilitating the de-protonation of base HIS: 272, PLpro may have catalytic characteristics that align further with other cysteine proteases [133,134]. The re-docking experiment produced a ligand binding mode that was remarkably similar to the binding mode of the attached inhibitor. As a result, Figure 2B illustrates a high resemblance in the residue interactions, highlighting the reliability of the utilized protocol for PLpro docking. Based on this crucial information, we narrowed down the pool of docked complexes to six, all of which had the necessary ligand–residue interactions involving the trio of these amino acids (see Table 4) according to our aforementioned set criteria. The chosen NPs bound to the PLpro protein are shown in Figure 9 below.

Table 4.
Ligands showing desired interactions for PLpro protein.
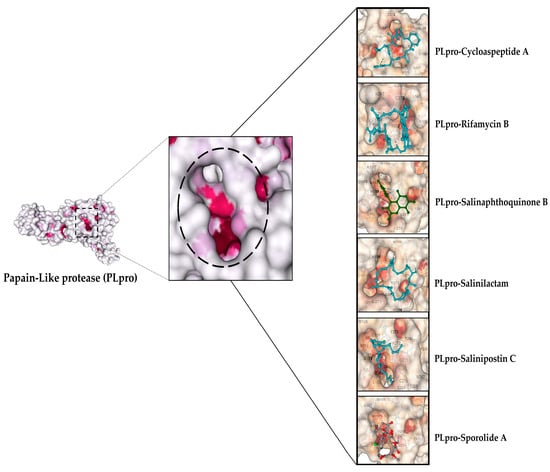
Figure 9.
Depicting the top ligands docked in the active site pocket of PLpro. The active site of the protein colored by B-factor and ligands are shown in different colors.
Table 4 below lists the interacting amino acids during the formation of the complex with the PLpro enzyme.
Cycloaspeptide A interacted with CYS: 111 and HIS: 272 by forming a single hydrogen (2.89 Å) and pi-pi T shaped (4.84 Å), respectively; however, it showed a weak interaction with ASP: 286. Rifamycin B interacted with CYS: 111 by making a hydrogen bond (2.21 Å). In the case of HIS: 272, a pi-pi T-shaped bond (4.34 Å), along with a carbon–hydrogen and a pi-donor hydrogen bond, was made (3.56 Å). Lastly, ASP: 286 interacted with Vander Waals forces. Salinaphthoquinone B strongly interacted with CYS: 111 by forming four bonds, out of which three were alkyl and/or pi-alkyl (4.64 Å, 4.96 Å, and 5.00 Å) and one was a hydrogen bond (2.18 Å). For HIS: 272, a pi-pi T-shaped bond (4.62 Å) was formed. Salinilactam interacted only with HIS: 272 and ASP: 286 by making a single pi-alkyl (4.34 Å) and hydrogen bond (2.57 Å), respectively. Salinipostin C interacted with CYS: 111 by forming one hydrogen (2.21 Å) and one alkyl bond (4.71 Å), whereas HIS: 272 was linked with a pi-alkyl bond (4.37 Å). Finally, sporolide A interacted with CYS: 111 and HIS: 272, forming a hydrogen (2.81 Å) and a pi-alkyl bond (4.34 Å) (see Table 4 and Figure 10).
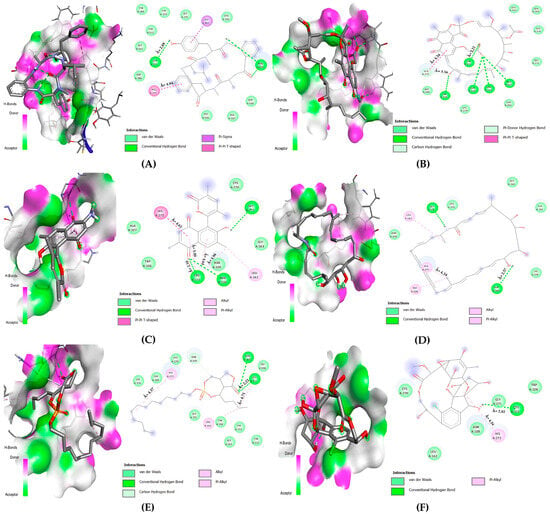
Figure 10.
Two-dimensional amino acid interaction maps. (A) PLpro-cycloaspeptide A complex, (B) PLpro-rifamycin B, (C) PLpro-salinaphthoquinone B, (D) PLpro-salinilactam, (E) PLpro-salinipostin C, (F) PLpro-sporolide A.
3.2.4. Ligand–Residue Non-Covalent Interactions for PLpro Complexes
All the six docked complexes were subjected to non-covalent interaction screening on the Protein Contact Atlas webserver (refer to Figure 11). The first complex, cycloaspeptide A, made direct non-covalent interactions with two key residues: HIS: 272 and ASP: 286. The carbon and oxygen atoms of cycloaspeptide A formed seven and four connections with HIS: 272, respectively. In the case of ASP: 286, only one atomic connection with carbon atom was observed. There was no direct non-covalent connection with residue CYS: 111 (see Figure 11A).
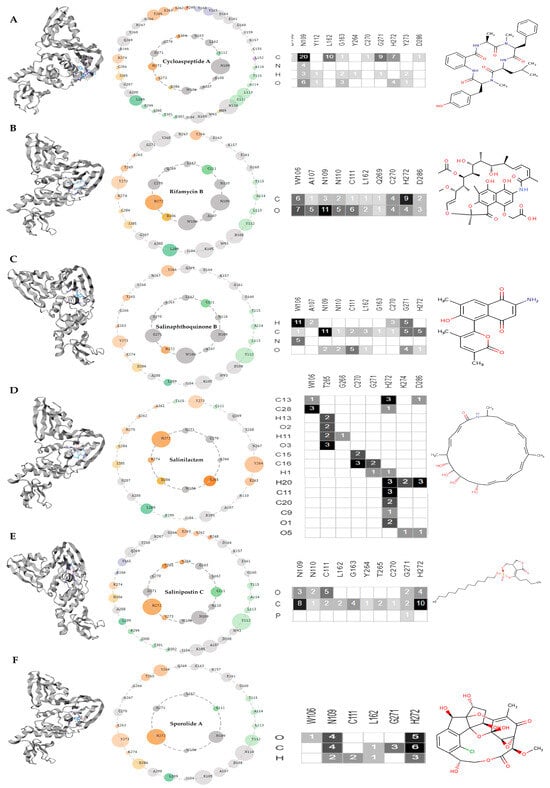
Figure 11.
Asteroid plots and ligand-residue matrices for PLpro protein. (A) PLpro-cycloaspeptide A complex, (B) PLpro-rifamycin B, (C) PLpro-salinaphthoquinone B, (D) PLpro-salinilactam, (E) PLpro-salinipostin C, (F) PLpro-sporolide A. The atomic scale interaction number percentage is represented by the circumference of the sphere. Residues are colored according to their respective secondary structures. The atoms of the ligand are displayed as rows in the ligand residue matrices, and the residues that are in contact with the ligand are displayed as columns. In the matrices, the number of atomic associations has also been presented. The shades of grey (from light to dark) depend on the number of atomic contacts made.
The second complex, rifamycin B, formed atomic connections with all three residues of importance. The carbon atom of rifamycin B formed one connection with residue CYS: 111 and nine connections with HIS: 272, whereas two associations with ASP: 286 were evident. On the other hand, the oxygen atom formed six contacts with CYS: 111, four contacts with HIS: 272, and three contacts with ASP: 286 (see Figure 11B).
The third complex, salinaphthoquinone B, formed two connections via the carbon atom and five contacts via the oxygen atom with residue CYS: 111. In the case of HIS: 272, five contacts were made by the carbon atom and one association was made by the oxygen atom. There was no direct non-covalent interaction with ASP: 286 (see Figure 11C).
The fourth complex, salinilactam, associated with HIS: 272 through C13, C28, H1, H20, C11, C20, C9, and O1 atoms, forming a total of 14 connections. Residue ASP: 286 was contacted non-covalently via C13, H20, and O5 atoms of salinilactam, forming four associations in total (see Figure 11D).
The fifth complex, salinipostin C, associated by forming 7 connections with CYS: 111 and 14 connections with HIS: 272 via carbon and oxygen atoms (see Figure 11E). Ultimately, sporolide A formed 2 contacts with CYS: 111 via the hydrogen atom, whereas HIS: 272 was connected non-covalently with carbon, hydrogen, and oxygen, forming 14 associations in total (see Figure 11F).
3.2.5. Simulations for PLpro-Ligand Complexes
The simulations were performed at 50, 125, and 500 cycles for all six docked complexes for two main reasons; one, to trace the residue fluctuation levels with increased simulation durations over time; two, to compare them to the RMSF plot values from the PLpro Apo-protein simulation. Residue CYS: 111 had a root mean square fluctuation value of Å = 1.50, HIS: 272 had a value of Å = 1.66, and ASP: 286 had a value of Å = 0.97 (see Figure 8A). It can be assumed that a relatively stable protein–ligand conformation is formed when there is a reduction in the fluctuation scores of the residues (see Figure 12).
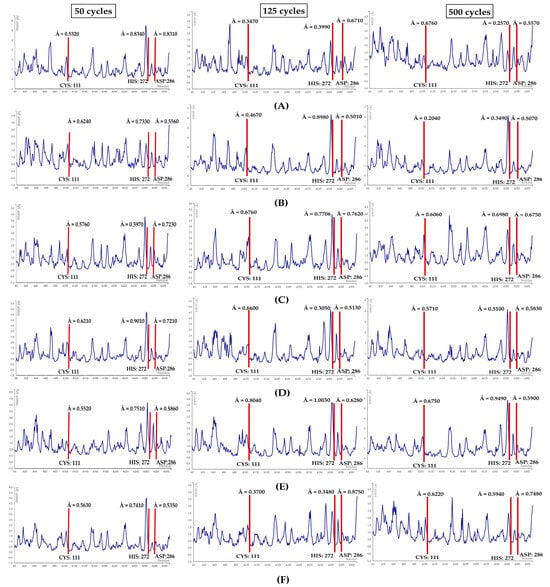
Figure 12.
RMSF plots for (A) PLpro-cycloaspeptide A complex, (B) PLpro-rifamycin B, (C) PLpro-salinaphthoquinone B, (D) PLpro-salinilactam, (E) PLpro-salinipostin C, (F) PLpro-sporolide A. Red lines highlight the position of the catalytic triad residues.
First, the three residues of the PLpro-cycloaspeptide A complex at 50 cycles showed RMSF values ranging from 0.53 to 0.83 Å. Further, at 125 cycles, the RMSF values for all the three residues dropped slightly. However, at 500 cycles the fluctuation score for CYS: 111 increased to 0.67 Å, whereas the values for HIS: 272 and ASP: 286 showed a further reduction (see Figure 12A).
Second, the PLpro-rifamycin B complex key residues displayed RMSF scores ranging from 0.55 to 0.73 Å at 50 cycles. There was a miniscule decline in fluctuations for the two residues—CYS: 111 (0.46 Å) and ASP: 286 (0.50 Å)—at 125 cycles. Further, at 500 cycles a downward trend in RMSF values was evident (see Figure 12B).
Third, the catalytic residues of the PLpro-salinaphthoquinone B complex at 50 cycles presented RMSF scores from 0.57 to 0.72 Å. A slight upward trend in fluctuations was observed at 125 cycles, followed by a minor decline at 500 cycles (see Figure 12C).
Fourth, the PLpro-salinilactam complex at 50 cycles demonstrated RMSF values of the three residues to be within 0.62 and 0.90 Å. Further, at 125 cycles, fluctuations reduced for HIS: 272 and ASP: 286, but an increase in fluctuations was observed in the instance of CYS: 111. At 500 cycles, an overall drop in fluctuations was witnessed (see Figure 12D).
Fifth, the concerned residues in the PLpro-salinipostin C complex showed fluctuations ranging from 0.55 to 0.75 Å at 50 cycles. There was a sharp rise in the fluctuations at 125 cycles for all the three residues, followed by a slight decline at 500 cycles (see Figure 12E).
Ultimately, the sixth, PLpro-sporolide A complex at 50 cycles demonstrated fluctuations from 0.53 Å to 0.74 Å for the three residues. In the case of 125 cycles, the fluctuations showed a sharp decline, with ASP: 286 being the exception. For the 500 cycle run, a minor rise in fluctuations occurred but not in the case of ASP: 286 (see Figure 12F).
The RMSF values for the crucial residues stayed well within the range of 0 to 3 Å for each of the six docked complexes of PLpro. Furthermore, it is essential to note that the RMSF values for the six complexes showed a substantial reduction when compared with the values obtained for the PLpro Apo structure, suggesting the anticipated stable conformations required for the enzyme inhibition.
In this study, Salinaphthoquinone B emerged as a putative dual inhibitor of the 3CLpro and PLpro proteins of SARS-CoV-2. Further MD simulations at 1000 cycles were performed on the two docked complexes (see Figure 13). The RMSF values remained less when compared to the simulated Apo-protein structures (see Figure 3A and Figure 4A). Moreover, no residue superceded the threshold value of 3.0 Å.
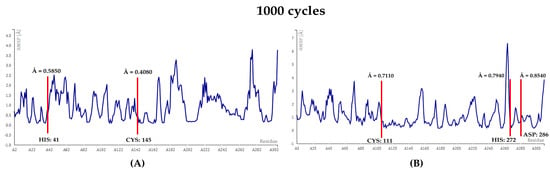
Figure 13.
RMSF plots at 1000 cycles. (A) 3CLpro-Salinaphthoquinone B, (B) PLpro-Salinaphthoquinone B.
3.3. Toxicity Evaluation
ProTox-II and StopTox webservers provided us with the estimation of potential toxicity resulting from the NPs.
3.3.1. ProTox-II Analysis
We received the predictions based on the resemblance of our query NPs’ functional group with the experimental data of in vitro and in vivo studies already included in the database’s contents. Also, the ProTox-II webserver uses an array of 33 models for the estimation of several toxicity endpoints, as presented below in Table 5.

Table 5.
Depicting the ProTox-II results for shortlisted ligands.
Salinipostins A and C have the potential to cause fatality upon oral exposure as they belong to class 2 (5 < LD50 ≤ 50). Being a class 3 member (50 < LD50 ≤ 300), salinosporamide C has the potential to be hazardous when taken orally. Salinilactam, sporolide A, lymphostin, neolymphostinol B, salinaphthoquinone E, saliniquinone F, and bromosalinosporamide were among the class 4 members (300 < LD50 ≤ 2000). Although they are not toxic, ingesting them might be harmful. Salinosporamide I, salinaphthoquinone B, and rifamycin B were found in class 5 (2000 < LD50 ≤ 5000); the likelihood of them being harmful is very low. Finally, a class 6 designation (LD50 > 5000), which denotes a probability for non-toxicity, was given to cycloaspeptide A.
All 14 NPs were inactive for hepatotoxicity. Only three natural products—lymphostin and salinipostins A and C—were found to be positively carcinogenic. Except for five NPs, the majority of them were determined to be active in terms of their propensity to induce immunotoxicity. Last but not least, merely three NPs—lymphostin, salinaphthoquinone B, and saliniquinone F—were shown to have mutagenic potential.
3.3.2. StopTox Analysis
The StopTox webserver that implements QSAR models, attempted to provide us with accurate estimations of the likelihood that the selected candidate molecules would impart acute toxicity, as shown in Table 6 below.

Table 6.
Acute toxicity for NPs.
All 14 NPs are probably non-toxic when inhaled. However, acute oral toxicity from salinosporamide C and salinipostins A and C are possible. For saliniquinone F, sporolide A, and once more for the two salinipostins, dermal toxicity was suggested. Only two NPs, sporolide A and saliniquinone F, were shown to not influence skin corrosion or eye discomfort. Other NPs, however, have the potential to irritate or corrode either the skin or eyes. Except for saliniquinone F and the two salinipostins, most NPs may not provoke skin sensitivity.
3.4. Drug-Likeness Evaluation
3.4.1. Pfizer’s/Lipinski’s Rule of Five
The above-mentioned NPs’ drug-like qualities were assessed using Pfizer’s/Lipinski’s rule of five (Ro5) concept. Both cycloaspeptide A and sporolide A violated the first criteria by going beyond the molecular weight threshold of 500 g/mol. Rifamycin B and sporolide A both violated the second criterion since they had more than ten hydrogen bond acceptors. Rifamycin B also breaks the third rule by having a sixth hydrogen bond donor. The two salinipostins further violate the fourth rule by going beyond the allowed consensus log p value. Salinipostin A and C also failed to pass the additional parameter, such as the number of rotatable bonds, by surpassing the maximum limit. In addition, the TPSA parameter was not passed by cycloaspeptide A, rifamycin B, or sporolide A (see Table 7).

Table 7.
Drug-likeness properties of NPs.
3.4.2. Swiss-ADME
Ten of the fourteen NPs were shown to be readily and moderately soluble in water, whereas four NPs were discovered to be poorly soluble. The lowest bioavailability scores were achieved by sporolide A and rifamycin B, with cycloaspeptide A coming in second at 0.17. Five of the fourteen NPs, including cycloaspeptide A, rifamycin B, salinipostin A, C, and sporolide A, had minimal GI absorption. All NPs with an absorption percentage (AB% > 50%) indicated high oral bioavailability, distribution, and circulation. The score for salinosporamide I was the highest at 89.90%, whereas sporolide A was the lowest at 45.26% (see Table 8).

Table 8.
Swiss-ADME results for NPs.
Four of the fourteen NPs—cycloaspeptide A, salinipostin A, salinipostin C, and sporolide A—cannot be passively absorbed by the GI tract. The blood–brain barrier can be passively crossed by salinosporamide C (see Figure 14).
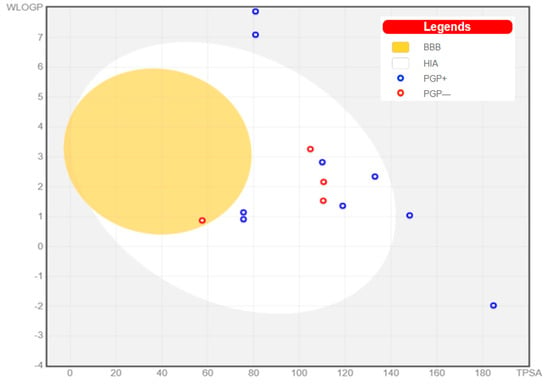
Figure 14.
Swiss-ADME boiled egg plot for NPs.
3.5. Probable Bioactivities
We were able to study the prospects of candidate NPs thanks to the PASSonline tool, a website that forecasts the likely bioactivities of molecules. The detailed outcome can be found in Table S11 mentioned in the supplemental information.
The bioactivity predictions indicated that the majority of the NPs used in this investigation may have antiviral properties. COVID-19-associated fungal infections, especially mucormycosis (CAM), may potentially be addressed, since these NPs have the propensity to be antifungals and antifungal enhancers. Due to this property, these NPs have the capability of inhibiting enzymes such as histidine kinase, fungal lipase, rhizopuspepsin, and alpha and beta glucuronidase, which were previously explored as possible therapeutic targets for mitigating mucormycosis [135]. Additionally, potential bioactivity toward the immune system was revealed by all fourteen NPs. By promoting the macrophage colony, interleukin receptors, NF kappa B transcription, and cytokine release, they may strengthen the immune system. On the other hand, these NPs also have immunosuppressive properties that might suppress the cytokine storm by competing with the C-C chemokine 2B, toll-like, and interleukin receptors. Moreover, they could potentially inhibit T cells, Janus kinase enzyme 2, cytokine, and histamine release. Antifungal, anticancer, antioxidant, antineoplastic alkaloids, antibiotic, antibacterial, antiparasitic, and antidiabetic properties were additional significant bioactivities to consider. Breathing difficulties in SARS-CoV-2 patients may be alleviated because of anti-infective, anti-asthmatic, respiratory analeptic, expectorant, and respiratory disease-treating properties of a few NPs.
4. Conclusions
Millions of individuals have died as a result of the COVID-19 outbreak [136], but the threat it still poses to human health necessitates the discovery of new drug-like molecules. It should be mentioned that in silico assessments could be a very fast method to evaluate other natural compounds with well-known antiviral activity, such as coumarins [137], peptides, and saccharides [138], for the need of COVID-19 therapeutics. The purpose of this study was to shed light on the Salinispora genus’s untapped antiviral potential, as it is typically primarily seen as a source of antibacterial substances. Under laboratory settings, these bacteria may be grown for bioactive substances, requiring less space, labor, and reasonable material expenses. In this paper, we proposed the idea of the repositioning of Salinispora natural products as potential inhibitors of SARS-CoV-2 3CLpro and PLpro enzymes.
To conclude, the docking investigations initially identified 35 NPs with strong affinities for both 3CLpro and PLpro scores ranging from −8.0 kcal/mol to −9.0 kcal/mol. Nine NPs screened based on binding residue studies criteria showed strong interactions with 3CLpros HIS: 41 and CYS: 145. Additionally, in the case of PLpro, 6 NPs were revealed out of 125 complexes that interacted desirably with CYS: 111, HIS: 272, and ASP: 286. The ligand–residue interaction investigations on the Protein Contact Atlas webserver additionally demonstrated that all of the catalytic amino acids were indeed forming non-covalent associations with all shortlisted molecules. Moreover, resulting from the MD simulations, all complexes were shown to be stable at 50, 125, and 500 cycles. Among the final shortlisted NPs, salinaphthoquinone B was a common natural product that showed highly desirable amino acid interactions with all catalytic residues for both 3CLpro and PLpro enzymes, and the complexes remained stable even at a 1000 cycle simulation run. Although detailed pharmacokinetic studies should be completed, our results have revealed a potential candidate, one more time highlighting in silico assessment as a good addition or inexpensive alternative to high throughput screening. We are suggesting further in vitro and/or in vivo studies to confirm the antiviral activities of our recommended natural products from the marine actinomycete genus Salinispora.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microbiolres14040130/s1, Table S1: List of natural products from genus salinispora involved in this work; Table S2: Canonical SMILES for selected natural products; Table S3: 3CL pro docking results and residue interactions category 1; Table S4: 3CL pro docking results and residue interaction category 2 (lower-moderate values); Table S5: 3CLpro docking results and residue interaction category 2 (upper-moderate values); Table S6: 3CLpro docking results and residue interactions category 3; Table S7: PLpro docking results and residue interactions category 1; Table S8: PLpro docking results and residue interactions category 2 (lower-moderate values); Table S9: PLpro docking results and residue interactions category 2 (upper-moderate values); Table S10: PLpro docking results and residue interactions category 3; Table S11: Predicted biological activities for candidate NPs.
Author Contributions
Conceptualization, O.P. and G.V.Z.; methodology, O.P.; software, O.P.; formal analysis, O.P.; investigation, O.P.; data curation, O.P.; writing—original draft preparation, O.P.; writing—review and editing, O.P., M.V.T. and G.V.Z.; supervision, G.V.Z. and M.V.T. All authors have read and agreed to the published version of the manuscript.
Funding
O.P. and G.V.Z. are thankful to the Ministry of Science and Education of RF (Agreement # 075-15-2022-1118 dated 29 June 2022) for funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and supplementary materials.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| Abbreviation | Full Name |
| MNPs | Marine natural products |
| NPs | Natural products |
| COVID-19 | Coronavirus disease of 2019 |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| FDA | Food and drug administration |
| RCSB PDB | Research Collaboratory for Structural Bioinformatics Protein Data Bank |
| PDB | Protein Data Bank |
| PDBQT | Protein Data Bank with partial charge Q and atom type T |
| SDF | Structure data file |
| MGL | Molecular Graphics Laboratory |
| 3CLpro | 3C-like protease |
| Mpro | Main protease |
| PLpro | Papain-like protease |
| HETATM | Hetero atom |
| RMSF | Root mean square fluctuation |
| RMSD | Root mean square deviation |
| GI | Gastrointestinal |
| TPSA | Topological polar surface area |
| ADME | Absorption, distribution, metabolism, excretion |
| SMILES | Simplified molecular-input line-entry system |
| QSAR | Quantitative structure–activity relationship |
| VdW | Van der Waals |
| PyMOL | Proprietary molecular visualization system |
| PASS | Prediction of activity spectra for substances |
| CAM | COVID-19-associated mucormycosis |
References
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef]
- Sohrabi, C.; Alsafi, Z.; O’Neill, N.; Khan, M.; Kerwan, A.; Al-Jabir, A.; Iosifidis, C.; Agha, R. World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). Int. J. Surg. 2020, 76, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed]
- She, J.; Jiang, J.; Ye, L.; Hu, L.; Bai, C.; Song, Y. 2019 novel coronavirus of pneumonia in Wuhan, China: Emerging attack and management strategies. Clin. Transl. Med. 2020, 9, 19. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Mishra, N.P.; Das, S.S.; Yadav, S.; Khan, W.; Afzal, M.; Alarifi, A.; Kenawy, E.R.; Ansari, M.T.; Hasnain, M.S.; Nayak, A.K. Global impacts of pre- and post-COVID-19 pandemic: Focus on socio-economic consequences. Sens. Int. 2020, 1, 100042. [Google Scholar] [CrossRef]
- Astuti, I.; Ysrafil. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An overview of viral structure and host response. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 407–412. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef]
- Li, Q.; Kang, C. Progress in Developing Inhibitors of SARS-CoV-2 3C-Like Protease. Microorganisms 2020, 8, 1250. [Google Scholar] [CrossRef]
- Suarez, D.; Diaz, N. SARS-CoV-2 Main Protease: A Molecular Dynamics Study. J. Chem. Inf. Model. 2020, 60, 5815–5831. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Telenti, A.; Arvin, A.; Corey, L.; Corti, D.; Diamond, M.S.; Garcia-Sastre, A.; Garry, R.F.; Holmes, E.C.; Pang, P.S.; Virgin, H.W. After the pandemic: Perspectives on the future trajectory of COVID-19. Nature 2021, 596, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.A.A.; Abdelrahman, A.H.M.; Hegazy, M.F. In-silico drug repurposing and molecular dynamics puzzled out potential SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2021, 39, 5756–5767. [Google Scholar] [CrossRef]
- Ibrahim, M.A.A.; Abdelrahman, A.H.M.; Allemailem, K.S.; Almatroudi, A.; Moustafa, M.F.; Hegazy, M.F. In Silico evaluation of prospective anti-COVID-19 drug candidates as potential SARS-CoV-2 main protease inhibitors. Protein J. 2021, 40, 296–309. [Google Scholar] [CrossRef]
- Pawar, A.Y. Combating devastating COVID-19 by drug repurposing. Int. J. Antimicrob. Agents 2020, 56, 105984. [Google Scholar] [CrossRef]
- Drozdzal, S.; Rosik, J.; Lechowicz, K.; Machaj, F.; Kotfis, K.; Ghavami, S.; Los, M.J. FDA approved drugs with pharmacotherapeutic potential for SARS-CoV-2 (COVID-19) therapy. Drug Resist. Updates 2020, 53, 100719. [Google Scholar] [CrossRef]
- Maltsev, O.V.; Kasyanenko, K.V.; Kozlov, K.V.; Zhdanov, K.V.; Lapikov, I.I. Prospects of using the nucleoside analogue riamilovir in patients with SARS-CoV-2 infection. Ter. Arkhiv 2022, 94, 1171–1176. [Google Scholar] [CrossRef]
- Marzi, M.; Vakil, M.K.; Bahmanyar, M.; Zarenezhad, E. Paxlovid: Mechanism of Action, Synthesis, and In Silico Study. Biomed. Res. Int. 2022, 2022, 7341493. [Google Scholar] [CrossRef]
- Bartha, F.A.; Juhász, N.; Marzban, S.; Han, R.; Röst, G. In Silico Evaluation of Paxlovid’s Pharmacometrics for SARS-CoV-2: A Multiscale Approach. Viruses 2022, 14, 1103. [Google Scholar] [CrossRef]
- Panahi, Y.; Gorabi, A.M.; Talaei, S.; Beiraghdar, F.; Akbarzadeh, A.; Tarhriz, V.; Mellatyar, H. An overview on the treatments and prevention against COVID-19. Virol. J. 2023, 20, 23. [Google Scholar] [CrossRef] [PubMed]
- Musarra-Pizzo, M.; Pennisi, R.; Ben-Amor, I.; Mandalari, G.; Sciortino, M.T. Antiviral Activity Exerted by Natural Products against Human Viruses. Viruses 2021, 13, 828. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.T.; Hsu, W.C.; Lin, C.C. Antiviral natural products and herbal medicines. J. Tradit. Complement. Med. 2014, 4, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Pokharkar, O.; Anumolu, H.; Zyryanov, G.V.; Tsurkan, M.V. Natural Products from Red Algal Genus Laurencia as Potential Inhibitors of RdRp and nsp15 Enzymes of SARS-CoV-2: An In Silico Perspective. Microbiol. Res. 2023, 14, 1020–1048. [Google Scholar] [CrossRef]
- Pokharkar, O.; Lakshmanan, H.; Zyryanov, G.V.; Tsurkan, M.V. Antiviral Potential of Antillogorgia americana and elisabethae Natural Products against nsp16–nsp10 Complex, nsp13, and nsp14 Proteins of SARS-CoV-2: An In Silico Investigation. Microbiol. Res. 2023, 14, 993–1019. [Google Scholar] [CrossRef]
- Banerjee, P.; Mandhare, A.; Bagalkote, V. Marine natural products as source of new drugs: An updated patent review (July 2018–July 2021). Expert Opin. Ther. Pat. 2022, 32, 317–363. [Google Scholar] [CrossRef]
- Mia, M.M.; Hasan, M.; Miah, M.M.; Hossain, M.A.S.; Islam, S.; Shanta, V. Inhibitory Potentiality of Secondary Metabolites Extracted from Marine Fungus Target on Avian Influenza Virus-A Subtype H5N8 (Neuraminidase) and H5N1 (Nucleoprotein): A Rational Virtual Screening. Vet. Anim. Sci. 2022, 15, 100231. [Google Scholar] [CrossRef]
- Yasuhara-Bell, J.; Lu, Y. Marine compounds and their antiviral activities. Antivir. Res. 2010, 86, 231–240. [Google Scholar] [CrossRef]
- Gogineni, V.; Schinazi, R.F.; Hamann, M.T. Role of Marine Natural Products in the Genesis of Antiviral Agents. Chem. Rev. 2015, 115, 9655–9706. [Google Scholar] [CrossRef]
- Park, J.Y.; Kim, J.H.; Kwon, J.M.; Kwon, H.J.; Jeong, H.J.; Kim, Y.M.; Kim, D.; Lee, W.S.; Ryu, Y.B. Dieckol, a SARS-CoV 3CL(pro) inhibitor, isolated from the edible brown algae Ecklonia cava. Bioorg. Med. Chem. 2013, 21, 3730–3737. [Google Scholar] [CrossRef]
- Lira, S.P.d.; Seleghim, M.H.R.; Williams, D.E.; Marion, F.; Hamill, P.; Jean, F.; Andersen, R.J.; Hajdu, E.; Berlinck, R.G.S. A SARS-coronovirus 3CL protease inhibitor isolated from the marine sponge Axinella cf. corrugata: Structure elucidation and synthesis. J. Braz. Chem. Soc. 2007, 18, 440–443. [Google Scholar] [CrossRef]
- Ibrahim, M.A.A.; Abdelrahman, A.H.M.; Mohamed, D.E.M.; Abdeljawaad, K.A.A.; Naeem, M.A.; Gabr, G.A.; Shawky, A.M.; Soliman, M.E.S.; Sidhom, P.A.; Paré, P.W.; et al. Chetomin, a SARS-CoV-2 3C-like Protease (3CLpro) Inhibitor: In Silico Screening, Enzyme Docking, Molecular Dynamics and Pharmacokinetics Analysis. Viruses 2023, 15, 250. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Castro, L.; Martínez-García, S.; Cancino-Diaz, J.C.; Maldonado, L.A.; Hernández-Guerrero, C.J.; Martínez-Díaz, S.F.; González-Acosta, B.; Quintana, E.T. Marine Sediment Recovered Salinispora sp. Inhibits the Growth of Emerging Bacterial Pathogens and other Multi-Drug-Resistant Bacteria. Pol. J. Microbiol. 2020, 69, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Patin, N.V.; Duncan, K.R.; Dorrestein, P.C.; Jensen, P.R. Competitive strategies differentiate closely related species of marine actinobacteria. ISME J. 2016, 10, 478–490. [Google Scholar] [CrossRef]
- Patin, N.V.; Schorn, M.; Aguinaldo, K.; Lincecum, T.; Moore, B.S.; Jensen, P.R. Effects of Actinomycete Secondary Metabolites on Sediment Microbial Communities. Appl. Environ. Microbiol. 2017, 83, e02676-16. [Google Scholar] [CrossRef] [PubMed]
- Patin, N.V.; Floros, D.J.; Hughes, C.C.; Dorrestein, P.C.; Jensen, P.R. The role of inter-species interactions in Salinispora specialized metabolism. Microbiology 2018, 164, 946–955. [Google Scholar] [CrossRef] [PubMed]
- Chhun, A. Antimicrobial Potential of the Marine Actinomycete Salinispora Tropica CNB-440 in Co-Culture: A Metabolomic, Proteomic and Genome Engineering Approach. Ph.D. Thesis, University of Warwick, Coventry, UK, 2020. [Google Scholar]
- Chhun, A.; Sousoni, D.; Aguiló-Ferretjans, M.D.M.; Song, L.; Corre, C.; Christie-Oleza, J.A. Phytoplankton trigger the production of cryptic metabolites in the marine actinobacterium Salinispora tropica. Microb. Biotechnol. 2021, 14, 291–306. [Google Scholar] [CrossRef]
- Fischer, C.; Feys, J.R. SARS-CoV-2 Mpro Inhibitors: Achieved Diversity, Developing Resistance and Future Strategies. Future Pharmacol. 2023, 3, 80–107. [Google Scholar] [CrossRef]
- Mestres, J.; Gregori-Puigjané, E. Conciliating binding efficiency and polypharmacology. Trends Pharmacol. Sci. 2009, 30, 470–474. [Google Scholar] [CrossRef]
- Wang, J.; Guo, Z.; Fu, Y.; Wu, Z.; Huang, C.; Zheng, C.; Shar, P.A.; Wang, Z.; Xiao, W.; Wang, Y. Weak-binding molecules are not drugs?-toward a systematic strategy for finding effective weak-binding drugs. Brief. Bioinform. 2017, 18, 321–332. [Google Scholar] [CrossRef]
- Home—Protein—NCBI. Available online: https://www.ncbi.nlm.nih.gov/protein/ (accessed on 10 June 2023).
- Wang, Y.; Xu, B.; Ma, S.; Wang, H.; Shang, L.; Zhu, C.; Ye, S. Discovery of SARS-CoV-2 3CLPro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions. Int. J. Mol. Sci. 2022, 23, 2392. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Qin, B.; Chen, P.; Zhu, K.; Hou, P.; Wojdyla, J.A.; Wang, M.; Cui, S. Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharm. Sin. B. 2021, 11, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Srivari, C.; Kurapati, S.; Reddy, G.; Mainkar, P. Total syntheses of arenamides A, B and C. Tetrahedron Asymmetry 2014, 25, 348–355. [Google Scholar] [CrossRef]
- Asolkar, R.N.; Freel, K.C.; Jensen, P.R.; Fenical, W.; Kondratyuk, T.P.; Park, E.J.; Pezzuto, J.M. Arenamides A-C, cytotoxic NFkappaB inhibitors from the marine actinomycete Salinispora arenicola. J. Nat. Prod. 2009, 72, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.G.; Miller, E.D.; Asolkar, R.N.; Jensen, P.R.; Fenical, W. Arenicolides A-C, 26-membered ring macrolides from the marine actinomycete Salinispora arenicola. J. Org. Chem. 2007, 72, 5025–5034. [Google Scholar] [CrossRef]
- Asolkar, R.N.; Kirkland, T.N.; Jensen, P.R.; Fenical, W. Arenimycin, an antibiotic effective against rifampin- and methicillin-resistant Staphylococcus aureus from the marine actinomycete Salinispora arenicola. J. Antibiot. 2010, 63, 37–39. [Google Scholar] [CrossRef]
- Bose, U.; Hodson, M.P.; Shaw, P.N.; Fuerst, J.A.; Hewavitharana, A.K. Two peptides, cycloaspeptide A and nazumamide A from a sponge associated marine actinobacterium Salinispora sp. Nat. Prod. Commun. 2014, 9, 545–546. [Google Scholar] [CrossRef]
- Lewer, P.; Graupner, P.R.; Hahn, D.R.; Karr, L.L.; Duebelbeis, D.O.; Lira, J.M.; Anzeveno, P.B.; Fields, S.C.; Gilbert, J.R.; Pearce, C. Discovery, synthesis, and insecticidal activity of cycloaspeptide E. J. Nat. Prod. 2006, 69, 1506–1510. [Google Scholar] [CrossRef]
- Banerjee, U.C.; Saxena, B.; Chisti, Y. Biotransformations of rifamycins: Process possibilities. Biotechnol. Adv. 1992, 10, 577–595. [Google Scholar] [CrossRef]
- Bonet, B.; Teufel, R.; Crüsemann, M.; Ziemert, N.; Moore, B.S. Direct capture and heterologous expression of Salinispora natural product genes for the biosynthesis of enterocin. J. Nat. Prod. 2015, 78, 539–542. [Google Scholar] [CrossRef]
- Bose, U.; Hodson, M.P.; Shaw, P.N.; Fuerst, J.A.; Hewavitharana, A.K. Bacterial production of the fungus-derived cholesterol-lowering agent mevinolin. Biomed. Chromatogr. 2014, 28, 1163–1166. [Google Scholar] [CrossRef] [PubMed]
- Bose, U.; Hewavitharana, A.K.; Ng, Y.K.; Shaw, P.N.; Fuerst, J.A.; Hodson, M.P. LC-MS-based metabolomics study of marine bacterial secondary metabolite and antibiotic production in Salinispora arenicola. Mar. Drugs. 2015, 13, 249–266. [Google Scholar] [CrossRef] [PubMed]
- Bose, U.; Hewavitharana, A.K.; Vidgen, M.E.; Ng, Y.K.; Shaw, P.N.; Fuerst, J.A.; Hodson, M.P. Discovering the recondite secondary metabolome spectrum of Salinispora species: A study of inter-species diversity. PLoS ONE 2014, 9, e91488. [Google Scholar] [CrossRef] [PubMed]
- Bose, U.; Ortori, C.A.; Sarmad, S.; Barrett, D.A.; Hewavitharana, A.K.; Hodson, M.P.; Fuerst, J.A.; Shaw, P.N. Production of N-acyl homoserine lactones by the sponge-associated marine actinobacteria Salinispora arenicola and Salinispora pacifica. FEMS Microbiol. Lett. 2017, 364, fnx002. [Google Scholar] [CrossRef][Green Version]
- Buchanan, G.O.; Williams, P.G.; Feling, R.H.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Sporolides A and B: Structurally unprecedented halogenated macrolides from the marine actinomycete Salinispora tropica. Org. Lett. 2005, 7, 2731–2734. [Google Scholar] [CrossRef] [PubMed]
- da Silva, A.B.; Pinto, F.C.L.; Silveira, E.R.; Costa-Lotufo, L.V.; Costa, W.S.; Ayala, A.P.; Canuto, K.M.; Barros, A.B.; Araújo, A.J.; Marinho Filho, J.D.B.; et al. 4-Hydroxy-pyran-2-one and 3-hydroxy-N-methyl-2-oxindole derivatives of Salinispora arenicola from Brazilian marine sediments. Fitoterapia 2019, 138, 104357. [Google Scholar] [CrossRef]
- da Silva, A.B.; Silveira, E.R.; Wilke, D.V.; Ferreira, E.G.; Costa-Lotufo, L.V.; Torres, M.C.M.; Ayala, A.P.; Costa, W.S.; Canuto, K.M.; de Araújo-Nobre, A.R.; et al. Antibacterial Salinaphthoquinones from a Strain of the Bacterium Salinispora arenicola Recovered from the Marine Sediments of St. Peter and St. Paul Archipelago, Brazil. J. Nat. Prod. 2019, 82, 1831–1838. [Google Scholar] [CrossRef]
- Duncan, K.R.; Crüsemann, M.; Lechner, A.; Sarkar, A.; Li, J.; Ziemert, N.; Wang, M.; Bandeira, N.; Moore, B.S.; Dorrestein, P.C.; et al. Molecular networking and pattern-based genome mining improves discovery of biosynthetic gene clusters and their products from Salinispora species. Chem. Biol. 2015, 22, 460–471. [Google Scholar] [CrossRef]
- Eustáquio, A.S.; Nam, S.J.; Penn, K.; Lechner, A.; Wilson, M.C.; Fenical, W.; Jensen, P.R.; Moore, B.S. The discovery of Salinosporamide K from the marine bacterium “Salinispora pacifica” by genome mining gives insight into pathway evolution. ChemBioChem 2011, 12, 61–64. [Google Scholar] [CrossRef]
- Farrell, D.J.; Putnam, S.D.; Biedenbach, D.J.; Moro, L.; Bozzella, R.; Celasco, G.; Jones, R.N. In vitro activity and single-step mutational analysis of rifamycin SV tested against enteropathogens associated with traveler’s diarrhea and Clostridium difficile. Antimicrob. Agents Chemother. 2011, 55, 992–996. [Google Scholar] [CrossRef]
- Barbie, P.; Kazmaier, U. Total Synthesis of Cyclomarin A, a Marine Cycloheptapeptide with Anti-Tuberculosis and Anti-Malaria Activity. Org. Lett. 2016, 18, 204–207. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.J.; Yao, Z.J. Total synthesis of cyclomarin C. Org. Lett. 2004, 6, 2721–2724. [Google Scholar] [CrossRef] [PubMed]
- Schultz, A.W.; Oh, D.C.; Carney, J.R.; Williamson, R.T.; Udwary, D.W.; Jensen, P.R.; Gould, S.J.; Fenical, W.; Moore, B.S. Biosynthesis and structures of cyclomarins and cyclomarazines, prenylated cyclic peptides of marine actinobacterial origin. J. Am. Chem. Soc. 2008, 130, 4507–4516. [Google Scholar] [CrossRef] [PubMed]
- Greunke, C.; Glöckle, A.; Antosch, J.; Gulder, T.A. Biocatalytic Total Synthesis of Ikarugamycin. Angew. Chem. Int. Ed. Engl. 2017, 56, 4351–4355. [Google Scholar] [CrossRef] [PubMed]
- Jomon, K.; Kuroda, Y.; Ajisaka, M.; Sakai, H. A new antibiotic, ikarugamycin. J. Antibiot. 1972, 25, 271–280. [Google Scholar] [CrossRef]
- Xu, J.; Mahmud, T.; Floss, H.G. Isolation and characterization of 27-O-demethylrifamycin SV methyltransferase provides new insights into the post-PKS modification steps during the biosynthesis of the antitubercular drug rifamycin B by Amycolatopsis mediterranei S699. Arch. Biochem. Biophys. 2003, 411, 277–288. [Google Scholar] [CrossRef]
- Laumer, J.M.; Kim, D.D.; Beak, P. Enantioselective synthesis of 2-substituted 2-phenylethylamines by lithiation-substitution sequences: Synthetic development and mechanistic pathway. J. Org. Chem. 2002, 67, 6797–6804. [Google Scholar] [CrossRef]
- Chen, X.; Chen, J.; Yan, Y.; Chen, S.; Xu, X.; Zhang, H.; Wang, H. Quorum sensing inhibitors from marine bacteria Oceanobacillus sp. XC22919. Nat. Prod. Res. 2019, 33, 1819–1823. [Google Scholar] [CrossRef]
- Liu, J.; Brabander, J.K. A concise total synthesis of saliniketal B. J. Am. Chem. Soc. 2009, 131, 12562–12563, Erratum in J. Am. Chem. Soc. 2010, 132, 8223. [Google Scholar] [CrossRef]
- Williams, P.G.; Asolkar, R.N.; Kondratyuk, T.; Pezzuto, J.M.; Jensen, P.R.; Fenical, W. Saliniketals A and B, bicyclic polyketides from the marine actinomycete Salinispora arenicola. J. Nat. Prod. 2007, 70, 83–88. [Google Scholar] [CrossRef]
- Paterson, I.; Razzak, M.; Anderson, E.A. Total synthesis of (-)-saliniketals A and B. Org. Lett. 2008, 10, 3295–3298. [Google Scholar] [CrossRef] [PubMed]
- Yadav, J.S.; Hossain, S.S.; Madhu, M.; Mohapatra, D.K. Formal total synthesis of (-)-saliniketals. J. Org. Chem. 2009, 74, 8822–8825. [Google Scholar] [CrossRef] [PubMed]
- Schlawis, C.; Harig, T.; Ehlers, S.; Guillen-Matus, D.G.; Creamer, K.E.; Jensen, P.R.; Schulz, S. Extending the Salinilactone Family. Chembiochem. 2020, 21, 1629–1632. [Google Scholar] [CrossRef]
- Murphy, B.T.; Narender, T.; Kauffman, C.A.; Woolery, M.; Jensen, P.R.; Fenical, W. Saliniquinones A-F, New Members of the Highly Cytotoxic Anthraquinone-γ-Pyrones from the Marine Actinomycete Salinispora arenicola. Aust. J. Chem. 2010, 63. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, S.; Adachi, K.; Matsuo, Y.; Nukina, M.; Shizuri, Y. Salinisporamycin, a novel metabolite from Salinispora arenicola. J. Antibiot. 2009, 62, 519–526, Erratum in J. Antibiot. 2009, 62, 537. [Google Scholar] [CrossRef]
- Chen, M.; Roush, W.R. Crotylboron-based synthesis of the polypropionate units of chaxamycins A/D, salinisporamycin, and rifamycin S. J. Org. Chem. 2013, 78, 3–8. [Google Scholar] [CrossRef]
- Villadsen, N.L.; Jacobsen, K.M.; Keiding, U.B.; Weibel, E.T.; Christiansen, B.; Vosegaard, T.; Bjerring, M.; Jensen, F.; Johannsen, M.; Tørring, T.; et al. Synthesis of ent-BE-43547A1 reveals a potent hypoxia-selective anticancer agent and uncovers the biosynthetic origin of the APD-CLD natural products. Nat. Chem. 2017, 9, 264–272. [Google Scholar] [CrossRef]
- Xu, M.; Hillwig, M.L.; Lane, A.L.; Tiernan, M.S.; Moore, B.S.; Peters, R.J. Characterization of an orphan diterpenoid biosynthetic operon from Salinispora arenicola. J. Nat. Prod. 2014, 77, 2144–2147. [Google Scholar] [CrossRef]
- Schlawis, C.; Kern, S.; Kudo, Y.; Grunenberg, J.; Moore, B.S.; Schulz, S. Structural Elucidation of Trace Components Combining GC/MS, GC/IR, DFT-Calculation and Synthesis-Salinilactones, Unprecedented Bicyclic Lactones from Salinispora Bacteria. Angew. Chem. Int. Ed. Engl. 2018, 57, 14921–14925. [Google Scholar] [CrossRef]
- Bruns, H.; Crüsemann, M.; Letzel, A.C.; Alanjary, M.; McInerney, J.O.; Jensen, P.R.; Schulz, S.; Moore, B.S.; Ziemert, N. Function-related replacement of bacterial siderophore pathways. ISME J. 2018, 12, 320–329. [Google Scholar] [CrossRef]
- Richter, T.K.; Hughes, C.C.; Moore, B.S. Sioxanthin, a novel glycosylated carotenoid, reveals an unusual subclustered biosynthetic pathway. Environ. Microbiol. 2015, 17, 2158–2171. [Google Scholar] [CrossRef]
- Lane, A.L.; Nam, S.J.; Fukuda, T.; Yamanaka, K.; Kauffman, C.A.; Jensen, P.R.; Fenical, W.; Moore, B.S. Structures and comparative characterization of biosynthetic gene clusters for cyanosporasides, enediyne-derived natural products from marine actinomycetes. J. Am. Chem. Soc. 2013, 135, 4171–4174. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.C.; Gontang, E.A.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Salinipyrones and pacificanones, mixed-precursor polyketides from the marine actinomycete Salinispora pacifica. J. Nat. Prod. 2008, 71, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Liu, J.; Carrasco, Y.P.; MacMillan, J.B.; Brabander, J.K. Rifamycin biosynthetic congeners: Isolation and total synthesis of rifsaliniketal and total synthesis of salinisporamycin and saliniketals A and B. J. Am. Chem. Soc. 2016, 138, 7130–7142. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.C.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J. Nat. Prod. 2007, 70, 515–520. [Google Scholar] [CrossRef]
- Macherla, V.R.; Mitchell, S.S.; Manam, R.R.; Reed, K.A.; Chao, T.H.; Nicholson, B.; Deyanat-Yazdi, G.; Mai, B.; Jensen, P.R.; Fenical, W.F.; et al. Structure-activity relationship studies of salinosporamide A (NPI-0052), a novel marine derived proteasome inhibitor. J. Med. Chem. 2005, 48, 3684–3687. [Google Scholar] [CrossRef]
- Miyanaga, A.; Janso, J.E.; McDonald, L.; He, M.; Liu, H.; Barbieri, L.; Eustáquio, A.S.; Fielding, E.N.; Carter, G.T.; Jensen, P.R.; et al. Discovery and assembly-line biosynthesis of the lymphostin pyrroloquinoline alkaloid family of mTOR inhibitors in Salinispora bacteria. J. Am. Chem. Soc. 2011, 133, 13311–13313. [Google Scholar] [CrossRef]
- Feling, R.H.; Buchanan, G.O.; Mincer, T.J.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Salinosporamide A: A highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew. Chem. Int. Ed. 2003, 42, 355–357. [Google Scholar] [CrossRef]
- Kudo, Y.; Awakawa, T.; Du, Y.L.; Jordan, P.A.; Creamer, K.E.; Jensen, P.R.; Linington, R.G.; Ryan, K.S.; Moore, B.S. Expansion of Gamma-Butyrolactone Signaling Molecule Biosynthesis to Phosphotriester Natural Products. ACS Chem. Biol. 2020, 15, 3253–3261. [Google Scholar] [CrossRef]
- Reed, K.A.; Manam, R.R.; Mitchell, S.S.; Xu, J.; Teisan, S.; Chao, T.H.; Deyanat-Yazdi, G.; Neuteboom, S.T.; Lam, K.S.; Potts, B.C. Salinosporamides D-J from the marine actinomycete Salinispora tropica, bromosalinosporamide, and thioester derivatives are potent inhibitors of the 20S proteasome. J. Nat. Prod. 2007, 70, 269–276. [Google Scholar] [CrossRef]
- Manam, R.R.; Macherla, V.R.; Tsueng, G.; Dring, C.W.; Weiss, J.; Neuteboom, S.T.; Lam, K.S.; Potts, B.C. Antiprotealide is a natural product. J. Nat. Prod. 2009, 72, 295–297. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.G.; Buchanan, G.O.; Feling, R.H.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. New cytotoxic salinosporamides from the marine Actinomycete Salinispora tropica. J. Org. Chem. 2005, 70, 6196–6203. [Google Scholar] [CrossRef] [PubMed]
- Groenhagen, U.; Leandrini, D.O.; Fielding, E.; Moore, B.S.; Schulz, S. Coupled Biosynthesis of Volatiles and salinosporamide A in Salinispora tropica. Chembiochem 2016, 17, 1978–1985. [Google Scholar] [CrossRef] [PubMed]
- Niewerth, D.; Jansen, G.; Riethoff, L.F.; Meerloo, V.J.; Kale, A.J.; Moore, B.S.; Assaraf, Y.G.; Anderl, J.L.; Zweegman, S.K.; Cloos, J. Antileukemic activity and mechanism of drug resistance to the marine Salinispora tropica proteasome inhibitor salinosporamide A (Marizomib). Mol. Pharmacol. 2014, 86, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Udwary, D.W.; Zeigler, L.; Asolkar, R.N.; Singan, V.; Lapidus, A.; Fenical, W.; Jensen, P.R.; Moore, B.S. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc. Natl. Acad. Sci. USA 2007, 104, 10376–10381. [Google Scholar] [CrossRef] [PubMed]
- Harig, T.; Schlawis, C.; Ziesche, L.; Pohlner, M.; Engelen, B.; Schulz, S. Nitrogen-Containing Volatiles from Marine Salinispora pacifica and Roseobacter-Group Bacteria. J. Nat. Prod. 2017, 80, 3289–3295. [Google Scholar] [CrossRef]
- Woo, C.M.; Beizer, N.E.; Janso, J.E.; Herzon, S.B. Isolation of lomaiviticins C-E, transformation of lomaiviticin C to lomaiviticin A, complete structure elucidation of lomaiviticin A, and structure-activity analyses. J. Am. Chem. Soc. 2012, 134, 15285–15288. [Google Scholar] [CrossRef]
- Oh, D.C.; Williams, P.G.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Cyanosporasides A and B, chloro- and cyano-cyclopenta[a]indene glycosides from the marine actinomycete “Salinispora pacifica”. Org. Lett. 2006, 8, 1021–1024. [Google Scholar] [CrossRef]
- Woo, C.M.; Gholap, S.L.; Herzon, S.B. Insights into lomaiviticin biosynthesis. Isolation and structure elucidation of (-)-homoseongomycin. J. Nat. Prod. 2013, 76, 1238–1241. [Google Scholar] [CrossRef]
- Roberts, A.A.; Schultz, A.W.; Kersten, R.D.; Dorrestein, P.C.; Moore, B.S. Iron acquisition in the marine actinomycete genus Salinispora is controlled by the desferrioxamine family of siderophores. FEMS Microbiol. Lett. 2012, 335, 95–103. [Google Scholar] [CrossRef]
- Castro, F.G.; Hahn, D.; Reimer, D.; Hughes, C.C. Thiol Probes to Detect Electrophilic Natural Products Based on Their Mechanism of Action. ACS Chem. Biol. 2016, 11, 2328–2336. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, S.; Kim, M.; Lee, C.; Yang, I.; Nam, S.J. Bioactive natural products from the genus salinospora: A review. Arch. Pharm. Res. 2020, 43, 1230–1258. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016, 44, D1202. [Google Scholar] [CrossRef] [PubMed]
- Pence, H.E.; Williams, A. ChemSpider: An Online Chemical Information Resource. J. Chem. Educ. 2010, 87, 1123–1124. [Google Scholar] [CrossRef]
- BIOVIA DS, Discovery, Studio. Available online: https://discover.3ds.com/discovery-studio-visualizer-download (accessed on 15 June 2023).
- The PyMOL Molecular Graphics System, Version 2.5.5, Schrödinger, LLC. Available online: https://pymol.org/ (accessed on 30 June 2023).
- Avogadro: An Open-Source Molecular Builder and Visualization Tool. Version 1.95. Available online: http://avogadro.cc/ (accessed on 10 June 2023).
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Kayikci, M.; Venkatakrishnan, A.J.; Scott-Brown, J.; Ravarani, C.N.J.; Flock, T.; Babu, M.M. Visualization and analysis of non-covalent contacts using the Protein Contacts Atlas. Nat. Struct. Mol. Biol. 2018, 25, 185–194. [Google Scholar] [CrossRef]
- Kuriata, A.; Gierut, A.M.; Oleniecki, T.; Ciemny, M.P.; Kolinski, A.; Kurcinski, M.; Kmiecik, S. CABS-flex 2.0: A web server for fast simulations of flexibility of protein structures. Nucleic Acids Res. 2018, 46, W338–W343. [Google Scholar] [CrossRef]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef]
- Borba, J.; Alves, V.; Overdahl, K.; Silva, A.; Hall, S.; Overdahl, E.; Braga, R.; Kleinstreuer, N.; Strickland, J.; Allen, D.; et al. STopTox: An In-Silico Alternative to Animal Testing for Acute Systemic and TOPical TOXicity. Environ. Health Perspect. 2022, 130, 27012. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 71, 42717. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.H.; Abraham, M.H.; Le, J.; Hersey, A.; Luscombe, C.N.; Beck, G.; Sherborne, B.; Cooper, I. Rate-limited steps of human oral absorption and QSAR studies. Pharm. Res. 2002, 19, 1446–1457. [Google Scholar] [CrossRef] [PubMed]
- Husain, A.; Ahmad, A.; Khan, S.A.; Asif, M.; Bhutani, R.; Al-Abbasi, F.A. Synthesis, molecular properties, toxicity and biological evaluation of some new substituted imidazolidine derivatives in search of potent anti-inflammatory agents. Saudi Pharm. J. 2016, 24, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Bhowmik, D.; Nandi, R.; Jagadeesan, R.; Kumar, N.; Prakash, A.; Kumar, D. Identification of potential inhibitors against SARS-CoV-2 by targeting proteins responsible for envelope formation and virion assembly using docking based virtual screening, and pharmacokinetics approaches. Infect. Genet. Evol. 2020, 84, 104451. [Google Scholar] [CrossRef]
- Lagunin, A.; Stepanchikova, A.; Filimonov, D.; Poroikov, V. PASS: Prediction of activity spectra for biologically active substances. Bioinformatics 2000, 16, 747–748. [Google Scholar] [CrossRef]
- Moustaqil, M.; Ollivier, E.; Chiu, H.P.; Van Tol, S.; Rudolffi-Soto, P.; Stevens, C.; Bhumkar, A.; Hunter, D.J.B.; Freiberg, A.N.; Jacques, D.; et al. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): Implications for disease presentation across species. Emerg. Microbes Infect. 2021, 10, 178–195. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, H.; Lin, Q.; Lyu, J.; Lu, L.; Chen, H.; Zhang, X.; Zhang, Y.; Chen, K. Progress on SARS-CoV-2 3CLpro Inhibitors: Inspiration from SARS-CoV 3CLpro Peptidomimetics and Small-Molecule Anti-Inflammatory Compounds. Drug Des. Devel Ther. 2022, 16, 1067–1082. [Google Scholar] [CrossRef]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.C.; Fadl, S.; Villanueva, A.J.; Rabeh, W.M. Catalytic Dyad Residues His41 and Cys145 Impact the Catalytic Activity and Overall Conformational Fold of the Main SARS-CoV-2 Protease 3-Chymotrypsin-Like Protease. Front. Chem. 2021, 9, 692168. [Google Scholar] [CrossRef] [PubMed]
- Calistri, A.; Munegato, D.; Carli, I.; Parolin, C.; Palù, G. The ubiquitin-conjugating system: Multiple roles in viral replication and infection. Cells 2014, 3, 386–417. [Google Scholar] [CrossRef]
- Mielech, A.M.; Kilianski, A.; Baez-Santos, Y.M.; Mesecar, A.D.; Baker, S.C. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology 2014, 450–451, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Lindner, H.A.; Lytvyn, V.; Qi, H.; Lachance, P.; Ziomek, E.; Ménard, R. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Arch. Biochem. Biophys. 2007, 466, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Barretto, N.; Jukneliene, D.; Ratia, K.; Chen, Z.; Mesecar, A.D.; Baker, S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005, 79, 15189–15198. [Google Scholar] [CrossRef]
- Gao, H.; Dai, R.; Su, R. Computer-aided drug design for the papain-like protease (PLpro) inhibitors against SARS-CoV-2. Biomed. Pharmacother. 2023, 159, 114247. [Google Scholar] [CrossRef]
- Osipiuk, J.; Azizi, S.A.; Dvorkin, S.; Endres, M.; Jedrzejczak, R.; Jones, K.A.; Kang, S.; Kathayat, R.S.; Kim, Y.; Lisnyak, V.G.; et al. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 2021, 12, 743. [Google Scholar] [CrossRef]
- Muhammad, M.; Habib, I.Y.; Yunusa, A.; Mikail, T.A.; ALhassan, A.J.; Alkhatib, A.J.; Sule, H.; Ismail, S.Y.; Liu, D. Identification of potential SARS-CoV-2 papain-like protease inhibitors with the ability to interact with the catalytic triad. AIMS Biophys. 2023, 10, 50–66. [Google Scholar] [CrossRef]
- Pokharkar, O.; Lakshmanan, H.; Zyryanov, G.; Tsurkan, M. In Silico Evaluation of Antifungal Compounds from Marine Sponges against COVID-19-Associated Mucormycosis. Mar. Drugs 2022, 20, 215. [Google Scholar] [CrossRef]
- Worldometer Coronavirus Statistics. Available online: https://www.worldometers.info/coronavirus/ (accessed on 31 January 2023).
- Sharapov, A.D.; Fatykhov, R.F.; Khalymbadzha, I.A.; Zyryanov, G.V.; Chupakhin, O.N.; Tsurkan, M.V. Plant Coumarins with Anti-HIV Activity: Isolation and Mechanisms of Action. Int. J. Mol. Sci. 2023, 24, 2839. [Google Scholar] [CrossRef] [PubMed]
- Satish, L.; Santra, S.; Tsurkan, M.V.; Werner, C.; Jana, M.; Sahoo, H. Conformational changes of GDNF-derived peptide induced by heparin, heparan sulfate, and sulfated hyaluronic acid-Analysis by circular dichroism spectroscopy and molecular dynamics simulation. Int. J. Biol. Macromol. 2021, 182, 2144–2150. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).