Poly(3-Hydroxybutyrate) Biosynthesis from [U-13C6]D-Glucose by Ralstonia eutropha NCIMB 11599 and Recombinant Escherichia coli
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Plasmid
2.2. Culture Conditions
2.3. Quantitative Analysis of P(3HB)
2.4. Polymer Extraction
2.5. Gel Permeation Chromatography
2.6. Nuclear Magnetic Resonance
2.7. Gas Chromatography-Mass Spectrometry
3. Results
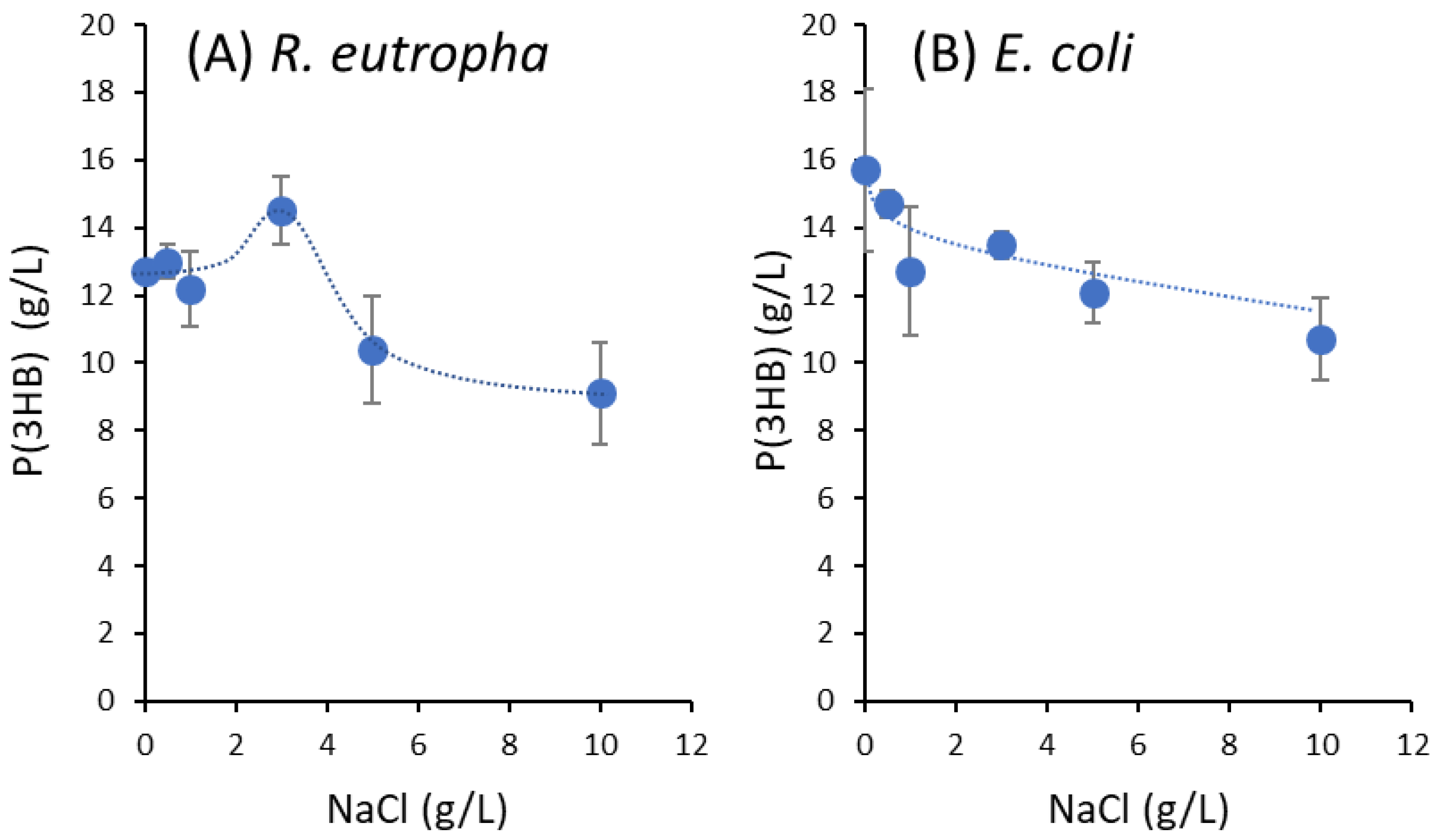
3.1. Effect of NaCl Concentration on P(3HB) Synthesis from Natural Glucose
3.2. P(3HB) Synthesis from 13C-Glucose by R. eutropha NCIMB 11599
3.3. P(3HB) Synthesis from 13C-Glucose by Recombinant E. coli JM109
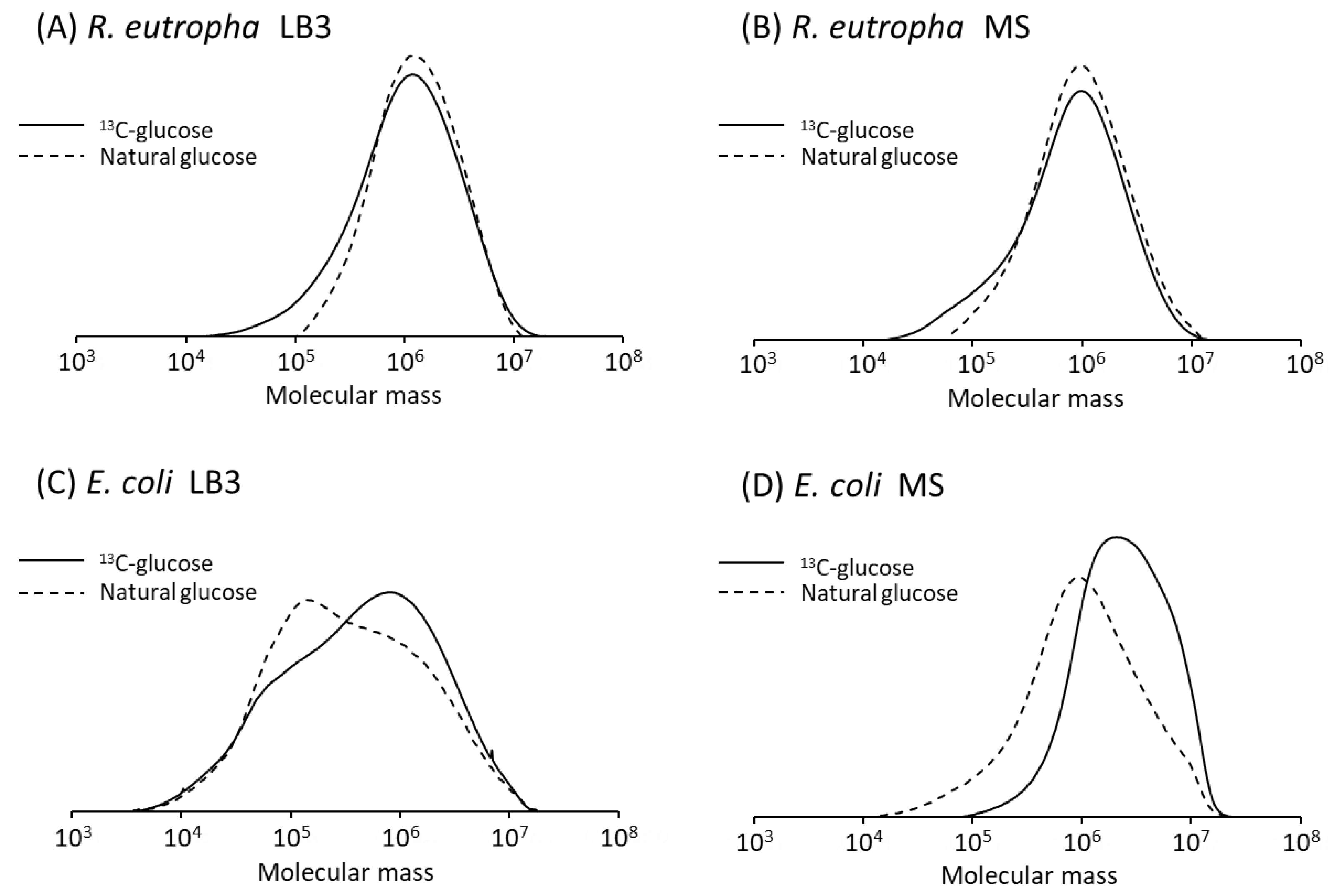
3.4. Molecular Weight of P(3HB)
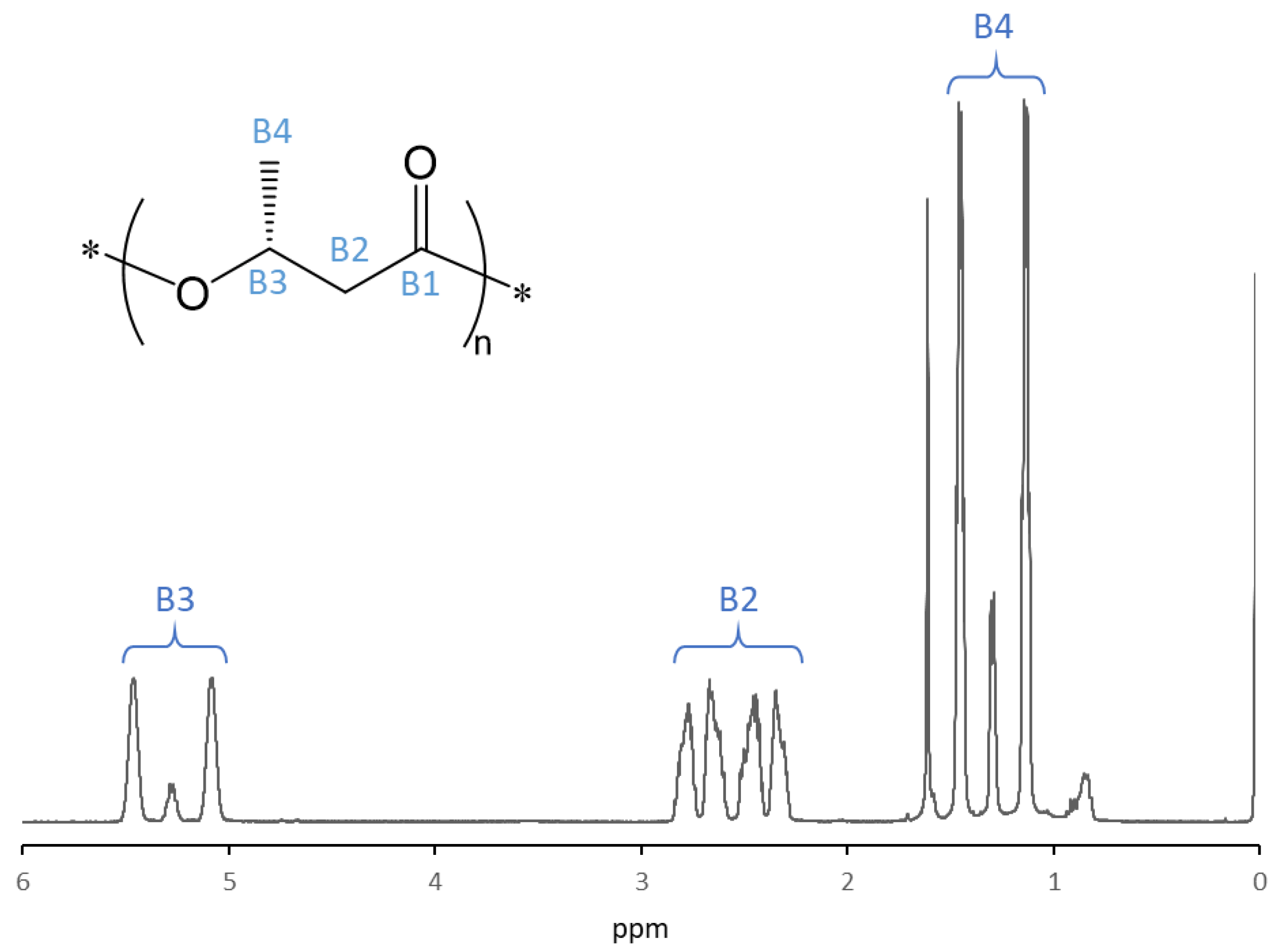
3.5. 1H NMR Analysis of P(3HB)
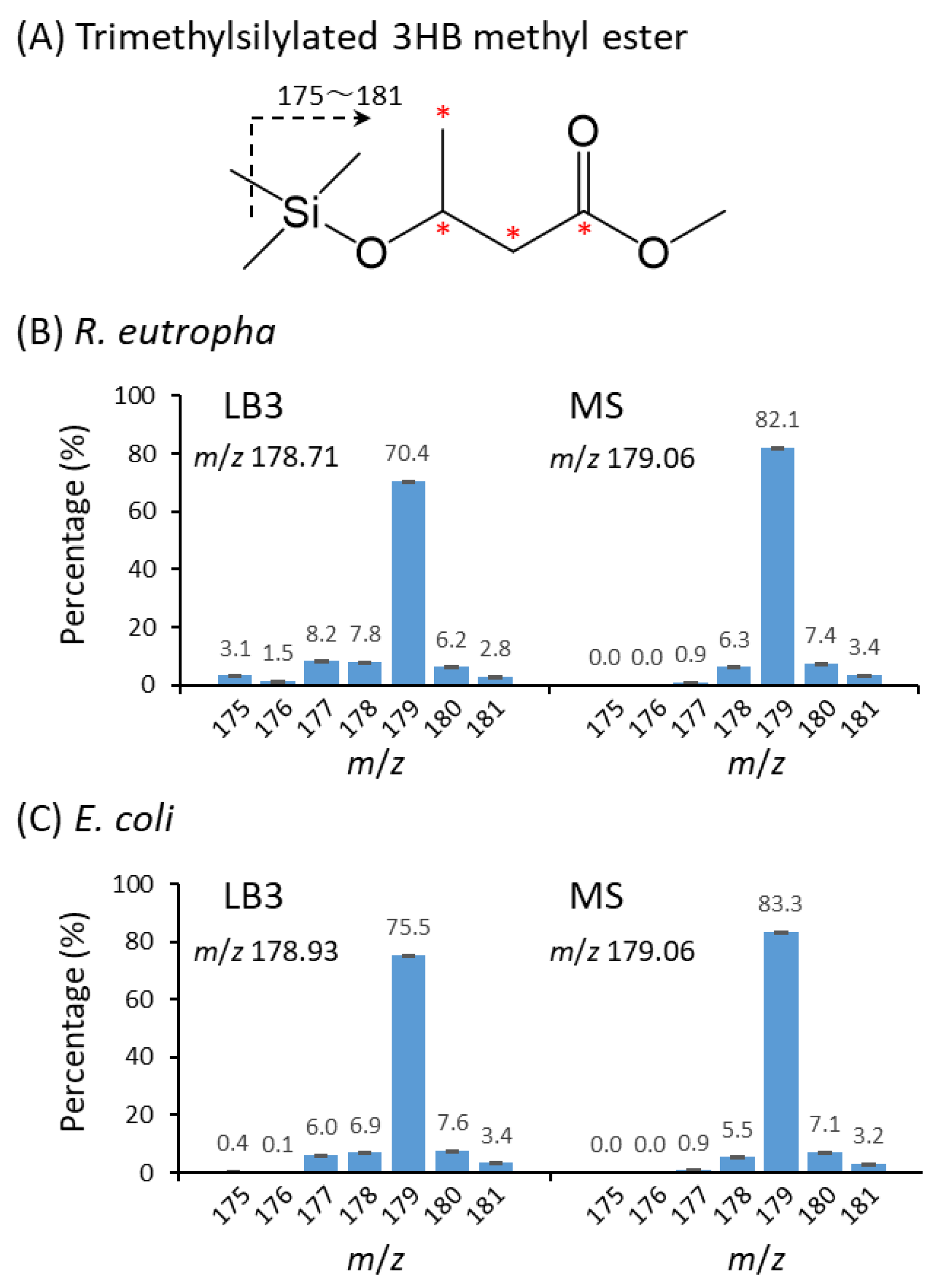
3.6. GC-MS Analysis of P(3HB)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sudesh, K.; Abe, H.; Doi, Y. Synthesis, structure and properties of polyhydroxyalkanoates: Biological polyesters. Prog. Polym. Sci. 2000, 25, 1503–1555. [Google Scholar] [CrossRef]
- Chen, G.Q.; Hajnal, I.; Wu, H.; Lv, L.; Ye, J. Engineering biosynthesis mechanisms for diversifying polyhydroxyalkanoates. Trends Biotechnol. 2015, 33, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Koller, M. Microbial polyHydroxyAlkanoate (PHA) biopolymers—Intrinsically natural. Bioengineering 2023, 10, 855. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Tachibana, Y.; Kasuya, K.I. Biodegradability of poly(3-hydroxyalkanoate) and poly(ε-caprolactone) via biological carbon cycles in marine environments. Polym. J. 2021, 53, 47–66. [Google Scholar] [CrossRef]
- Narancic, T.; Verstichel, V.; Chaganti, S.R.; Morales-Gamez, L.; Kenny, S.T.; De Wilde, B.; Padamati, R.B.; O’Connor, K.E. Biodegradable plastic blends create new possibilities for end-of-life management of plastics but they are not a panacea for plastic pollution. Environ. Sci. Technol. 2018, 52, 10441–10452. [Google Scholar] [CrossRef]
- Goudriaan, M.; Morales, V.H.; van der Meer, M.T.J.; Mets, A.; Ndhlovu, R.T.; van Heerwaarden, J.; Simon, S.; Heuer, V.B.; Hinrichs, K.U.; Niemann, H. A stable isotope assay with 13C-labeled polyethylene to investigate plastic mineralization mediated by Rhodococcus ruber. Mar. Pollut. Bull. 2023, 186, 114369. [Google Scholar] [CrossRef]
- Zumstein, M.T.; Schintlmeister, A.; Nelson, T.F.; Baumgartner, R.; Woebken, D.; Wagner, M.; Kohler, H.E.; McNeill, K.; Sander, M. Biodegradation of synthetic polymers in soils: Tracking carbon into CO2 and microbial biomass. Sci. Adv. 2018, 4, eaas9024. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Shiraki, M.; Tatsumichi, M. Study on development of the evaluation method of biodegradable plastics by detecting 13CO2. Kobunshi Ronbunshu 1993, 50, 781–783. [Google Scholar] [CrossRef][Green Version]
- Yang, Y.; Yang, J.; Wu, W.M.; Zhao, J.; Song, Y.; Gao, L.; Yang, R.; Jiang, L. Biodegradation and mineralization of polystyrene by plastic-eating mealworms: Part 1. Chemical and physical characterization and isotopic tests. Environ. Sci. Technol. 2015, 49, 12080–12086. [Google Scholar] [CrossRef]
- Vaksmaa, A.; Polerecky, L.; Dombrowski, N.; Kienhuis, M.V.; Posthuma, I.; Gerritse, J.; Boekhout, T.; Niemann, H. Polyethylene degradation and assimilation by the marine yeast Rhodotorula mucilaginosa. ISME Commun. 2023, 3, 68. [Google Scholar] [CrossRef]
- Radajewski, S.; Ineson, P.; Parekh, N.R.; Murrell, J.C. Stable-isotope probing as a tool in microbial ecology. Nature 2000, 403, 646–649. [Google Scholar] [CrossRef] [PubMed]
- Dumont, M.G.; Murrell, J.C. Stable isotope probing—Linking microbial identity to function. Nat. Rev. Microbiol. 2005, 3, 499–504. [Google Scholar] [CrossRef]
- Neufeld, J.D.; Dumont, M.G.; Vohra, J.; Murrell, J.C. Methodological considerations for the use of stable isotope probing in microbial ecology. Microb. Ecol. 2007, 53, 435–442. [Google Scholar] [CrossRef]
- Orita, I.; Iwazawa, R.; Nakamura, S.; Fukui, T. Identification of mutation points in Cupriavidus necator NCIMB 11599 and genetic reconstitution of glucose-utilization ability in wild strain H16 for polyhydroxyalkanoate production. J. Biosci. Bioeng. 2012, 113, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Agus, J.; Kahar, P.; Abe, H.; Doi, Y.; Tsuge, T. Molecular weight characterization of poly[(R)-3-hydroxybutyrate] synthesized by genetically engineered strains of Escherichia coli. Polym. Degrad. Stabil. 2006, 91, 1138–1146. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Lee, E.Y.; Choi, C.Y. Gas chromatography—Mass spectrometric analysis and its application to a screening procedure for novel bacterial polyhydroxyalkanoic acids containing long chain saturated and unsaturated monomers. J. Ferment. Bioeng. 1995, 80, 408–414. [Google Scholar] [CrossRef]
- Ashby, R.D.; Solaiman, D.K.Y.; Foglia, T.A. The synthesis of short-and medium-chain-length poly(hydroxyalkanoate) mixtures from glucose- or alkanoic acid-grown Pseudomonas oleovorans. J. Ind. Microbiol. Biotechnol. 2002, 28, 147–153. [Google Scholar] [CrossRef]
- Doi, Y.; Kawaguchi, Y.; Nakamura, Y.; Kunioka, M. Nuclear magnetic resonance studies of poly(3-hydroxybutyrate) and polyphosphate metabolism in Alcaligenes eutrophus. Appl. Environ. Microbiol. 1989, 55, 2932–2938. [Google Scholar] [CrossRef]
- Saegusa, H.; Shiraki, M.; Kanai, C.; Saito, T. Cloning of an intracellular poly[D(−)-3-hydroxybutyrate] depolymerase gene from Ralstonia eutropha H16 and characterization of the gene product. J. Bacteriol. 2001, 183, 94–100. [Google Scholar] [CrossRef]
- Tsuge, T. Fundamental factors determining the molecular weight of polyhydroxyalkanoate during biosynthesis. Polym. J. 2016, 48, 1051–1057. [Google Scholar] [CrossRef]
- Huijberts, G.N.; de Rijk, T.C.; de Waard, P.; Eggink, G. 13C nuclear magnetic resonance studies of Pseudomonas putida fatty acid metabolic routes involved in poly(3-hydroxyalkanoate) synthesis. J. Bacteriol. 1994, 176, 1661–1666. [Google Scholar] [CrossRef][Green Version]
- Fukui, T.; Kato, M.; Matsusaki, H.; Iwata, T.; Doi, Y. Morphological and 13C-nuclear magnetic resonance studies for polyhydroxyalkanoate biosynthesis in Pseudomonas sp. 61-3. FEMS. Microbiol. Lett. 1998, 164, 219–225. [Google Scholar] [CrossRef]
- Saika, A.; Ushimaru, K.; Mizuno, S.; Tsuge, T. Genome-based analysis and gene dosage studies provide new insight into 3-hydroxy-4-methylvalerate biosynthesis in Ralstonia eutropha. J. Bacteriol. 2015, 197, 1350–1359. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shah, D.T.; Berger, P.A.; Tran, M.; Asrar, J.; Madden, L.A.; Anderson, A.J. Synthesis of poly(3-hydroxybutyrate) by Ralstonia eutropha in the presence of 13C-labeled ethylene glycol. Macromolecules 2000, 33, 6624–6626. [Google Scholar] [CrossRef]
- Liu, C.; Nangle, S.N.; Colón, B.C.; Silver, P.A.; Nocera, D.G. 13C-Labeling the carbon-fixation pathway of a highly efficient artificial photosynthetic system. Faraday Discuss. 2017, 198, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Ludevese-Pascual, G.; Ahmed, F.; De Troch, M.; Amar, E.; Laranja, J.L.; Bode, S.; Boeckx, P.; Bossier, P.; De Schryver, P. Determination of poly-β-hydroxybutyrate assimilation by postlarval whiteleg shrimp, Litopenaeus vannamei using stable 13C isotope tracing. J. World Aquac. Soc. 2021, 52, 184–194. [Google Scholar] [CrossRef]
- Lütte, S.; Pohlmann, A.; Zaychikov, E.; Schwartz, E.; Becher, J.R.; Heumann, H.; Friedrich, B. Autotrophic production of stable-isotope-labeled arginine in Ralstonia eutropha strain H16. Appl. Environ. Microbiol. 2012, 78, 7884–7890. [Google Scholar] [CrossRef]
- Fraser-Smith, E.C.; Austin, M.A.; Reed, L.L. Utilization of amino acids as a source of nitrogen by Hydrogenomonas eutropha. J. Bacteriol. 1969, 97, 457–459. [Google Scholar] [CrossRef] [PubMed]
- Iskandaryan, M.; Blbulyan, S.; Sahakyan, M.; Vassilian, A.; Trchounian, K.; Poladyan, A. L-amino acids affect the hydrogenase activity and growth of Ralstonia eutropha H16. AMB Express 2023, 13, 33. [Google Scholar] [CrossRef]
- Sezonov, G.; Joseleau-Petit, D.; d’Ari, R. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 2007, 189, 8746–8749. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Nakaya, A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002, 30, 42–46. [Google Scholar] [CrossRef]
- Mizuno, S.; Sakurai, T.; Nabasama, M.; Kawakami, K.; Hiroe, A.; Taguchi, S.; Tsuge, T. The influence of medium composition on the microbial secretory production of hydroxyalkanoate oligomers. J. Gen. Appl. Microbiol. 2021, 67, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, T.; Mizuno, S.; Miyahara, Y.; Hiroe, A.; Taguchi, S.; Tsuge, T. Optimization of culture conditions for secretory production of 3-hydroxybutyrate oligomers using recombinant Escherichia coli. Front. Bioeng. Biotechnol. 2022, 10, 829134. [Google Scholar] [CrossRef] [PubMed]
- Utsunomia, C.; Matsumoto, K.I.; Taguchi, S. Microbial secretion of D-lactate-based oligomers. ACS Sustain. Chem. Eng. 2017, 5, 2360–2367. [Google Scholar] [CrossRef]
- Nduko, J.M.; Taguchi, S. Microbial production of biodegradable lactate-based polymers and oligomeric building blocks from renewable and waste resources. Front. Bioeng. Biotechnol. 2021, 8, 618077. [Google Scholar] [CrossRef] [PubMed]
- Pohlmann, A.; Fricke, W.F.; Reinecke, F.; Kusian, B.; Liesegang, H.; Cramm, R.; Eitinger, T.; Ewering, C.; Pötter, M.; Schwartz, E.; et al. Genome sequence of the bioplastic-producing “Knallgas” bacterium Ralstonia eutropha H16. Nat. Biotechnol. 2006, 24, 1257–1262. [Google Scholar] [CrossRef]
- Panich, J.; Fong, B.; Singer, S.W. Metabolic engineering of Cupriavidus necator H16 for sustainable biofuels from CO2. Trends Biotechnol. 2021, 39, 412–424. [Google Scholar] [CrossRef]
- Heinrich, D.; Raberg, M.; Steinbüchel, A. Studies on the aerobic utilization of synthesis gas (syngas) by wild type and recombinant strains of Ralstonia eutropha H16. Microb. Biotechnol. 2018, 11, 647–656. [Google Scholar] [CrossRef]
- Brigham, C. Perspectives for the biotechnological production of biofuels from CO2 and H2 using Ralstonia eutropha and other ‘Knallgas’ bacteria. Appl. Microbiol. Biotechnol. 2019, 103, 2113–2120. [Google Scholar] [CrossRef]
- Garcia-Gonzalez, L.; Mozumder, M.S.I.; Dubreuil, M.; Volcke, E.I.; De Wever, H. Sustainable autotrophic production of polyhydroxybutyrate (PHB) from CO2 using a two-stage cultivation system. Catal. Today 2015, 257, 237–245. [Google Scholar] [CrossRef]
- Miyahara, Y.; Wang, C.T.; Ishii-Hyakutake, M.; Tsuge, T. Continuous supply of non-combustible gas mixture for safe autotrophic culture to produce polyhydroxyalkanoate by hydrogen-oxidizing bacteria. Bioengineering 2022, 9, 586. [Google Scholar] [CrossRef]
- Shimizu, R.; Dempo, Y.; Nakayama, Y.; Nakamura, S.; Bamba, T.; Fukusaki, E.; Fukui, T. New insight into the role of the Calvin cycle: Reutilization of CO2 emitted through sugar degradation. Sci. Rep. 2015, 5, 11617. [Google Scholar] [CrossRef]
- Thorbecke, R.; Yamamoto, M.; Miyahara, Y.; Oota, M.; Mizuno, S.; Tsuge, T. The gene dosage effect of carbonic anhydrase on the biosynthesis of poly(3-hydroxybutyrate) under autotrophic and mixotrophic culture conditions. Polym. J. 2021, 53, 209–213. [Google Scholar] [CrossRef]
- Schwartz, E.; Voigt, B.; Zühlke, D.; Pohlmann, A.; Lenz, O.; Albrecht, D.; Schwarze, A.; Kohlmann, Y.; Krause, C.; Hecker, M.; et al. A proteomic view of the facultatively chemolithoautotrophic lifestyle of Ralstonia eutropha H16. Proteomics 2009, 9, 5132–5142. [Google Scholar] [CrossRef] [PubMed]
- Strittmatter, C.S.; Eggers, J.; Biesgen, V.; Pauels, I.; Becker, F.; Steinbüchel, A. The reliance of glycerol utilization by Cupriavidus necator on CO2 fixation and improved glycerol catabolism. Appl. Microbiol. Biotechnol. 2022, 106, 2541–2555. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, C.G.; Friedrich, B.; Bowien, B. Formation of enzymes of autotrophic metabolism during heterotrophic growth of Alcaligenes eutrophus. Microbiology 1981, 122, 69–78. [Google Scholar] [CrossRef]

| Strain | Culture Medium | Type of Glucose | Initial Glucose (g/L) | Dry Cell wt. (g/L) | P(3HB) Content (wt%) | P(3HB) (g/L) | Glucose Consumption (g/L) | P(3HB) Yield (g-polymer/ g-glc) |
|---|---|---|---|---|---|---|---|---|
| R. eutropha | LB3 | Natural | 40 | 16.3 ± 0.8 | 82 ± 6 | 12.6 ± 3.5 | 38.6 ± 0.3 | 0.38 ± 0.03 |
| LB3 | [U-13C6] | 40 | 16.2 ± 0.6 | 64 ± 1 | 10.0 ± 0.3 | 38.2 ± 0.0 | 0.26 ± 0.01 | |
| MS | Natural | 20 | 5.4 ± 1.7 | 84 ± 2 | 4.5 ± 0.1 | 15.8 ± 0.1 | 0.29 ± 0.00 | |
| MS | [U-13C6] | 20 | 5.7 ± 0.1 | 82 ± 0 | 4.7 ± 0.1 | 16.7 ± 0.1 | 0.28 ± 0.00 | |
| E. coli | LB3 | Natural | 40 | 15.7 ± 0.2 | 83 ± 4 | 13.5 ± 0.4 | 38.2 ± 0.2 | 0.35 ± 0.01 |
| LB3 | [U-13C6] | 40 | 16.5 ± 0.1 | 63 ± 4 | 10.4 ± 0.7 | 39.9 ± 0.1 | 0.26 ± 0.01 | |
| MS | Natural | 20 | 2.2 ± 0.0 | 53 ± 1 | 1.2 ± 0.0 | 12.9 ± 0.1 | 0.09 ± 0.00 | |
| MS | [U-13C6] | 20 | 2.6 ± 0.1 | 57 ± 6 | 1.4 ± 0.1 | 13.1 ± 0.3 | 0.11 ± 0.01 |
| Strain | Culture Medium | Type of Glucose | Molecular Weight a | 13C-Labeling Ratio (atom%) | ||||
|---|---|---|---|---|---|---|---|---|
| Mw (×106) | Mw/Mn | by 1H-NMR b | by GC-MS c | |||||
| B3 | B4 | Average | ||||||
| R. eutropha | LB3 | Natural | 1.87 | 2.08 | - | - | - | - |
| LB3 | [U-13C6] | 1.77 | 3.41 | 88.8 | 87.2 | 88.0 | 88.8 ± 1.4 | |
| MS | Natural | 1.52 | 2.45 | - | - | - | - | |
| MS | [U-13C6] | 1.36 | 3.37 | 98.1 | 97.1 | 97.6 | 97.6 ± 0.0 | |
| E. coli | LB3 | Natural | 1.04 | 8.67 | - | - | - | - |
| LB3 | [U-13C6] | 1.27 | 9.87 | 93.8 | 94.7 | 94.3 | 94.4 ± 0.3 | |
| MS | Natural | 2.06 | 3.31 | - | - | - | - | |
| MS | [U-13C6] | 3.60 | 2.30 | 100 | 90.1 | 95.1 | 97.6 ± 0.1 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sivashankari, R.M.; Miyahara, Y.; Tsuge, T. Poly(3-Hydroxybutyrate) Biosynthesis from [U-13C6]D-Glucose by Ralstonia eutropha NCIMB 11599 and Recombinant Escherichia coli. Microbiol. Res. 2023, 14, 1894-1906. https://doi.org/10.3390/microbiolres14040129
Sivashankari RM, Miyahara Y, Tsuge T. Poly(3-Hydroxybutyrate) Biosynthesis from [U-13C6]D-Glucose by Ralstonia eutropha NCIMB 11599 and Recombinant Escherichia coli. Microbiology Research. 2023; 14(4):1894-1906. https://doi.org/10.3390/microbiolres14040129
Chicago/Turabian StyleSivashankari, Ramamoorthi M., Yuki Miyahara, and Takeharu Tsuge. 2023. "Poly(3-Hydroxybutyrate) Biosynthesis from [U-13C6]D-Glucose by Ralstonia eutropha NCIMB 11599 and Recombinant Escherichia coli" Microbiology Research 14, no. 4: 1894-1906. https://doi.org/10.3390/microbiolres14040129
APA StyleSivashankari, R. M., Miyahara, Y., & Tsuge, T. (2023). Poly(3-Hydroxybutyrate) Biosynthesis from [U-13C6]D-Glucose by Ralstonia eutropha NCIMB 11599 and Recombinant Escherichia coli. Microbiology Research, 14(4), 1894-1906. https://doi.org/10.3390/microbiolres14040129






