Antibiotic-Resistant Bacteria in Environmental Water Sources from Southern Chile: A Potential Threat to Human Health
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Bacterial Isolation and Preservation
2.3. Identification of GNB
2.4. Antibiotic Susceptibility Testing
2.5. Molecular Detection of Antibiotic-Resistance Genes (ARGs)
3. Results
3.1. Distribution and Prevalence of GNB from Environmental Surface Waters
3.2. Resistance Profile of Environmental GNB
3.3. Colistin and β-Lactam Resistance Genes in Environmental GNB
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Despotovic, M.; De Nies, L.; Busi, S.B.; Wilmes, P. Reservoirs of antimicrobial resistance in the context of One Health. Curr. Opin. Microbiol. 2023, 73, 102291. [Google Scholar] [CrossRef] [PubMed]
- Fursova, N.K.; Kislichkina, A.A.; Khokhlova, O.E. Plasmids Carrying Antimicrobial Resistance Genes in Gram-Negative Bacteria. Microorganisms 2022, 10, 1678. [Google Scholar] [CrossRef] [PubMed]
- WHO, World Health Organization. Thirteenth General Programme of Work. 2019. Available online: www.iniscommunication.com (accessed on 5 June 2023).
- Von Wintersdorff, C.J.; Penders, J.; Van Niekerk, J.M.; Mills, N.D.; Majumder, S.; Van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Thomsen, L.E.; Olsen, J.E. Antimicrobial-induced horizontal transfer of antimicrobial resistance genes in bacteria: A mini-review. J. Antimicrob. Chemother. 2022, 77, 556–567. [Google Scholar] [CrossRef]
- Kelly, A.M.; Mathema, B.; Larson, E.L. Carbapenem-resistant Enterobacteriaceae in the community: A scoping review. Int. J. Antimicrob. Agents 2017, 50, 127–134. [Google Scholar] [CrossRef]
- Olaitan, A.O.; Morand, S.; Rolain, J.M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 1–18. [Google Scholar] [CrossRef]
- Gutiérrez, C.; Zenis, J.; Legarraga, P.; Cabrera-Pardo, J.R.; García, P.; Bello-Toledo, H.; Opazo-Capurro, A.; González-Rocha, G. Genetic analysis of the first mcr-1 positive Escherichia coli isolate collected from an outpatient in Chile. Braz. J. Infect. Dis. 2019, 23, 203–206. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, L.; Wang, J.; Yassin, A.K.; Butaye, P.; Kelly, P.; Gong, J.; Guo, W.; Li, J.; Li, M.; et al. Molecular detection of colistin resistance genes (mcr-1, mcr-2 and mcr-3) in nasal/oropharyngeal and anal/cloacal swabs from pigs and poultry. Sci. Rep. 2018, 8, 3705. [Google Scholar] [CrossRef]
- Hernando-Amado, S.; Coque, T.M.; Baquero, F.; Martínez, J.L. Defining and combating antibiotic resistance from one health and global health perspectives. Nat. Microbiol. 2019, 4, 1432–1442. [Google Scholar] [CrossRef]
- Surleac, M.; Czobor Barbu, I.; Paraschiv, S.; Popa, L.I.; Gheorghe, I.; Marutescu, L.; Popa, M.; Sarbu, I.; Talapan, D.; Nita, M.; et al. Whole genome sequencing snapshot of multi-drug resistant Klebsiella pneumoniae strains from hospitals and receiving wastewater treatment plants in Southern Romania. PLoS ONE 2020, 15, e0228079. [Google Scholar] [CrossRef] [PubMed]
- Mulchandani, R.; Wang, Y.; Gilbert, M.; Van Boeckel, T.P. Global trends in antimicrobial use in food-producing animals: 2020 to 2030. PLoS Glob. Public Health 2023, 3, e0001305. [Google Scholar] [CrossRef] [PubMed]
- Fresia, P.; Antelo, V.; Salazar, C.; Giménez, M.; D’Alessandro, B.; Afshinnekoo, E.; Mason, C.; Gonnet, G.H.; Iraola, G. Urban metagenomics uncover antibiotic resistance reservoirs in coastal beach and sewage waters. Microbiome 2019, 7, 35. [Google Scholar] [CrossRef]
- Bueno, I.; Verdugo, C.; Jimenez-Lopez, O.; Alvarez, P.P.; Gonzalez-Rocha, G.; Lima, C.A.; Travis, D.A.; Wass, B.; Zhang, Q.; Ishii, S.; et al. Role of wastewater treatment plants on environmental abundance of Antimicrobial Resistance Genes in Chilean rivers. Int. J. Hyg. Environ. Health 2020, 223, 56–64. [Google Scholar] [CrossRef]
- Amos, G.C.; Zhang, L.; Hawkey, P.M.; Gaze, W.H.; Wellington, E.M. Functional metagenomic analysis reveals rivers are a reservoir for diverse antibiotic resistance genes. Vet. Microbiol. 2014, 171, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Gavidia, C.; Barría, C.; Rivas, L.; García, P.; Alvarez, F.P.; González-Rocha, G.; Opazo-Capurro, A.; Araos, R.; Munita, J.M.; Cortes, S.; et al. Isolation of Ciprofloxacin and Ceftazidime Resistant Enterobacterales from Vegetables and River Water Is Strongly Associated with the Season and the Sample Type. Front. Microbiol. 2021, 12, 604567. [Google Scholar] [CrossRef]
- CLSI, Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 32nd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022. [Google Scholar]
- Jofré, M.; Barrera, B.; Silva, F.; Berrocal, L. Evaluation of sensidiscs elution for the determination of susceptibility to colistin in multi-resistant gram-negative bacilli. Rev. Chil. Infectol. 2020, 37, 87–88. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Rebelo, A.R.; Bortolaia, V.; Kjelgaard, J.; Pedersen, S.; Leekitcharoenphon, P.; Hansen, I.M.; Guerra, B.; Malorny, B.; Borowiak, M.; Hammerl, J.A.; et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 2018, 23, 1560–7917. [Google Scholar] [CrossRef]
- Salgado-Caxito, M.; Benavides, J.A.; Munita, J.M.; Rivas, L.; García, P.; Listoni, F.J.; Moreno-Switt, A.I.; Paes, A.C. Risk factors associated with faecal carriage of extended-spectrum cephalosporin-resistant Escherichia coli among dogs in Southeast Brazil. Prev. Vet. Med. 2021, 190, 105316. [Google Scholar] [CrossRef]
- Poirel, L.; Berçot, B.; Millemann, Y.; Bonnin, R.A.; Pannaux, G.; Nordmann, P. Carbapenemase-producing Acinetobacter spp. in cattle, France. Emerg. Infect. Dis. 2012, 18, 523–525. [Google Scholar] [CrossRef]
- Shin, H.; Kim, Y.; Han, S.; Hur, H.G. Resistome Study in Aquatic Environments. J. Microbiol. Biotechnol. 2023, 33, 277–287. [Google Scholar] [CrossRef]
- Mathlouthi, N.; Al-Bayssari, C.; Bakour, S.; Rolain, J.M.; Chouchani, C. Prevalence and emergence of carbapenemases-producing Gram-negative bacteria in Mediterranean basin. Crit. Rev. Microbiol. 2017, 43, 43–61. [Google Scholar] [CrossRef] [PubMed]
- Lupo, A.; Haenni, M.; Madec, J.Y. Antimicrobial Resistance in Acinetobacter spp. and Pseudomonas spp. Microbiol. Spectr. 2018, 6, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Brooke, J.S. Advances in the Microbiology of Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 2021, 34, e0003019. [Google Scholar] [CrossRef] [PubMed]
- Breidenstein, E.B.; De la Fuente-Núñez, C.; Hancock, R.E. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, Z.; Li, J.; Lei, L.; Yin, W.; Li, M.; Wu, C.; Walsh, T.R.; Wang, Y.; Wang, S.; et al. Presence of VIM-positive Pseudomonas species in chickens and their surrounding environment. Antimicrob. Agents Chemother. 2017, 61, e00167-17. [Google Scholar] [CrossRef]
- Fariñas, M.C.; Martínez-Martínez, L. Multiresistant Gram-negative bacterial infections: Enterobacteria, Pseudomonas aeruginosa, Acinetobacter baumannii and other non-fermenting Gram-negative bacilli. Enferm. Infecc. Microbiol. Clin. 2013, 31, 402–409. [Google Scholar] [CrossRef]
- Qin, S.; Xiao, W.; Zhou, C.; Pu, Q.; Deng, X.; Lan, L.; Liang, H.; Song, X.; Wu, M. Pseudomonas aeruginosa: Pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct. Target. Ther. 2022, 7, 199. [Google Scholar] [CrossRef]
- Remuzgo-Martínez, S.; Lázaro-Díez, M.; Mayer, C.; Aranzamendi-Zaldumbide, M.; Padilla, D.; Calvo, J.; Marco, F.; Martínez-Martínez, L.; Icardo, J.M.; Otero, A.; et al. Biofilm Formation and Quorum-Sensing-Molecule Production by Clinical Isolates of Serratia liquefaciens. Appl. Environ. Microbiol. 2015, 81, 3306–3315. [Google Scholar] [CrossRef]
- Koczura, R.; Mokracka, J.; Makowska, N. Environmental Isolate of Rahnella aquatilis Harbors Class 1 Integron. Curr. Microbiol. 2016, 72, 64–67. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, W.J.; Song, Y.; Park, S.Y.; Kim, M.J. Bacteremia due to Rahnella aquatilis in a Patient with a Chemoport. Infect. Chemother. 2019, 51, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Guevarra, R.B.; Magez, S.; Peeters, E.; Chung, M.S.; Kim, K.H.; Radwanska, M. Comprehensive genomic analysis reveals virulence factors and antibiotic resistance genes in Pantoea agglomerans KM1, a potential opportunistic pathogen. PLoS ONE 2021, 16, e0239792. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira DM, P.; Forde, B.M.; Kidd, T.J.; Harris PN, A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- Ruppé, É.; Woerther, P.L.; Barbier, F. Mechanisms of antimicrobial resistance in Gram-negative bacilli. Ann. Intensive Care 2015, 5, 61. [Google Scholar] [CrossRef]
- Stachurová, T.; Piková, H.; Bartas, M.; Semerád, J.; Svobodová, K.; Malachová, K. Beta-lactam resistance development during the treatment processes of municipal wastewater treatment plants. Chemosphere 2021, 280, 130749. [Google Scholar] [CrossRef]
- Laborda, P.; Hernando-Amado, S.; Martínez, J.L.; Sanz-García, F. Antibiotic Resistance in Pseudomonas. Adv. Exp. Med. Biol. 2022, 1386, 117–143. [Google Scholar] [CrossRef]
- Said, B.L.; Jouini, A.; Klibi, N.; Dziri, R.; Alonso, C.A.; Boudabous, A.; Ben Slama, K.; Torres, C. Detection of extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae in vegetables, soil and water of the farm environment in Tunisia. Int. J. Food Microbiol. 2015, 203, 86–92. [Google Scholar] [CrossRef]
- Caltagirone, M.; Nucleo, E.; Spalla, M.; Zara, F.; Novazzi, F.; Marchetti, V.M.; Piazza, A.; Bitar, I.; De Cicco, M.; Paolucci, S.; et al. Occurrence of extended spectrum β-lactamases, KPC-type, and MCR-1.2-producing Enterobacteriaceae from wells, river water, and wastewater treatment plants in Oltrepò Pavese area, northern Italy. Front. Microbiol. 2017, 8, 2232. [Google Scholar] [CrossRef]
- Marcelino, V.R.; Wille, M.; Hurt, A.C.; González-Acuña, D.; Klaassen, M.; Schlub, T.E.; Eden, J.-S.; Shi, M.; Iredell, J.R.; Sorrell, T.C.; et al. Meta-transcriptomics reveals a diverse antibiotic resistance gene pool in avian microbiomes. BMC Biol. 2019, 17, 31. [Google Scholar] [CrossRef]
- Benavides, J.A.; Salgado-Caxito, M.; Opazo-Capurro, A.; González Muñoz, P.; Piñeiro, A.; Otto Medina, M.; Rivas, L.; Munita, J.; Millán, J. ESBL-producing Escherichia coli carrying CTX-M genes circulating among livestock, dogs, and wild mammals in small-scale farms of Central Chile. Antibiotics 2021, 10, 510. [Google Scholar] [CrossRef] [PubMed]
- Cevidanes, A.; Esperón, F.; Cataldo SDi Neves, E.; Sallaberry-Pincheira, N.; Millán, J. Antimicrobial resistance genes in Andean foxes inhabiting anthropized landscapes in Central Chile. Sci. Total Environ. 2020, 724, 138247. [Google Scholar] [CrossRef] [PubMed]
- Sacristán, I.; Esperón, F.; Acuña, F.; Aguilar, E.; García, S.; López, M.J.; Cevidanes, A.; Neves, E.; Cabello, J.; Hidalgo-Hermoso, E.; et al. Antibiotic resistance genes as landscape anthropization indicators: Using a wild felid as sentinel in Chile. Sci. Total Environ. 2020, 703, 134900. [Google Scholar] [CrossRef]
- Raphael, E.; Riley, L.W. Infections Caused by Antimicrobial Drug-Resistant Saprophytic Gram-Negative Bacteria in the Environment. Front. Med. 2017, 4, 183. [Google Scholar] [CrossRef] [PubMed]
- Nappier, S.P.; Liguori, K.; Ichida, A.M.; Stewart, J.R.; Jones, K.R. Antibiotic Resistance in Recreational Waters: State of the Science. Int. J. Environ. Res. Public. Health 2020, 17, 8034. [Google Scholar] [CrossRef]
- Delgado-Gardea, M.C.; Tamez-Guerra, P.; Gomez-Flores, R.; Zavala-Díaz de la Serna, F.J.; Eroza-de la Vega, G.; Nevárez-Moorillón, G.V.; Pérez-Recoder, M.C.; Sánchez-Ramírez, B.; González-Horta, M.D.C.; Infante-Ramírez, R. Multidrug-Resistant Bacteria Isolated from Surface Water in Bassaseachic Falls National Park, Mexico. Int. J. Environ. Res. Public. Health 2016, 13, 597. [Google Scholar] [CrossRef]
- Franklin, A.M.; Brinkman, N.E.; Jahne, M.A.; Keely, S.P. Twenty-first century molecular methods for analyzing antimicrobial resistance in surface waters to support One Health assessments. J. Microbiol. Methods 2021, 184, 106174. [Google Scholar] [CrossRef]
- Hubeny, J.; Korzeniewska, E.; Ciesielski, S.; Płaza, G.; Harnisz, M. The Resistome of ESKAPEE Pathogens in Untreated and Treated Wastewater: A Polish Case Study. Biomolecules 2022, 12, 1160. [Google Scholar] [CrossRef]
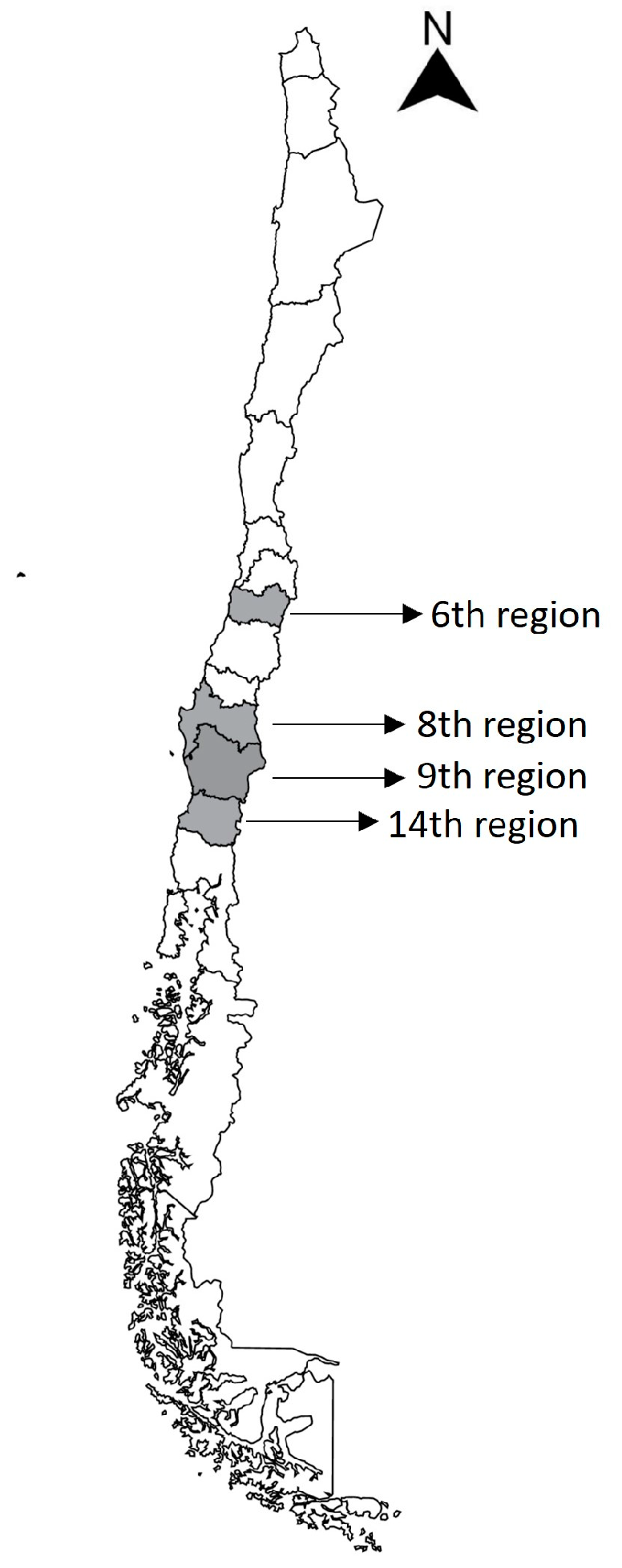
| N° Correlative Water Sample | Origin of Sample | Sampling Location | Region | Latitude | Longitude | Growth in Blood Agar | Growth in MacConkey Agar | Re-Isolated Strains Name |
|---|---|---|---|---|---|---|---|---|
| 1 | Cachapoal river | Doñihue, Chile | 6th | −34.217.836 | −70.892.976 | yes | yes | 1.4–1.5 |
| 2 | Cachapoal river | Doñihue, Chile | 6th | −34.214.474 | −70.896.846 | yes | yes | 2.1–2.2 |
| 3 | Cachapoal river | Doñihue, Chile | 6th | −34.213.610 | −70.906.275 | yes | yes | 3.1–3.2 |
| 4 | Cachapoal river | Doñihue, Chile | 6th | −34.229.342 | −70.938.954 | yes | yes | 4.2 |
| 5 | Ñuble river | San Nicolás, Chile | 8th | −36.550.236 | −72.094.312 | yes | none | none |
| 6 | Cachapoal river | Doñihue, Chile | 6th | −34.211.686 | −70.873.574 | yes | yes | 6.2 |
| 7 | Cachapoal river | Doñihue, Chile | 6th | −34.209.675 | −70.861.052 | yes | yes | 7.1–7.2 |
| 8 | Bellavista waterfall | Pucón, Chile | 9th | −39.219.463 | −71.844.840 | none | none | none |
| 9 | Coñaripe watershed | Coñaripe, Chile | 14th | −39.582.347 | −72.025.156 | yes | yes | 9.1–9.2 |
| 10 | Calafquen lake | Licán Ray, Chile | 9th | −39.492.032 | −72.161.685 | yes | yes | 10.2 |
| 11 | Voipir river | Villarrica, Chile | 9th | −39.280.841 | −72.306.407 | yes | none | none |
| 12 | Bío-Bío river | San Carlos de Purén, Chile | 8th | −37.599.828 | −72.274.804 | yes | yes | 12.1 |
| 13 | Ñuble river | San Nicolás, Chile | 8th | −36.551.936 | −72.090.913 | yes | yes | 13.1–13.2 |
| 14 | Bío-Bío river | San Carlos de Purén, Chile | 8th | −37.603.116 | −72.268.453 | yes | yes | none |
| 15 | Ñuble river | San Nicolás, Chile | 8th | −36.548.835 | −72.096.363 | yes | yes | 15.1–15.2 |
| 16 | Mulchen-Bureo river | Mulchén, Chile | 8th | −37.719.543 | −72.260.213 | yes | yes | none |
| 17 | Caburgua lake | Caburgua, Chile | 9th | −39.193.653 | −71.796.384 | yes | yes | 17.1–17.2–17.3 |
| 18 | Calafquen lake | Coñaripe, Chile | 14th | −39.565.623 | −72.019.635 | yes | yes | 18.1–18.2 |
| 19 | Villarrica lake | Villarrica, Chile | 9th | −39.275.456 | −72.227.820 | yes | yes | 19.1–19.2–19.3 |
| 20 | Caburgua lagoon | Pucón, Chile | 9th | −39.240.204 | −71.831.649 | yes | none | none |
| 21 | Villarrica lake | Villarrica, Chile | 9th | −39.283.296 | −72.207.432 | yes | none | none |
| Strain | Identification | Resistance Profile | BLEE Genes |
|---|---|---|---|
| 1.4 | Pseudomonas aeruginosa/fluorescens/putida/mosselli | AMP–CFZ–FEP–COL | NF |
| 1.5 | Pseudomonas aeruginosa/fluorescens/putida/mosselli | AMP–CFZ–FEP–COL | NF |
| 2.1 | Pseudomonas aeruginosa/fluorescens/putida/mosselli | AMP–CFZ–COL | NF |
| 2.2 | Pseudomonas aeruginosa/fluorescens/putida/mosselli | AMP–CFZ–MEM–COL | NF |
| 3.1 | Comamonas aquatica | CIP | NF |
| 3.2 | Enterobacter cloacae | AMP–CFZ–CAZ–FEP–IPM–MEM–CIP | blaCTX–M–blaTEM |
| 4.2 | Pseudomonas aeruginosa/fluorescens/putida/mosselli | AMP–CFZ–COL | NF |
| 6.2 | Pseudomonas aeruginosa/fluorescens/putida/mosselli | AMP–CFZ | NF |
| 7.1 | Acinetobacter pittii/calcoaceticus/haemolyticus | AMP–CFZ–COL | NF |
| 7.2 | Acinetobacter pittii/calcoaceticus/haemolyticus | AMP–CFZ–COL | NF |
| 9.1 | Acinetobacter pittii/calcoaceticus/haemolyticus | AMP–CFZ–CAZ | blaTEM |
| 9.2 | Rahnella aquatilis | AMP–CFZ | NF |
| 10.2 | Pseudomonas aeruginosa/fluorescens/putida/mosselli | AMP–CFZ | NF |
| 12.1 | Stenotrophomonas maltophilia | AMP–CFZ–CAZ–FEP | NF |
| 13.1 | Pantoea agglomerans | AMP–CFZ–FEP | blaCTX–M |
| 13.2 | Pseudomonas aeruginosa/fluorescens/putida/mosselli | AMP–CFZ | NF |
| 15.1 | Stenotrophomonas maltophilia | AMP–CFZ | NF |
| 15.2 | Pseudomonas aeruginosa/fluorescens/putida/mosselli | AMP–CFZ | NF |
| 17.1 | Acinetobacter pittii/calcoaceticus/haemolyticus | AMP–CFZ–CAZ | NF |
| 17.2 | Stenotrophomonas maltophilia | AMP–CFZ–CAZ | NF |
| 17.3 | Acinetobacter pittii/calcoaceticus/haemolyticus | AMP–CFZ–FEP–COL | NF |
| 18.1 | Acinetobacter pittii/calcoaceticus/haemolyticus | AMP–CFZ–CAZ | NF |
| 18.2 | Acinetobacter pittii/calcoaceticus/haemolyticus | AMP–CFZ–MEM–COL | NF |
| 19.1 | Enterobacter cloacae | AMP–CFZ–CAZ–FEP–IPM–MEM–CIP | blaCTX–M–blaTEM |
| 19.2 | Rahnella aquatilis | AMP–CFZ–FEP | NF |
| 19.3 | Serratia marcescens | AMP–CFZ–FEP–COL | NF |
| Resistance Phenotype | % (Frequency) |
|---|---|
| Ampicillin | 96.2% (25/26) |
| Cefazoline | 96.2% (25/26) |
| Ceftazidime | 26.9% (7/26) |
| Cefepime | 34.6% (9/26) |
| Imipenem | 7.7% (2/26) |
| Meropenem | 15.4% (4/26) |
| Ciprofloxacin | 11.5% (3/26) |
| Colistin | 38.5% (10/26) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jofré Bartholin, M.; Barrera Vega, B.; Berrocal Silva, L. Antibiotic-Resistant Bacteria in Environmental Water Sources from Southern Chile: A Potential Threat to Human Health. Microbiol. Res. 2023, 14, 1764-1773. https://doi.org/10.3390/microbiolres14040121
Jofré Bartholin M, Barrera Vega B, Berrocal Silva L. Antibiotic-Resistant Bacteria in Environmental Water Sources from Southern Chile: A Potential Threat to Human Health. Microbiology Research. 2023; 14(4):1764-1773. https://doi.org/10.3390/microbiolres14040121
Chicago/Turabian StyleJofré Bartholin, Matías, Boris Barrera Vega, and Liliana Berrocal Silva. 2023. "Antibiotic-Resistant Bacteria in Environmental Water Sources from Southern Chile: A Potential Threat to Human Health" Microbiology Research 14, no. 4: 1764-1773. https://doi.org/10.3390/microbiolres14040121
APA StyleJofré Bartholin, M., Barrera Vega, B., & Berrocal Silva, L. (2023). Antibiotic-Resistant Bacteria in Environmental Water Sources from Southern Chile: A Potential Threat to Human Health. Microbiology Research, 14(4), 1764-1773. https://doi.org/10.3390/microbiolres14040121






