Short-Term PE Generation Processes in the Soils of a Farmer Plots Network in the Madagascar Highlands: Actors and Drivers
Abstract
:1. Introduction
2. Materials and Methods
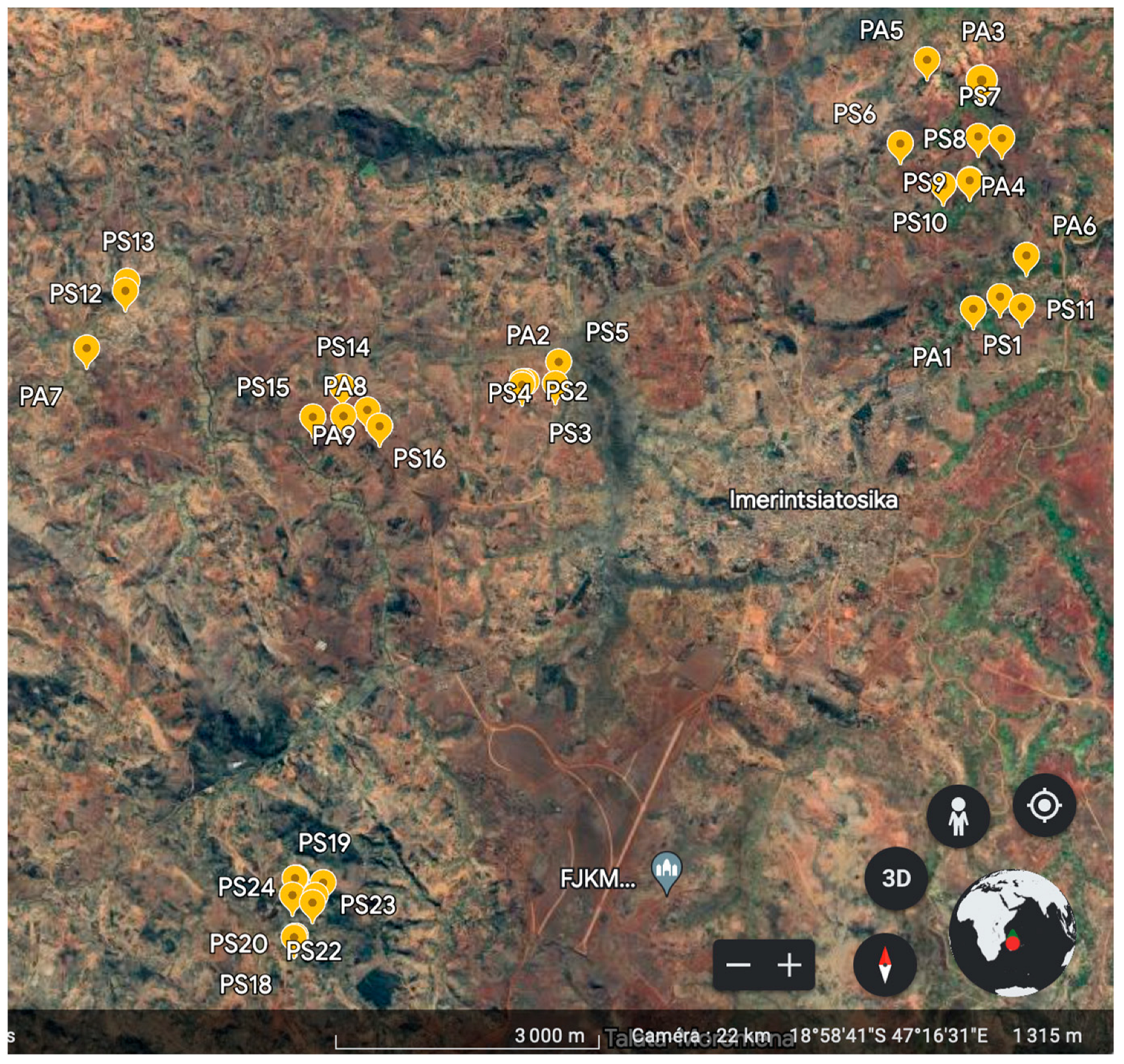
2.1. Soil Sampling Strategy
2.2. Soil Characterization
2.3. Molecular Analyses of Soil Bacterial and Fungal Communities
2.4. Soil Microcosm Set-Up for CO2 Measurements
2.5. Statistics
3. Results
3.1. Soil Characteristics
3.2. Microbial Community Properties
3.3. Potential Mineralization Activities
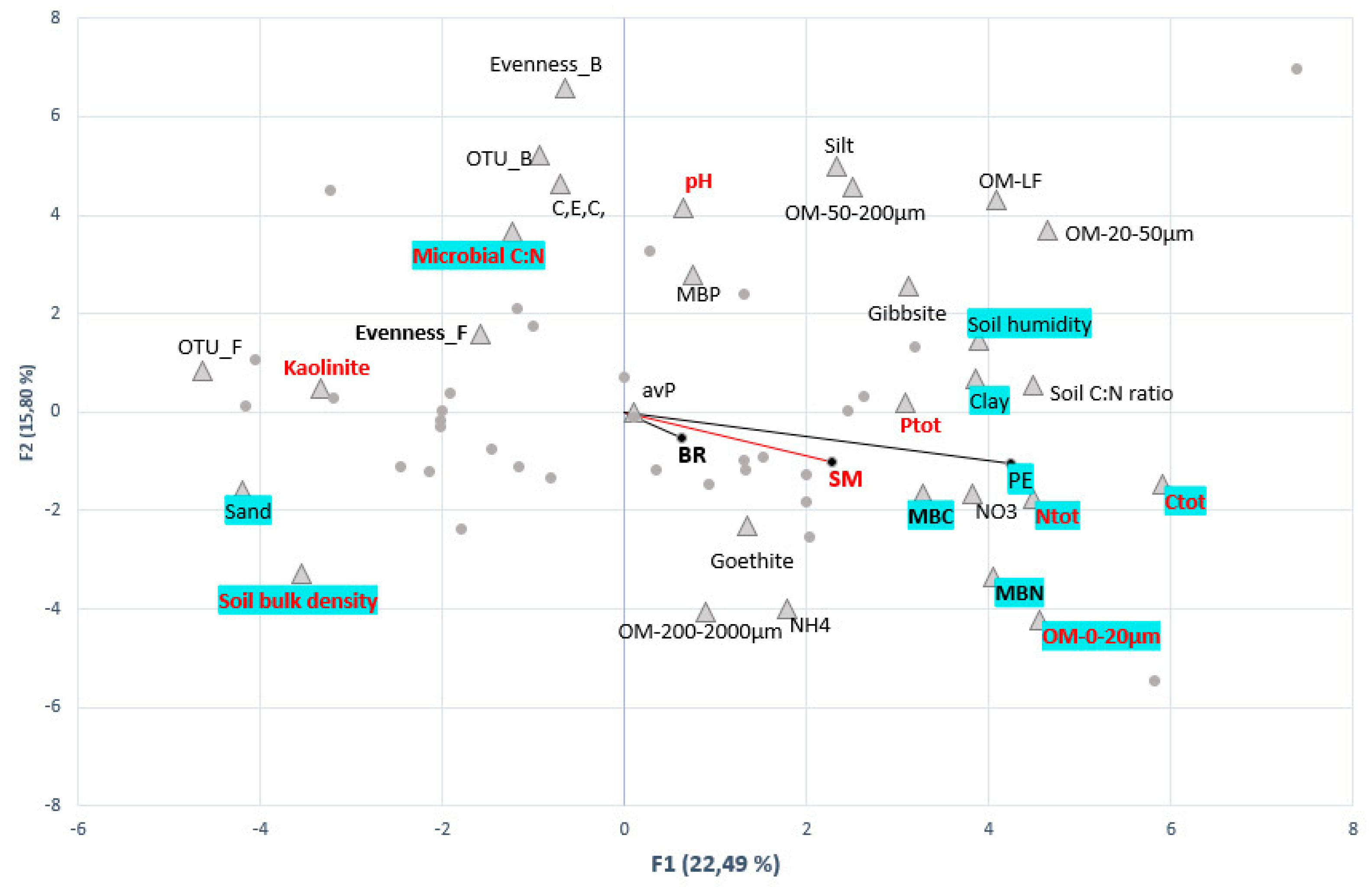
3.4. Links between Agricultural, Edaphic, Biotic Soil Parameters and C-Cycling Activities
3.4.1. Biotic and Abiotic Drivers of C-Mineralization Activities
3.4.2. Microbial Populations Linked to C-mineralization Activities
3.4.3. Determinants of Microbial Functional Groups
3.4.4. Influence of Agricultural Practices on Soil Characteristics and C-Mineralization Processes
4. Discussion
4.1. C-Cycling Activities in Agricultural Ferralsols
4.2. Microbial Actors of C Dynamic in Agricultural Ferralsols
4.3. How Agricultural System Components Drive C-Cycling Activities?
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paustian, K.; Lehmann, J.; Ogle, S.; Reay, D.; Robertson, G.P.; Smith, P. Climate-smart soils. Nature 2016, 532, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Niang, I.; Ruppel, O.C.; Abdrabo, M.A.; Essel, A.; Lennard, C.; Padgham, J.; Urquhart, P. Africa. In Climate Change: Impacts, Adaptation, and Vulnerability. Part B: Regional Aspects; Contribution of Working Group II to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2014; pp. 1199–1265. [Google Scholar]
- FAO. World Agriculture: Towards 2015/2030; Summary Report; Food and Agriculture Organization of the United Nations (FAO): Rome, Italy, 2002; p. 97. [Google Scholar]
- Waldrop, M.P.; Balser, T.C.; Firestone, M.K. Linking microbial community composition to function in a tropical soil. Soil Biol. Biochem. 2000, 32, 1837–1846. [Google Scholar] [CrossRef]
- Moorhead, D.L.; Sinsabaugh, R.L. A Theoretical Model Of Litter Decay And Microbial Interaction. Ecol. Monogr. 2006, 76, 151–174. [Google Scholar] [CrossRef]
- Neill, C.; Gignoux, J. Soil organic matter decomposition driven by microbial growth: A simple model for a complex network of interactions. Soil Biol. Biochem. 2006, 38, 803–811. [Google Scholar] [CrossRef]
- Six, J.; Frey, S.D.; Thiet, R.K.; Batten, K.M. Bacterial and Fungal Contributions to Carbon Sequestration in Agroecosystems. Soil Sci. Soc. Am. J. 2006, 70, 555. [Google Scholar] [CrossRef]
- Schmidt, M.W.I.; Torn, M.S.; Abiven, S.; Dittmar, T.; Guggenberger, G.; Janssens, I.A.; Kleber, M.; Kögel-Knabner, I.; Lehmann, J.; Manning, D.A.C.; et al. Persistence of soil organic matter as an ecosystem property. Nature 2011, 478, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Louis, B.P.; Maron, P.-A.; Menasseri-Aubry, S.; Sarr, A.; Lévêque, J.; Mathieu, O.; Jolivet, C.; Leterme, P.; Viaud, V. Microbial Diversity Indexes Can Explain Soil Carbon Dynamics as a Function of Carbon Source. PLoS ONE 2016, 11, e0161251. [Google Scholar] [CrossRef]
- Bernard, L.; Basile-Doelsch, I.; Derrien, D.; Fanin, N.; Fontaine, S.; Guenet, B.; Karimi, B.; Marsden, C.; Maron, P. Advancing the mechanistic understanding of the priming effect on soil organic matter mineralization. Funct. Ecol. 2022, 36, 1355–1377. [Google Scholar] [CrossRef]
- Fontaine, S.; Mariotti, A.; Abbadie, L. The priming effect of organic matter: A question of microbial competition? Soil Biol. Biochem. 2003, 35, 837–843. [Google Scholar] [CrossRef]
- Keiluweit, M.; Bougoure, J.J.; Nico, P.S.; Pett-Ridge, J.; Weber, P.K.; Kleber, M. Mineral protection of soil carbon counteracted by root exudates. Nat. Clim. Change 2015, 5, 588–595. [Google Scholar] [CrossRef]
- Chen, R.; Senbayram, M.; Blagodatsky, S.; Myachina, O.; Dittert, K.; Lin, X. Soil C and N availability determine the priming effect: Microbial N mining and stoichiometric decomposition theories. Glob. Change Biol. 2014, 20, 2356–2367. [Google Scholar] [CrossRef] [PubMed]
- Clarholm, M.; Skyllberg, U.; Rosling, A. Organic acid induced release of nutrients from metal-stabilized soil organic matter—The unbutton model. Soil Biol. Biochem. 2015, 84, 168–176. [Google Scholar] [CrossRef]
- Jilling, A.; Keiluweit, M.; Gutknecht, J.L.M.; Grandy, A.S. Priming mechanisms providing plants and microbes access to mineral-associated organic matter. Soil Biol. Biochem. 2021, 158, 108265. [Google Scholar] [CrossRef]
- Li, H.; Bölscher, T.; Winnick, M.; Tfaily, M.M.; Cardon, Z.G.; Keiluweit, M. Simple plant and microbial exudates destabilize mineral-associated organic matter via multiple pathways. Environ. Sci. Technol. 2021, 55, 3389–3398. [Google Scholar] [CrossRef]
- Sano, M.; Tanaka, T.; Ohara, H.; Aso, Y. Itaconic acid derivatives: Structure, function, biosynthesis, and perspectives. Appl. Microbiol. Biotechnol. 2020, 104, 9041–9051. [Google Scholar] [CrossRef]
- Thevenot, M.; Dignac, M.F.; Rumpel, C. Fate of lignins in soils: A review. Soil Biol. Biochem. 2010, 42, 1200–1211. [Google Scholar] [CrossRef]
- Uroz, S.; Kelly, L.C.; Turpault, M.P.; Lepleux, C.; Frey-Klett, P. The mineralosphere concept: Mineralogical control of the distribution and function of mineral-associated bacterial communities. Trends Microbiol. 2015, 23, 751–762. [Google Scholar] [CrossRef]
- Adeleke, R.; Nwangburuka, C.; Oboirien, B. Origins, roles and fate of organic acids in soils: A review. S. Afr. J. Bot. 2017, 108, 393–406. [Google Scholar] [CrossRef]
- Lehmann, J.; Kleber, M. The contentious nature of soil organic matter. Nature 2015, 528, 60–68. [Google Scholar] [CrossRef]
- Blagodatskaya, E.; Khomyakov, N.; Myachina, O.; Bogomolova, I.; Blagodatsky, S.; Kuzyakov, Y. Microbial interactions affect sources of priming induced by cellulose. Soil Biol. Biochem. 2014, 74, 39–49. [Google Scholar] [CrossRef]
- Derrien, D.; Plain, C.; Courty, P.-E.; Gelhaye, L.; Moerdijk-Poortvliet, T.C.; Thomas, F.; Versini, A.; Zeller, B.; Koutika, L.-S.; Boschker, H.T.; et al. Does the addition of labile substrate destabilise old soil organic matter? Soil Biol. Biochem. 2014, 76, 149–160. [Google Scholar] [CrossRef]
- Balesdent, J.; Mariotti, A.; Guillet, B. Natural 13C abundance as a tracer for studies of soil organic matter dynamics. Soil Biol. Biochem. 1987, 19, 25–30. [Google Scholar] [CrossRef]
- Cotrufo, M.F.; Wallenstein, M.D.; Boot, C.M.; Denef, K.; Paul, E. The Microbial Efficiency-Matrix Stabilization (MEMS) framework integrates plant litter decomposition with soil organic matter stabilization: Do labile plant inputs form stable soil organic matter? Glob. Change Biol. 2013, 19, 988–995. [Google Scholar] [CrossRef]
- Tardy, V.; Spor, A.; Mathieu, O.; Lévèque, J.; Terrat, S.; Plassart, P.; Regnier, T.; Bardgett, R.D.; van der Putten, W.H.; Roggero, P.P.; et al. Shifts in microbial diversity through land use intensity as drivers of carbon mineralization in soil. Soil Biol. Biochem. 2015, 90, 204–213. [Google Scholar] [CrossRef]
- Malik, A.A.; Martiny, J.B.; Brodie, E.L.; Martiny, A.C.; Treseder, K.K.; Allison, S.D. Defining trait-based microbial strategies with consequences for soil carbon cycling under climate change. ISME J. 2020, 14, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Razanamalala, K.; Razafimbelo, T.; Maron, P.-A.; Ranjard, L.; Chemidlin, N.; Lelièvre, M.; Dequiedt, S.; Ramaroson, V.H.; Marsden, C.; Becquer, T.; et al. Microbial diversity drives Priming effect along climate gradients of the Madagascar Highlands. ISME J. 2018, 12, 451–462. [Google Scholar] [CrossRef]
- IUSS Working Group WRB. World reference base for soil resources 2014 international soil classification system. In World Soil Resources Reports; Food and Agriculture Organization of the United Nations: Rome, Italy, 2014; Volume 106, 203p. [Google Scholar]
- Pansu, M.; Gautheyrou, J. Handbook of Soil Analysis: Mineralogical, Organic and Inorganic Methods; Springer Science & Business: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Gavinelli, E.; Feller, C.; Larré-Larrouy, M.C.; Bacye, B.; Djegui, N.; Nzila, J.D.D. A routine method to study soil organic matter by particle-size fractionation: Examples for tropical soils. Commun. Soil Sci. Plant Anal. 1995, 26, 1749–1760. [Google Scholar] [CrossRef]
- Ramaroson, V.H.; Becquer, T.; Sá, S.O.; Razafimahatratra, H.; Delarivière, J.L.; Blavet, D.; Vendrame, P.R.; Rabeharisoa, L.; Rakotondrazafy, A.F. Mineralogical analysis of ferralitic soils in Madagascar using NIR spectroscopy. CATENA 2017, 168, 102–109. [Google Scholar] [CrossRef]
- Ciesielski, H.; Sterckeman, T.; Santerne, M.; Willery, J.P. Determination of cation exchange capacity and exchangeable cations in soils by means of cobalt hexamine trichloride. Effects of experimental conditions. Agronomy 1997, 17, 1–7. [Google Scholar] [CrossRef]
- Mulvaney, M.L. Nitrogen —Inorganic forms. In Methods of Soil Analysis Part 3. Chemical Methods; Klute, A., Ed.; American Society of Agronomy: Madison, WI, USA, 1996; p. 1130. [Google Scholar]
- Jackson, M.L. Soil Chemical Analysis; Prentice Hall: Englewood Cliffs, NJ, USA, 1958. [Google Scholar]
- Kouno, K.; Tuchiya, Y.; Ando, T. Measurement of Soil Microbial Biomass Phosphorus by an Anion-Exchange Membrane Method. Soil Biol. Biochem. 1995, 27, 1353–1357. [Google Scholar] [CrossRef]
- Amer, F.; Bouldin, D.R.; Black, C.A.; Duke, F.R. Characterization of soil phosphorus by anion exchange resin adsorption and P32-equilibration. Plant Soil 1955, 6, 391–408. [Google Scholar] [CrossRef]
- Vance, E.D.; Brookes, P.C.; Jenkinson, D.S. An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 1987, 19, 703–707. [Google Scholar] [CrossRef]
- Jenkinson, D.S.; Brookes, P.C.; Powlson, D.S. Measuring soil microbial biomass. Soil Biol. Biochem. 2004, 36, 5–7. [Google Scholar] [CrossRef]
- Plassart, P.; Terrat, S.; Thomson, B.; Griffiths, R.; Dequiedt, S.; Lelievre, M.; Regnier, T.; Nowak, V.; Bailey, M.; Lemanceau, P.; et al. Evaluation of the ISO Standard 11063 DNA Extraction Procedure for Assessing Soil Microbial Abundance and Community Structure. PLoS ONE 2012, 7, e44279. [Google Scholar] [CrossRef] [PubMed]
- Terrat, S.; Christen, R.; Dequiedt, S.; Lelièvre, M.; Nowak, V.; Regnier, T.; Bachar, D.; Plassart, P.; Wincker, P.; Jolivet, C.; et al. Molecular biomass and MetaTaxogenomic assessment of soil microbial communities as influenced by soil DNA extraction procedure. Microbiol. Biotechnol. 2012, 5, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Prévost-Bouré, N.C.; Christen, R.; Dequiedt, S.; Mougel, C.; Lelièvre, M.; Jolivet, C.; Shahbazkia, H.R.; Guillou, L.; Arrouays, D.; Ranjard, L. Validation and application of a PCR primer set to quantify fungal communities in the soil environment by real-time quantitative PCR. PLoS ONE 2011, 6, e24166. [Google Scholar] [CrossRef]
- Djemiel, C.; Dequiedt, S.; Karimi, B.; Cottin, A.; Girier, T.; El Djoudi, Y.; Wincker, P.; Lelièvre, M.; Mondy, S.; Prévost-Bouré, N.C.; et al. BIOCOM-PIPE: A new user-friendly metabarcoding pipeline for the characterization of microbial diversity from 16S, 18S and 23S rRNA gene amplicons. BMC Bioinf. 2020, 21, 492. [Google Scholar] [CrossRef]
- Bernard, L.; Chapuis-Lardy, L.; Razafimbelo, T.; Razafindrakoto, M.; Pablo, A.-L.; Legname, E.; Poulain, J.; Brüls, T.; O′Donohue, M.; Brauman, A.; et al. Endogeic earthworms shape bacterial functional communities and affect organic matter mineralization in a tropical soil. ISME J. 2012, 6, 213–222. [Google Scholar] [CrossRef]
- Fanin, N.; Mooshammer, M.; Sauvadet, M.; Meng, C.; Alvarez, G.; Bernard, L.; Bertrand, I.; Blagodatskaya, E.; Bon, L.; Fontaine, S.; et al. Soil enzymes in response to climate warming: Mechanisms and feedbacks. Funct. Ecol. 2022, 36, 1378–1395. [Google Scholar] [CrossRef]
- Blagodatskaya, E.; Kuzyakov, Y. Mechanisms of real and apparent priming effects and their dependence on soil microbial biomass and community structure: Critical review. Biol. Fert. Soil 2008, 45, 115–131. [Google Scholar] [CrossRef]
- Raminoarison, M.; Razafimbelo, T.; Rakotoson, T.; Becquer, T.; Blanchart, E.; Trap, J. Multiple-nutrient limitation of upland rainfed rice in Ferralsols: A greenhouse nutrient-omission trial. J. Plant Nut. 2020, 43, 270–284. [Google Scholar] [CrossRef]
- Liang, C.; Schimel, J.P.; Jastrow, J.D. The importance of anabolism in microbial control over soil carbon storage. Nat. Microbiol. 2017, 2, 17105 . [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Zhang, J.; Zhang, L.; Yang, M.; He, J.Z. Long-term fertilization regimes affect bacterial community structure and diversity of an agricultural soil in northern China. J. Soil Sediments 2008, 8, 43–50. [Google Scholar] [CrossRef]
- Recorbet, G.; Picard, C.; Normand, P.; Simonet, P. Kinetics of the persistence of chromosomal DNA from genetically engineered Escherichia coli introduced into soil. Appl. Environ. Microbiol. 1993, 59, 4289–4294. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, Y.; Chang, S.X.; Xu, Q.; Li, Y.; Ma, Z.; Qin, H.; Cai, Y. Linking enhanced soil nitrogen mineralization to increased fungal decomposition capacity with Moso bamboo invasion of broadleaf forests. Sci. Total Environ. 2021, 771, 144779. [Google Scholar] [CrossRef]
- Cleveland, C.C.; Nemergut, D.R.; Schmidt, S.K.; Townsend, A.R. Increases in soil respiration following labile carbon additions linked to rapid shifts in soil microbial community composition. Biogeochemistry 2007, 82, 229–240. [Google Scholar] [CrossRef]
- Wang, J.; Ran, Q.; Du, X.; Wu, S.; Wang, J.; Sheng, D.; Chen, Q.; Du, Z.; Li, Y.-Z. Two new Polyangium species, P. aurulentum sp. nov. and P. jinanense sp. nov., isolated from a soil sample. Syst. Appl. Microbiol. 2021, 44, 126274. [Google Scholar] [CrossRef]
- Schoch, C.L.; Sung, G.-H.; López-Giráldez, F.; Townsend, J.P.; Miadlikowska, J.; Hofstetter, V.; Robbertse, B.; Matheny, P.B.; Kauff, F.; Wang, Z.; et al. The Ascomycota Tree of Life: A Phylum-wide Phylogeny Clarifies the Origin and Evolution of Fundamental Reproductive and Ecological Traits. Syst. Biol. 2009, 58, 224–239. [Google Scholar] [CrossRef]
- Christensen, M. A View of Fungal Ecology. Mycologia 1989, 81, 1–19. [Google Scholar] [CrossRef]
- Fontaine, S.; Henault, C.; Aamor, A.; Bdioui, N.; Bloor, J.; Maire, V.; Mary, B.; Revaillot, S.; Maron, P. Fungi mediate long term sequestration of carbon and nitrogen in soil through their priming effect. Soil Biol. Biochem. 2011, 43, 86–96. [Google Scholar] [CrossRef]
- De Chaves, M.G.; Silva, G.G.Z.; Rossetto, R.; Edwards, R.A.; Tsai, S.M.; Navarrete, A.A. Acidobacteria subgroups and their metabolic potential for carbon degradation in sugarcane soil amended with vinasse and nitrogen fertilizers. Front. Microbiol. 2019, 10, 1680. [Google Scholar] [CrossRef] [PubMed]
- Shange, R.S.; Ankumah, R.O.; Ibekwe, A.M.; Zabawa, R.; Dowd, S.E. Distinct soil bacterial communities revealed under a diversely managed agroecosystem. PLoS ONE 2012, 7, e40338. [Google Scholar] [CrossRef]
- Lueders, T.; Wagner, B.; Claus, P.; Friedrich, M.W. Stable isotope probing of rRNA and DNA reveals a dynamic methylotroph community and trophic interactions with fungi and protozoa in oxic rice field soil. Environ. Microbiol. 2004, 6, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; May, T.W.; Smith, M.E. Ectomycorrhizal lifestyle in fungi: Global diversity, distribution, and evolution of phylogenetic lineages. Mycorrhiza 2010, 20, 217–263. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, G.T.; Bates, S.T.; Eilers, K.G.; Lauber, C.L.; Caporaso, J.G.; Walters, W.A.; Knight, R.; Fierer, N. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol. Biochem. 2011, 43, 1450–1455. [Google Scholar] [CrossRef]
- Thoumazeau, A.; Bessou, C.; Renevier, M.-S.; Trap, J.; Marichal, R.; Mareschal, L.; Decaëns, T.; Bottinelli, N.; Jaillard, B.; Chevallier, T.; et al. Biofunctool®: A new framework to assess the impact of land management on soil quality. Part A: Concept and validation of the set of indicators. Ecol. Indic. 2019, 97, 100–110. [Google Scholar] [CrossRef]
- Obriot, F.; Stauffer, M.; Goubard, Y.; Cheviron, N.; Peres, G.; Eden, M.; Revallier, A.; Vieublé-Gonod, L.; Houot, S. Multi-criteria indices to evaluate the effects of repeated organic amendment applications on soil and crop quality. Agric. Ecosyst. Environ. 2016, 232, 165–178. [Google Scholar] [CrossRef]
- Trap, J.; Bonkowski, M.; Plassard, C.; Villenave, C.; Blanchart, E. Ecological importance of soil bacterivores for ecosystem functions. Plant Soil 2016, 398, 1–24. [Google Scholar] [CrossRef]
- Soares, M.; Rousk, J. Microbial growth and carbon use efficiency in soil: Links to fungal-bacterial dominance, SOC-quality and stoichiometry. Soil Biol. Biochem. 2019, 131, 195–205. [Google Scholar] [CrossRef]
- Jensen, E.S.; Peoples, M.B.; Boddey, R.M.; Gresshoff, P.M.; Hauggaard-Nielsen, H.; Alves, B.J.; Morrison, M.J. Legumes for mitigation of climate change and the provision of feedstock for biofuels and biorefineries. A review. Agron. Sustain. Dev. 2012, 32, 329–364. [Google Scholar] [CrossRef]
- Bulluck III, L.R.; Brosius, M.; Evanylo, G.K.; Ristaino, J.B. Organic and synthetic fertility amendments influence soil microbial, physical and chemical properties on organic and conventional farms. Appl. Soil Ecol. 2002, 19, 147–160. [Google Scholar] [CrossRef]
- Pérez-Piqueres, A.; Edel-Hermann, V.; Alabouvette, C.; Steinberg, C. Response of soil microbial communities to compost amendments. Soil Biol. Biochem. 2006, 38, 460–470. [Google Scholar] [CrossRef]
- Andriarimalala, J.H.; Rakotozandriny, J.N.; Andriamandroso, A.L.H.; Penot, E.; Naudin, K.; Dugué, P.; Tillard, E.; Decruyenaere, V.; Salgado, P. Creating synergies between conservation agriculture and cattle production in crop–livestock farms: A study case in the lake Alaotra region of Madagascar. Exp. Agric. 2013, 49, 352–365. [Google Scholar] [CrossRef]
- Albrecht, A.; Kandji, S.T. Carbon sequestration in tropical agroforestry systems. Agric. Ecosyst. Environ. 2003, 99, 15–27. [Google Scholar] [CrossRef]

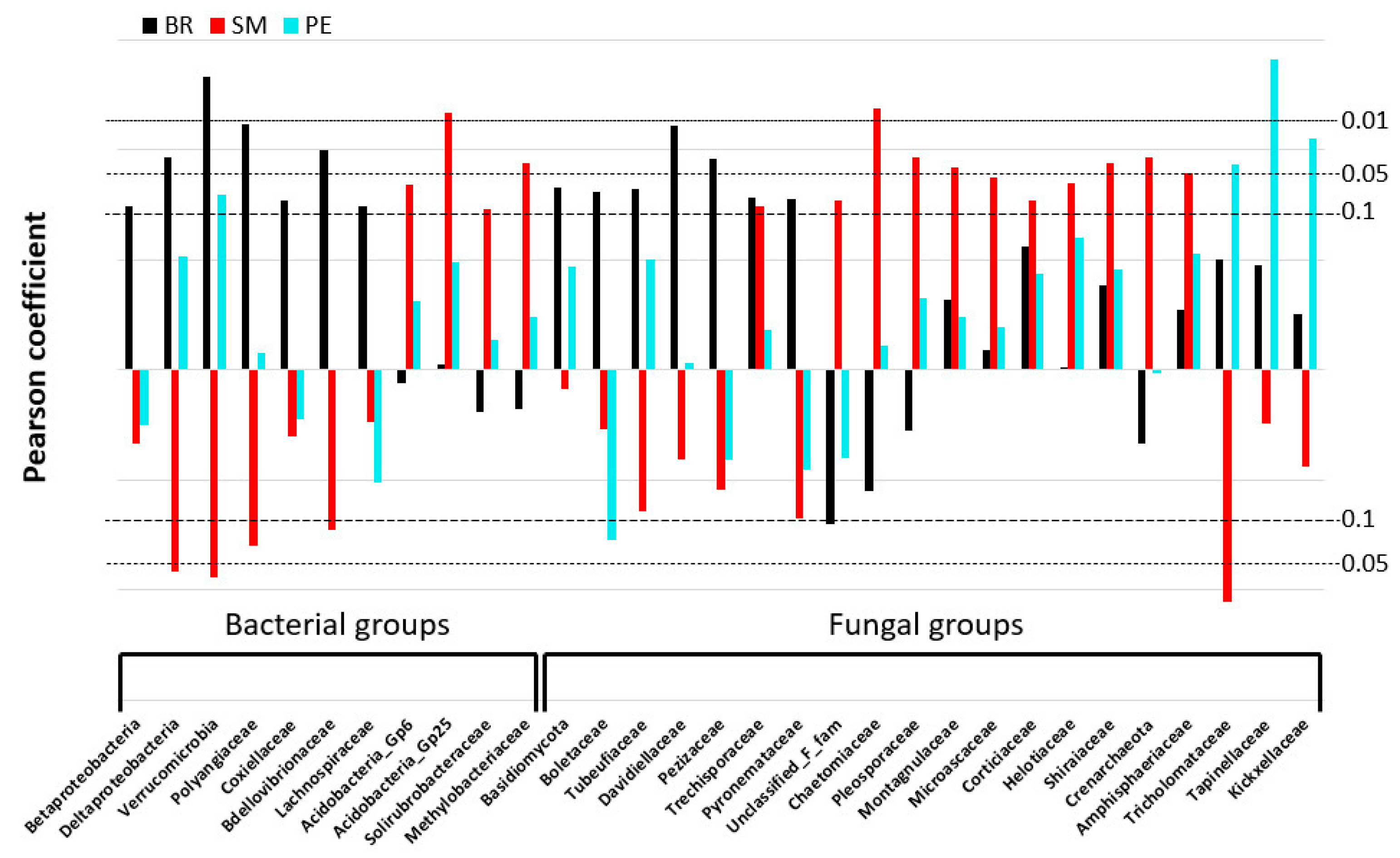

| Phylogenetic Groups (BR, SM, PE) | Abiotic Variables | Biotic Variables |
|---|---|---|
| Archaebacteria | ||
| Crenarchaetoa (SM) | MBC (−0.36); C:Nmic (−0.34) | |
| Bacteria | ||
| Acidobacteria_gp6 (SM) | Silt (0.33); Ctot (0.31); OM 0–20 µm (0.31) | OTU-B (0.40) |
| Acidobacteria_gp25 (SM) | Kt (−0.37); Ctot (0.37); Ntot (0.37); Ptot (0.37); OM 0–20 µm (0.43) | |
| Solirubrobacteraceae (SM) | Kt (−0.32); OM 0–20 µm (0.33) | OTU-B (0.32) |
| Methylobacteriaceae (SM) | Kt (−0.32); | |
| Deltaproteobacteria (BR, −SM) | Kt (0.36); Fe203cbd (−0.38); pH (0.37); C.E.C (0.49); Ctot (−0.29); OM 0–20 µm (−0.36) | MBC (0.36); C:Nmic (0.34); OTU-B (0.34) |
| Polyangiaceae (BR, −SM) | SBD (0.39); Kt (0.3); pH (0.38); Ctot (−0.39); Ntot (−0.30); OM 0–20 µm (−0.40); NO3 (−0.30) | C:Nmic (0.32) |
| Verrucomicrobia (BR, PE, −SM) | SBD (0.40); Kt (0.30); C.E.C (0.30); OM 200–2000 µm (0.41); NH4 (0.48) | MBC (0.72); MBN (0.51); Evenness F (−0.39) |
| Betaproteobacteria (BR) | Ctot (−0.29); OM 200–2000 µm (−0.37); OM 0–20 µm (−0.29) | Evenness F (−0.5) |
| Coxiellaceae (BR) | Kt (0.32), Gb (−0.30); | |
| Lachnospiraceae (BR) | Ctot (−0.32); OM 0–20 µm (−0.34); NO3 (−0.34); AvP (0.40) | |
| Bdellovibrionaceae (BR) | SBD (−0.30); Fe203cbd (−0.34); OM 0–20 µm (−0.35) | |
| Fungi | ||
| Unclassified F families(SM) | Silt (−0.38); SH (−0.39); pH (−0.44); Soil C:N (−0.51); OM-LF (−0.64); OM 20–50 (−0.55), NH4 (−0.33) | MBC (−0.53); MBN (−0.34), OTU-F (0.68) |
| Chaetomiaceae (SM) | Kt (−0.35); Fe203cbd (0.33); C.E.C (−0.33); Ntot (0.40); OM-LF (−0.33); OM 0–20 µm (0.36); | MBC (−0.40); OTU-F (0.36) |
| Pleosporaceae (SM) | Kt (−0.45); C.E.C (−0.35); | C:Nmic (−0.30) |
| Montagnulaceae (SM) | Kt (−0.40); Fe203cbd (0.38); Ctot (0.30); Ntot (0.30); Ptot (0.36); OM 0–20 µm (0.36) | |
| Microascaceae (SM) | Kt (−0.34); Ntot (0.35); Ptot (0.39); 0M 0–20 µm (0.34) | |
| Corticiaceae (SM) | Ntot (0.38); Ptot (0.50); OM 0–20 µm (0.29) | |
| Helotiaceae (SM) | SBD (−0.30); Kt (0.36); Fe203cbd (−0.33); OM 20–50 µm (−0.36) | OTU-F (0.32) |
| Shiraiaceae (SM) | OM 20–50 µm (−0.31) | |
| Amphisphaeriaceae (SM) | Ntot (0.31); Ptot (0.40); OM 0–20 µm (0.34) | |
| Trechisporaceae (BR, SM) | Ptot (0.34) | |
| Basidiomycota (BR) | Evenness F (−0.71) | |
| Boletaceae (BR) | NO3 (−0.42); AvP (0.40) | MBP (0.48); C:Nmic (0.52) |
| Tubeufiaceae (BR) | SBD (0.40); Fe203cbd (−0.32); Ptot (−0.36); NH4 (0.37) | MBC (0.60); MBN (0.52); Evenness B (−0.45) |
| Davidiellaceae (BR) | SBD (0.40); Fe203cbd (−0.40) | |
| Pezizaceae (BR) | NO3 (−0.30) | MBP (0.55); C:Nmic (0.54) |
| Pyronemataceae (BR) | SBD (0.41); SH (−0.37); Ptot (−0.30); | Evenness F (−0.30) |
| Tricholomataceae (PE) | pH (0.41); Ctot (0.31); OM 200–2000 µm (0.47); NH4 (0.62) | MBC (0.63); MBN (0.54); Evenness B (−0.41); OTU-F (−0.35); |
| Tapinellaceae (PE) | Clay (0.51); SH (0.33); OM 20–50 µm (0.30); NH4 (0.35) | MBC (0.49); MBN (0.52); Evenness F (−0.42) |
| Kickxellaceae (PE) | Clay (0.36); SBD (−0.30); | Evenness F (−0.32) |
| Culture | Fertilization | Trees NB | |||
|---|---|---|---|---|---|
| Variables | Off-Season | Season | Off-Season | Season | |
| Soil humidity | Compost (0.30) | ||||
| PH | Tuber (0.31) Cereal (−0.29) | Compost (−0.33) CM (−0.37) | |||
| C.E.C | Cereal (−0.36) | Legume (0.33) | |||
| Ctot | Tuber (−0.30) | Compost (0.33) CM (−0.38) | |||
| Ntot | Compost (0.30) CM (−0.38) | ||||
| Om-200–2000µm | Tuber (0.32) | Legume (0.39) | Compost (0.65) | (0.48) | |
| Om-0–20µm | Compost (0.47) CM (−0.34) | ||||
| NH4 | Compost (0.48) | ||||
| AvP | Cereal (0.29) | ||||
| MBN | Compost (0.38) | ||||
| MBP | Cereal (0.36) | ||||
| OTU_B | Compost (0.31) | ||||
| OTU_F | Tuber (−0.31) | ||||
| Acidobacteria_gp6 (SM) | Legume (0.38) | ||||
| Acidobacteria_gp25 (SM) | Legume (0.32) | ||||
| Solirubrobacteraceae (SM) | Legume (0.35) | ||||
| Methylobacteriaceae (SM) | Legume (0.40) | ||||
| Deltaproteobacteria (BR) | (−0.32) | ||||
| Polyangiaceae (BR) | Tuber (0.34) Fallow (−0.3) | ||||
| Verrucomicrobia (BR, PE) | Compost (0.39) | ||||
| Betaproteobacteria (BR) | Cereal (0.32) Fallow (−0.4) | ||||
| Bdellovibrionaceae (BR) | Cereal (0.32) | Fallow (0.6) | |||
| Chaetomiaceae (SM) | CM (−0.3) | ||||
| Pleosporaceae (SM) | Tuber (−0.36) Cereal (0.41) | CM (−0.35) | |||
| Montagnulaceae (SM) | Cereal (0.29) | ||||
| Microascaceae (SM) | CM (−0.36) | ||||
| Helotiaceae (SM) | Cereal (−0.29) | Compost (−0.32) | |||
| Shiraiaceae (SM) | Tuber (0.31) Fallow (−0.31) | ||||
| Amphisphaeriaceae (SM) | Legume (0.38) | ||||
| Trechisporaceae (BR, SM) | |||||
| Basidiomycota (BR) | |||||
| Boletaceae (BR) | Cereal (0.30) | Cereal (0.31) | |||
| Tubeufiaceae (BR) | Fallow (0.39) | Fallow (0.38) | |||
| Pezizaceae (BR) | Cereal (0.44) | Tuber-Cereal (0.44) | Compost (0.35) | ||
| Pyronemataceae (BR) | Fallow (0.34) | Fallow (0.60) | CM (−0.31) | ||
| Tricholomataceae (PE) | Legume (0.32) | ||||
| Tapinellaceae (PE) | Legume (0.34) | ||||
| Kickxellaceae (PE) | Legume (0.31) | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Razanamalala, K.; Razafimbelo, T.; Maron, P.-A.; Ranjard, L.; Chemidlin Prévost-Bouré, N.; Dequiedt, S.; Becquer, T.; Trap, J.; Blanchart, E.; Bernard, L. Short-Term PE Generation Processes in the Soils of a Farmer Plots Network in the Madagascar Highlands: Actors and Drivers. Microbiol. Res. 2023, 14, 1702-1719. https://doi.org/10.3390/microbiolres14040117
Razanamalala K, Razafimbelo T, Maron P-A, Ranjard L, Chemidlin Prévost-Bouré N, Dequiedt S, Becquer T, Trap J, Blanchart E, Bernard L. Short-Term PE Generation Processes in the Soils of a Farmer Plots Network in the Madagascar Highlands: Actors and Drivers. Microbiology Research. 2023; 14(4):1702-1719. https://doi.org/10.3390/microbiolres14040117
Chicago/Turabian StyleRazanamalala, Kanto, Tantely Razafimbelo, Pierre-Alain Maron, Lionel Ranjard, Nicolas Chemidlin Prévost-Bouré, Samuel Dequiedt, Thierry Becquer, Jean Trap, Eric Blanchart, and Laetitia Bernard. 2023. "Short-Term PE Generation Processes in the Soils of a Farmer Plots Network in the Madagascar Highlands: Actors and Drivers" Microbiology Research 14, no. 4: 1702-1719. https://doi.org/10.3390/microbiolres14040117
APA StyleRazanamalala, K., Razafimbelo, T., Maron, P.-A., Ranjard, L., Chemidlin Prévost-Bouré, N., Dequiedt, S., Becquer, T., Trap, J., Blanchart, E., & Bernard, L. (2023). Short-Term PE Generation Processes in the Soils of a Farmer Plots Network in the Madagascar Highlands: Actors and Drivers. Microbiology Research, 14(4), 1702-1719. https://doi.org/10.3390/microbiolres14040117








