In Vitro Cytotoxicity and Hemolysis Effect of Poly-Gamma-Glutamic Acid Nano-Polymer Biosynthesized Using Some Isolates of Bacillus spp.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Screening of Bacillus Isolates for the Ability to Synthesize ɣ-PGA NPs
2.2. Isolation and Purification of ɣ-PGA NPs
2.3. Characterization of the Biosynthesized ɣ-PGA NPs
2.4. 16S rRNA Gene Sequencing for the Molecular Identification of the Selected Isolates
2.5. Optimization of the Biosynthesized ɣ-PGA NPs
2.6. Characterization of the Optimized Biosynthesized ɣ-PGA NPs
2.7. In Vitro Hemolysis Assay
2.8. In Vitro Cytotoxicity
2.8.1. Toxicity Tests in PBMCs
2.8.2. Cytotoxicity by MTT Assay
2.9. Statistical Analysis
3. Results and Discussion
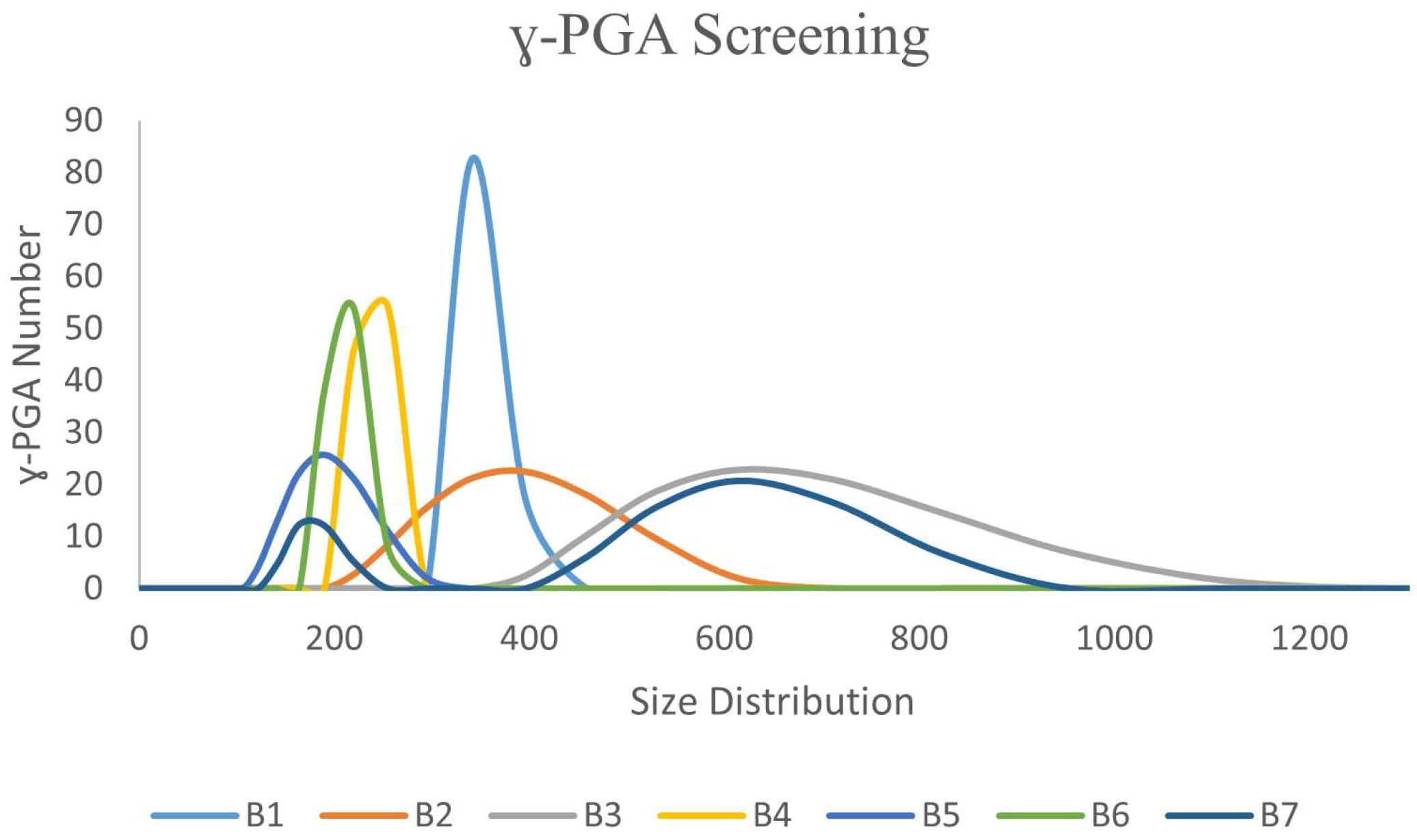
3.1. Ability of Bacillus spp. Isolates to Biosynthesis ɣ-PGA NPs
3.2. Molecular Identification and 16S rDNA Sequence-Based Phylogenetic Analysis of the Isolates
3.3. Optimization of the Conditions of ɣ-PGA NPs Biosynthesis
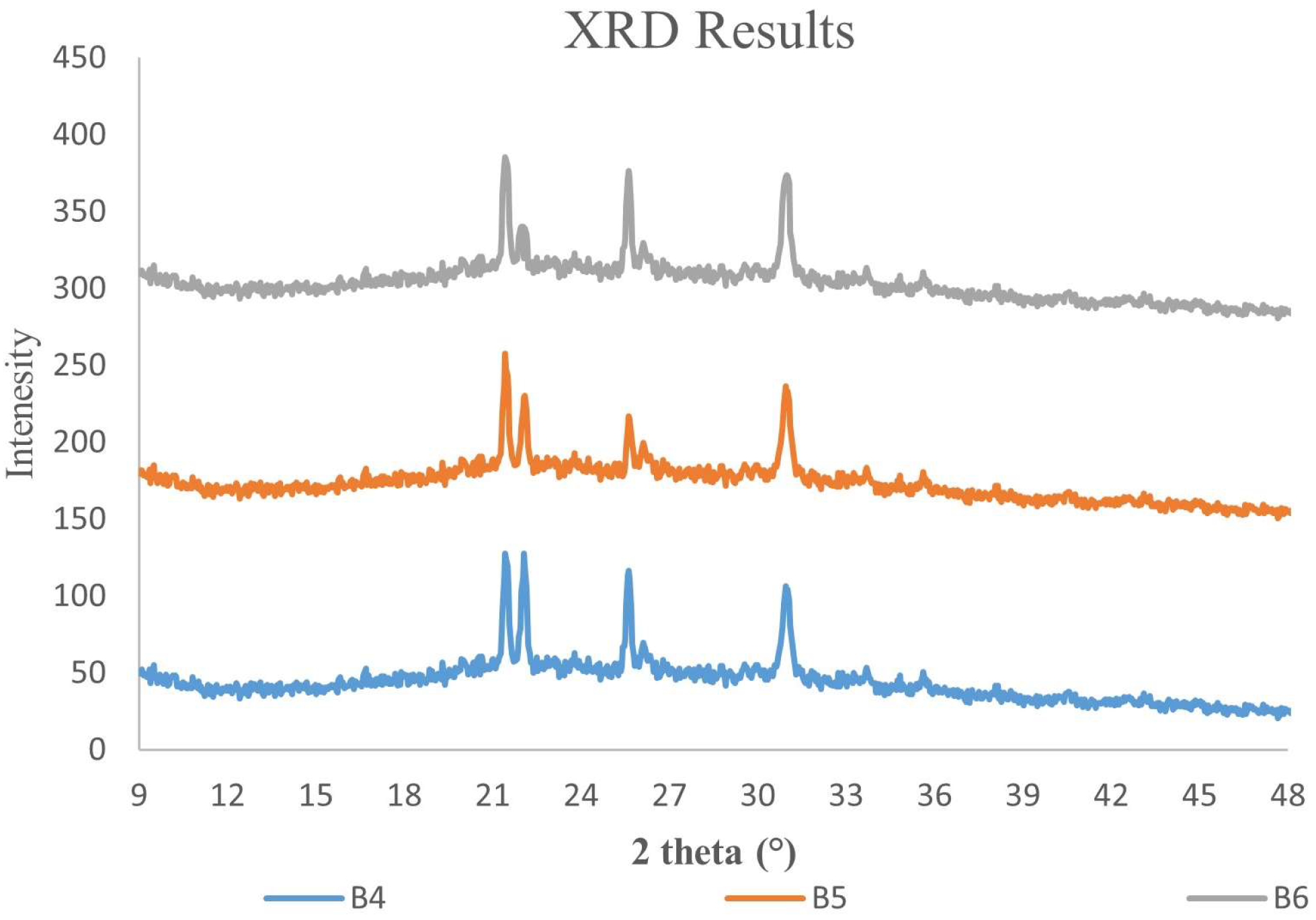
3.4. X-ray Diffraction and Microstructure Analysis of ɣ-PGA NPs
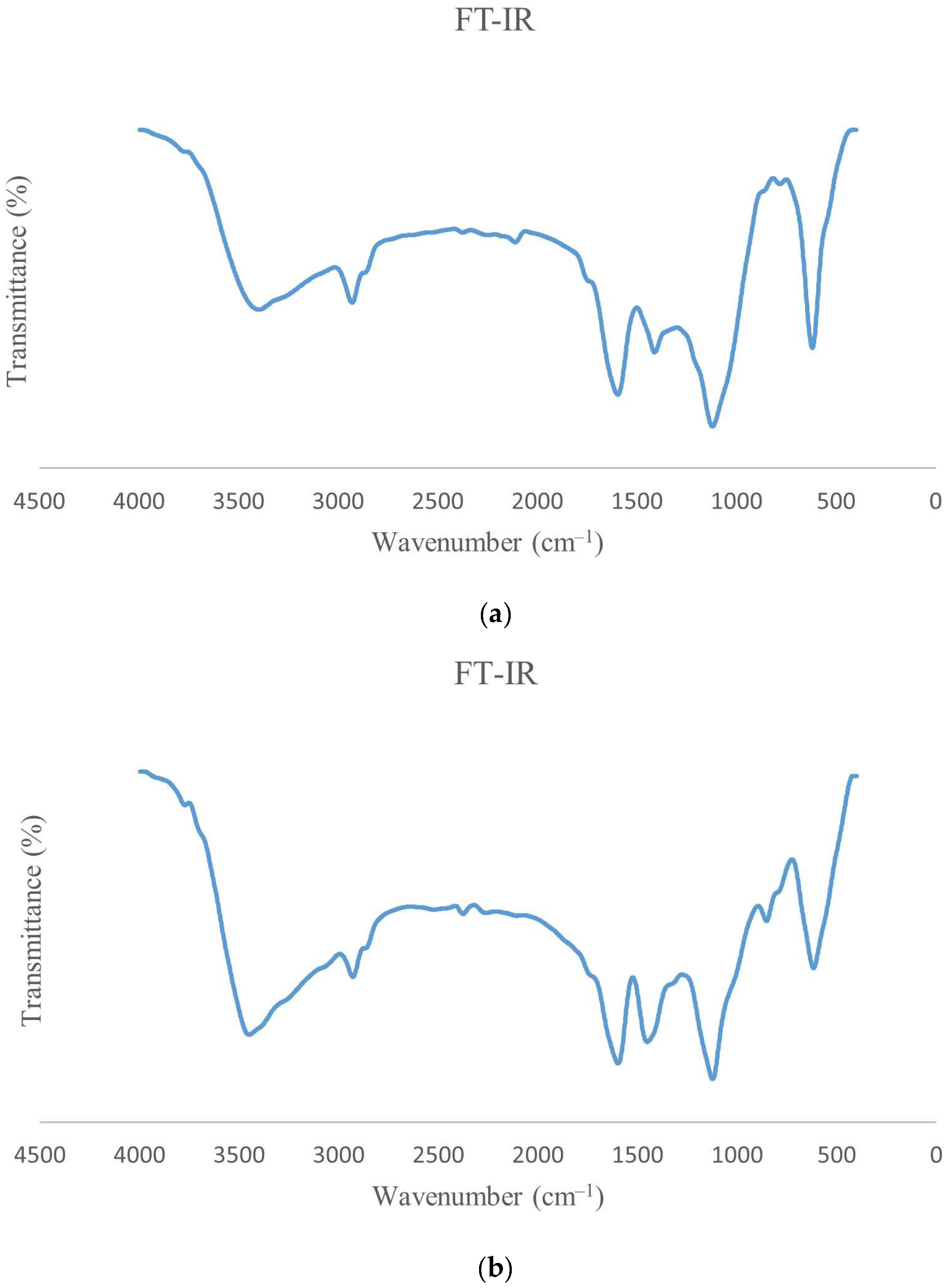
3.5. FT-IR of ɣ-PGA NPs
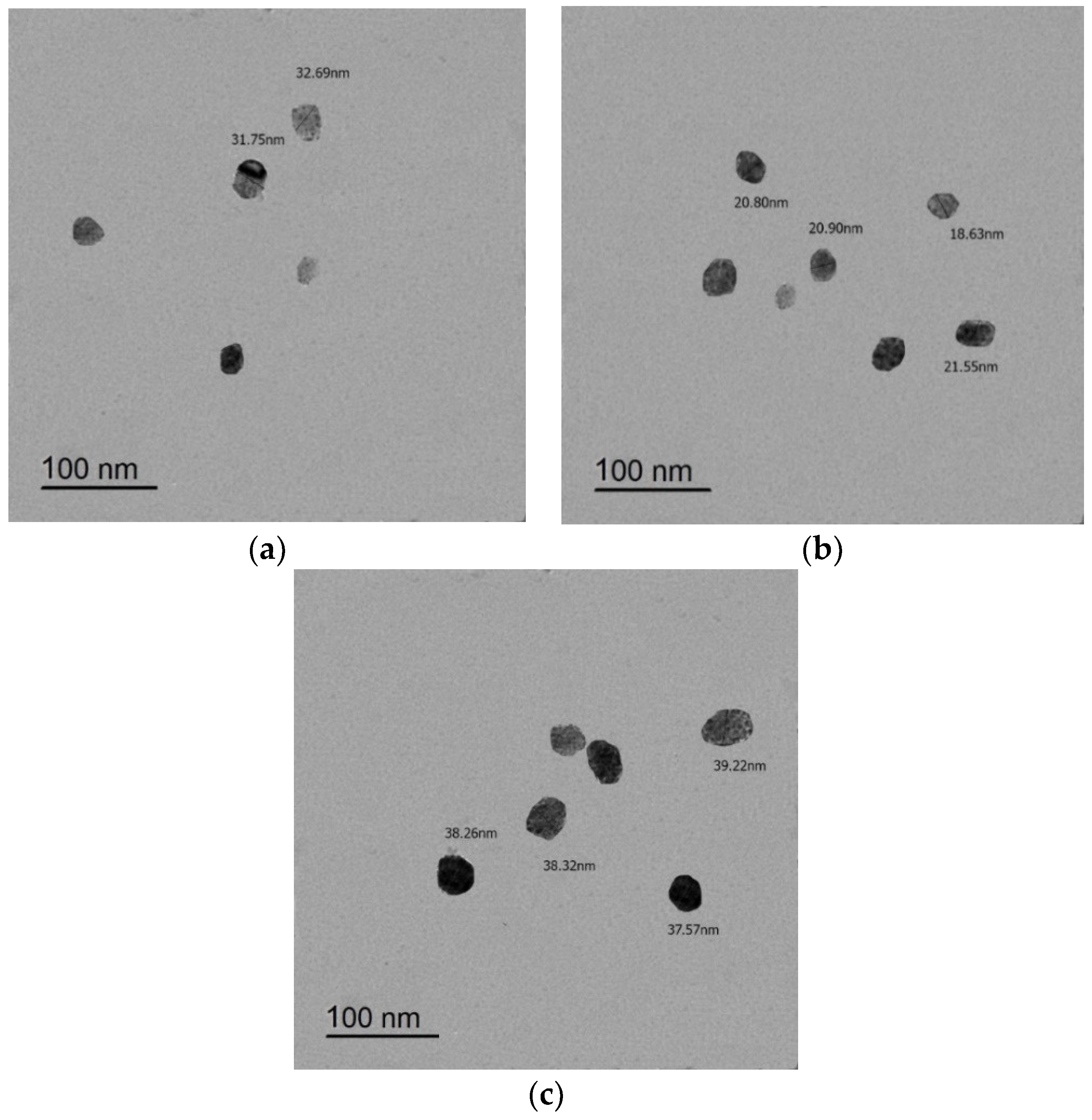
3.6. TEM Imaging
3.7. NMR Results
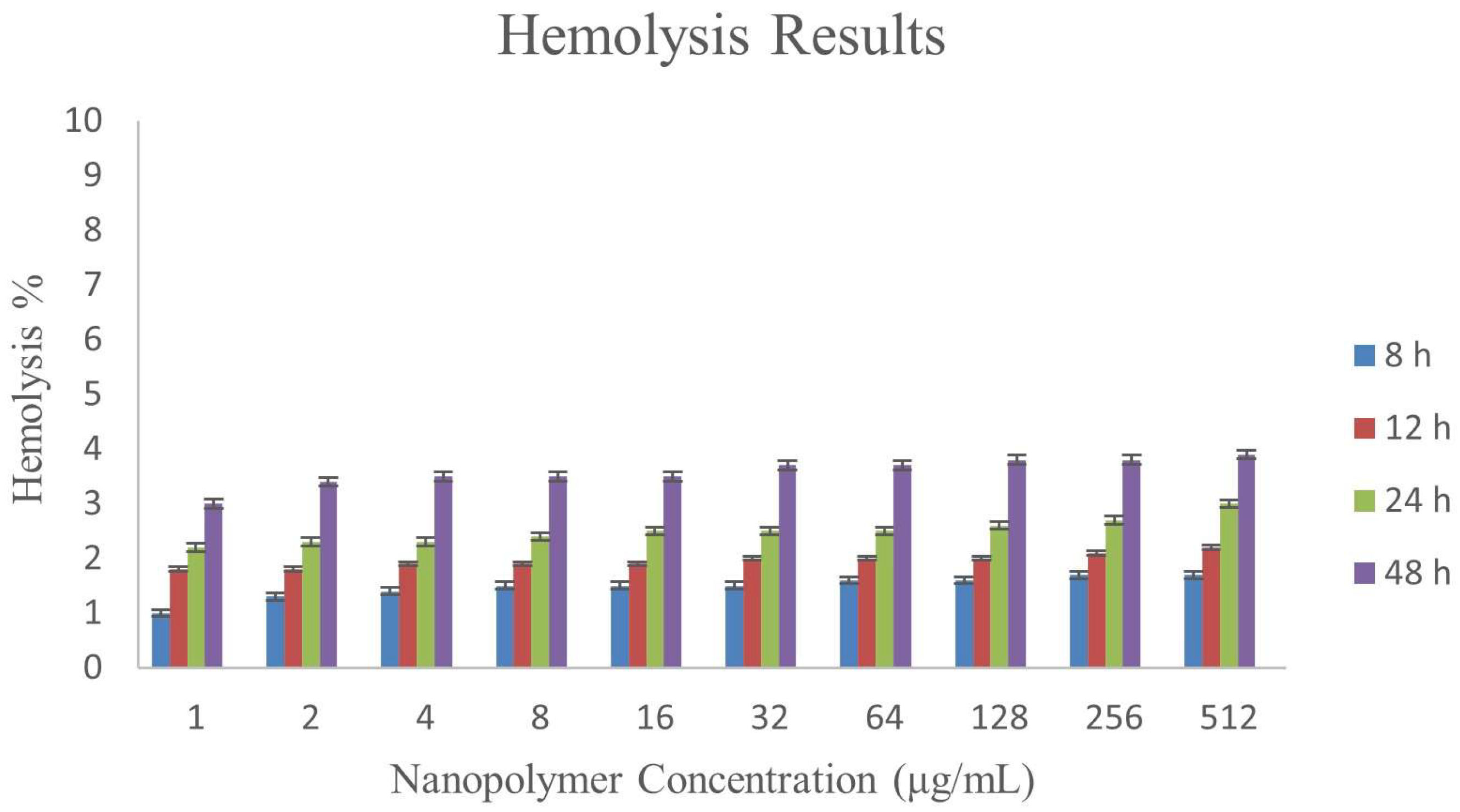
3.8. Hemolysis Assay
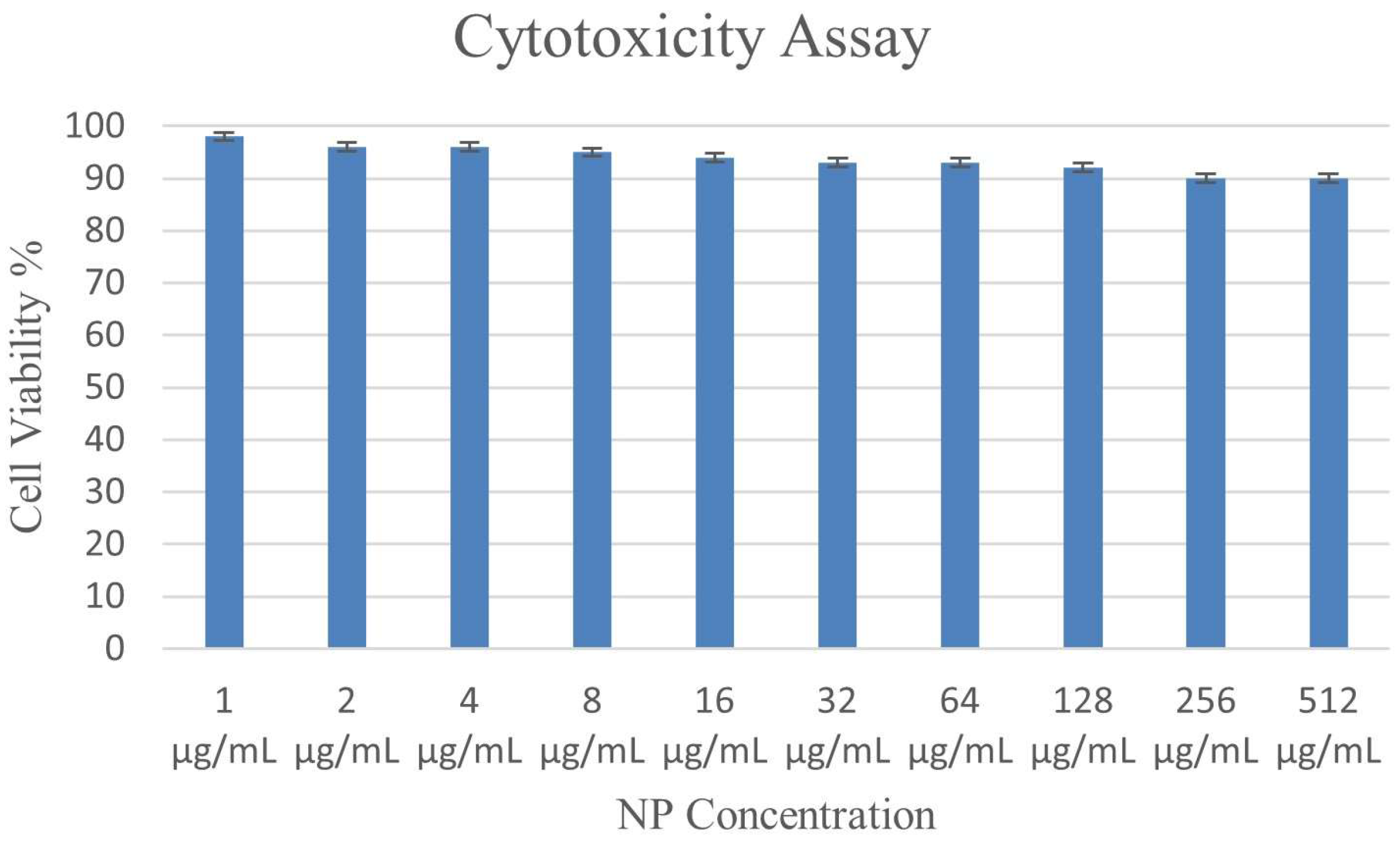
3.9. Impact of the ɣ-PGA NPs on Cell Viability in PBMC
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Doppalapudi, S.; Jain, A.; Khan, W.; Domb, A.J. Biodegradable polymers—An overview. Polym. Adv. Technol. 2014, 25, 427–435. [Google Scholar] [CrossRef]
- Shih, I.L.; Van, Y.T.; Yeh, L.C.; Lin, H.G.; Chang, Y.N. Production of a biopolymer flocculant from Bacillus licheniformis and its flocculation properties. Bioresour. Technol. 2001, 78, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Buescher, J.M.; Margaritis, A. Microbial biosynthesis of polyglutamic acid biopolymer and applications in the biopharmaceutical, biomedical and food industries. Crit. Rev. Biotechnol. 2007, 27, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Ogunleye, A.; Bhat, A.; Irorere, U.V.; Hill, D.; Williams, C.; Radecka, I. Poly-ɣ-glutamic acid: Production, properties and applications. Microbiology 2015, 161, 1–17. [Google Scholar] [CrossRef]
- Cao, M.; Geng, W.; Liu, L.; Song, C.; Xie, H.; Guo, W.; Jin, Y.; Wang, S. Glutamic acid independent production of poly-ɣ-glutamic acid by Bacillus amyloliquefaciens LL3 and cloning of pgs BCA genes. Bioresour. Technol. 2010, 201, 4251–4257. [Google Scholar]
- Akagi, T.; Baba, M.; Akashi, M. Preparation of nanoparticles by the self-organization of polymers consisting of hydrophobic and hydrophilic segments: Potential applications. Polymer 2007, 48, 6729–6747. [Google Scholar] [CrossRef]
- Bodnár, M.; Kjøniksen, A.; Molnár, R.; Hartmann, J.; Daróczi, L.; Nyström, B.; Borbély, B. Nanoparticles formed by complexation of poly-gamma-glutamic acid with lead ions. J. Hazard. Mater. 2008, 153, 1185–1192. [Google Scholar] [CrossRef]
- Candela, T.; Fouet, A. Poly-gamma-glutamate in bacteria. Mol. Microbiol. 2006, 60, 1091–1098. [Google Scholar] [CrossRef]
- Ko, Y.H.; Gross, R.A. Effects of glucose and glycerol on ɣ-poly(glutamic acid) formation by Bacillus licheniformis ATCC 9945a. Biotechnol. Bioeng. 1998, 57, 430–437. [Google Scholar] [CrossRef]
- Ashiuchi, M.; Kamei, T.; Baek, D.-H.; Shin, S.-Y.; Sung, M.-H.; Soda, K.; Yagi, T.; Misono, H. Isolation of Bacillus subtilis (chungkookjang), a poly-ɣ-glutamate producer with high genetic competence. Appl. Microbiol. Biotechnol. 2001, 57, 764–769. [Google Scholar] [CrossRef]
- Shih, I.L.; Van, Y.T. The production of poly-(ɣ-glutamic acid) from microorganisms and its various applications. Bioresour. Technol. 2001, 79, 207–225. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Choi, J.; Choi, Y.; Nakamura, H.; Shimanouchi, K.; Horiuchi, T.; Misono, H.V.T.; Soda, K.; Ashiuchi, M.; Sunga, M. Synthesis of super-high-molecular-weight poly-ɣ-glutamic acid by Bacillus subtilis subsp. Chungkookjang. J. Mol. Catal. B Enzym. 2005, 35, 128–133. [Google Scholar] [CrossRef]
- Kang, H.S.; Park, S.-H.; Lee, Y.-G.; Son, T.-I. Polyelectrolyte Complex Hydrogel Composed of Chitosan and Poly(g-Glutamic Acid) for Biological Application: Preparation, Physical Properties, and Cytocompatibility. J. Appl. Polym. Sci. 2010, 116, 2658–2667. [Google Scholar] [CrossRef]
- Hsieh, C.Y.; Tsai, S.P.; Wang, D.M.; Chang, Y.N.; Hsieh, H.J. Preparation of ɣ-PGA/chitosan composite tissue engineering matrices. Biomaterials 2005, 26, 5617–5623. [Google Scholar] [CrossRef]
- Lin, Y.H.; Chung, C.K.; Chen, C.T.; Liang, H.F.; Chen, S.C.; Sung, H.W. Preparation and Characterization of Nanoparticles Shelled with Chitosan for Oral Insulin Delivery. Biomacromolecules 2007, 8, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.K.; Zayed, M.A.; El-dek, S.I.; Hady, M.A.; El Sherbiny, D.H.; Uskoković, V. Nanofibrous ε-polycaprolactone scaffolds containing Ag-doped magnetite nanoparticles: Physicochemical characterization and biological testing for wound dressing applications in vitro and in vivo. Bioact. Mater. 2021, 6, 2070–2088. [Google Scholar] [CrossRef]
- Mosquera, J.; García, I.; Liz-Marzán, L.M. Cellular Uptake of Nanoparticles versus Small Molecules: A Matter of Size. Acc. Chem. Res. 2018, 51, 2305–2313. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Saeed, K.; Khan, I. Nanoparticles: Properties, applications and toxicities. Arab. J. Chem. 2019, 12, 908–931. [Google Scholar] [CrossRef]
- Deenadayalan, A.; Maddineni, P.; Raja, A. Comparison of whole blood and PBMC assays for T-cell functional analysis. BMC Res. Notes 2013, 6, 120. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.-H.; Yan, G.-H.; Chai, O.H.; Song, C.H. Inhibitory effects of curcumin on passive cutaneous anaphylactoid response and compound 48/80-induced mast cell activation. Anat. Cell Biol. 2010, 43, 36. [Google Scholar] [CrossRef]
- Xu, G.; Zha, J.; Cheng, H.; Ibrahim, M.H.; Yang, F.; Dalton, H.; Cao, R.; Zhu, Y.; Fang, J.; Chi, K.; et al. Engineering Corynebacterium glutamicum for the de novo biosynthesis of tailored poly-ɣ-glutamic acid. Metab. Eng. 2019, 56, 39–49. [Google Scholar] [CrossRef]
- Wei, X.; Tian, G.; Ji, Z.; Chen, S. A new strategy for enhancement of poly-ɣ-glutamic acid production by multiple physicochemical stresses in Bacillus licheniformis. J. Chem. Technol. Biotechnol. 2015, 90, 709–713. [Google Scholar] [CrossRef]
- Qiu, Y.; Zhang, Y.; Zhu, Y.; Sha, Y.; Xu, Z.; Feng, X.; Li, S.; Xu, H. Improving poly-(ɣ-glutamic acid) production from a glutamic acid-independent strain from inulin substrate by consolidated bioprocessing. Bioprocess Biosyst. Eng. 2019, 42, 1711–1720. [Google Scholar] [CrossRef]
- El-Sayed, E.M.; Salah, T.A.; Husseiny, S.M. Biosynthesized silver nanoparticles from new bacterial isolate and studying their effect on dermatophyte fungi. J. Sci. Res. Sci. 2017, 34, 123–141. [Google Scholar]
- Pisani, S.; Dorati, R.; Scocozza, F.; Mariotti, C.; Chiesa, E.; Bruni, G.; Genta, I.; Auricchio, F.; Conti, M.; Conti, B. Preliminary investigation on a new natural based poly(gamma-glutamic acid)/Chitosan bioink. J. Biomed. Mater. Res. Part B Appl. Biomater. 2020, 108, 2718–2732. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Cui, S.; Yang, Q.; Song, X.; Wang, D.; Hu, J.; Zhou, Y.; Liu, Y. AgNPs-incorporated nanofiber mats: Relationship between AgNPs size/content, silver release, cytotoxicity, and antibacterial activity. Mater. Sci. Eng. C 2021, 118, 111331. [Google Scholar] [CrossRef] [PubMed]
- Jett, B.D.; Hatter, K.L.; Huycke, M.M.; Gilmore, M.S. Simplified agar plate method for quantifying viable bacteria. BioTechniques 1997, 23, 648–650. [Google Scholar] [CrossRef]
- Goto, A.; Kunioka, M. Biosynthesis and Hydrolysis of Poly (ɣ-glutamic acid) from Bacillus subtilis IF03335. Biosci. Biotechnol. Biochem. 2014, 56, 1031–1035. [Google Scholar] [CrossRef]
- Jeevan, P.; Ramya, K.; Rena, A.E. Extracellular biosynthesis of silver nanoparticles by culture supernatant of Pseudomonas aeruginosa. Indian J. Biotechnol. 2012, 11, 72–76. [Google Scholar]
- Singh, V.; Chaudhary, D.; Mani, I. Molecular characterization and modeling of secondary structure of 16S rRNA from Aeromonas veronii. Int. J. Appl. Biol. Pharm. Technol. 2012, 3, 253–260. [Google Scholar]
- Prasad, P.; Turner, M.S. What bacteria are living in my food? An open-ended practical series involving identification of unknown foodborne bacteria using molecular techniques. Biochem. Mol. Biol. Educ. 2011, 39, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.; Banerjee, G.; Gupta, M.K.; Saxena, S.; Ramteke, P.W. Assessment of phylogenetic affiliation using 16S rRNA gene sequence analysis for Pseudomonas aeruginosa in patients of lower respiratory tract infection. Indian J. Med. Res. 2013, 138, 557–559. [Google Scholar] [PubMed]
- Chen, Y.; Yan, X.; Zhao, J.; Feng, H.; Li, P.; Tong, Z.; Yang, Z.; Li, S.; Yang, J.; Jin, S. Preparation of the chitosan/poly(glutamic acid)/alginate polyelectrolyte complexing hydrogel and study on its drug releasing property. J. Carbohydr. Polym. 2018, 191, 8–16. [Google Scholar] [CrossRef]
- Khalil, I.R.; Irorere, V.U.; Radecka, I.; Burns, A.T.; Kowalczuk, M.; Mason, J.L.; Khechara, M.P. Poly-ɣ-glutamic acid: Biodegradable polymer for potential protection of beneficial viruses. Materials 2016, 9, 28. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Naqvi, H.N.; Ahmad, M. Comparative study of the cytotoxic and genotoxic potentials of zinc oxide and titanium dioxide nanoparticles. Toxicol. Rep. 2015, 2, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Rodríguez, N.L.; Tavárez, S.; González-Sánchez, Z.I. In vitro toxicity assessment of zinc and nickel ferrite nanoparticles in human erythrocytes and peripheral blood mononuclear cell. Toxicol. Vitr. 2019, 57, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Yu, Y.; Li, Y.; Yu, Y.; Duan, J.; Zou, Y.; Li, Q.; Sun, Z. Oxidative Damage and Energy Metabolism Disorder Contribute to the Hemolytic Effect of Amorphous Silica Nanoparticles. Nanoscale Res. Lett. 2016, 11, 57. [Google Scholar] [CrossRef]
- Aimola, I.A.; Inuwa, H.M.; Nok, A.J.; Mamman, A.I.; Habila, N.; Muhammad, A.; Ndidi, U.S.; Ignatius, B.; Jande, P.L.; Isaac, L.C.; et al. Isolation of peripheral blood mononuclear cells using glycerol density gradient. Cell Biochem. Biophys. 2014, 68, 583–585. [Google Scholar] [CrossRef]
- Hamburger, A.W.; Dunn, F.E.; Tencer, K.L. Isolation of peripheral blood mononuclear cells using glycerol density gradient, Separation on percoll density gradients of cells derived from malignant ascites of mice. JNCI J. Natl. Cancer Inst. 1983, 70, 157–160. [Google Scholar] [CrossRef]
- Rushdy, A; Gomaa, E. Antimicrobial compounds produced by probiotic Lactobacillus brevis isolated from dairy products. Ann. Microbiol. 2013, 63, 81–90. [Google Scholar] [CrossRef]
- Yu, N.A.B.; Mirgorod, A.; Fedosyuk, V.M. Physico-Chemical Properties of Nanoparticles Functionalized by Polypyrrole. J. Nano-Electron. Phys. 2013, 5, 5–7. [Google Scholar]
- Panda, P.K.; Yang, J.M.; Chang, Y.H. Preparation and characterization of ferulic acid-modified water soluble chitosan and poly (ɣ-glutamic acid) polyelectrolyte films through layer-by-layer assembly towards protein adsorption. Int. J. Biol. Macromol. 2021, 171, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Jeong, Y.I.; Choi, K.C. Hair dye-incorporated poly-ɣ-glutamic acid/glycol chitosan nanoparticles based on ion-complex formation. Int. J. Nanomed. 2011, 6, 2879–2888. [Google Scholar] [CrossRef]
- Liu, C.; Peng, Y.; Zhou, F.; Yin, Y.; Huang, X.; Wang, L.; Wang, W.; Zhou, W.; Tang, D. Large-scale synthesis and quantitative characterization of size-controllable potassium tungsten bronze nanowires. J. Phys. D. Appl. Phys. 2018, 51, 095305. [Google Scholar] [CrossRef]
- Kar, M.; Pauline, M.; Sharma, K.; Kumaraswamy, G.; Sen Gupta, S. Synthesis of poly-l-glutamic acid grafted silica nanoparticles and their assembly into macroporous structures. Langmuir 2011, 27, 12124–12133. [Google Scholar] [CrossRef]
- Pereira, C.L.; Antunes, J.C.; Gonçalves, R.M.; Ferreira-Da-Silva, F.; Barbosa, M.A. Biosynthesis of poly-ɣ-glutamic acid and its application in the development of chitosan/poly-ɣ-glutamic acid nanoparticles for drug delivery. J. Mater. Sci. Mater. Med. 2012, 23, 1583–1591. [Google Scholar] [CrossRef]
- Hajdu, I.; Bodnár, M.; Filipcsei, G.; Hartmann, J.F.; Daróczi, L.; Zrínyi, M.; Borbély, J. Nanoparticles prepared by self-assembly of Chitosan and poly-ɣ-glutamic acid. Colloid Polym. Sci. 2008, 286, 343–350. [Google Scholar] [CrossRef]
- Nguyen, Q.T.; Kwak, C.; Lee, W.S.; Kim, J.; Jeong, J.; Sung, M.H.; Yang, J.; Poo, H. Poly-ɣ-glutamic acid complexed with alum induces cross-protective immunity of pandemic H1N1 vaccine. Front. Immunol. 2019, 10, 1604. [Google Scholar] [CrossRef]
- Anju, A.J.; Binod, P.; Pandey, A. Production and characterization of microbial poly-ɣ-glutamic acid from renewable resources. Indian J. Exp. Biol. 2017, 55, 405–410. [Google Scholar]
- Aseichev, A.V.; Azizova, O.A.; Beckman, E.M.; Skotnikova, O.I.; Dudnik, L.B.; Shcheglovitova, O.N.; Sergienko, V.I. Effects of gold nanoparticles on erythrocyte hemolysis. Bull. Exp. Biol. Med. 2014, 156, 495–498. [Google Scholar] [CrossRef]
- Paula, A.J.; Martinez, D.S.T.; Júnior, R.T.A.; Filho, A.G.S.; Alves, O.L.A. Suppression of the hemolytic effect of mesoporous silica nanoparticles after protein corona interaction: Independence of the surface microchemical environment. J. Braz. Chem. Soc. 2012, 23, 1807–1814. [Google Scholar] [CrossRef]
- Lu, B.; Lu, F.; Ran, L.; Yu, K.; Xiao, Y.; Li, Z.; Dai, F.; Wu, D.; Lan, G. Imidazole-molecule-capped chitosan–gold nanocomposites with enhanced antimicrobial activity for treating biofilm-related infections. J. Colloid Interface Sci. 2018, 531, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.M.; Feris, K.; Bell, J.; Wingett, D.G.; Hanley, C.; Punnoose, A. Selective toxicity of zinc oxide nanoparticles to prokaryotic and eukaryotic systems. Appl. Phys. Lett. 2007, 90, 10–13. [Google Scholar] [CrossRef] [PubMed]
- Hanley, C.; Layne, J.; Punnoose, A.; Reddy, K.; Coombs, I.; Coombs, A.; Feris, K.; Wingett, D. Preferential killing of cancer cells and activated human T cells using ZnO nanoparticles. Nanotechnology 2008, 19, 295103. [Google Scholar] [CrossRef]
- Tsai, W.B.; Lai, H.Y.; Lee, J.L.; Lo, C.W.; Chen, W.S. Enhancement of the cytotoxicity and selectivity of doxorubicin to hepatoma cells by synergistic combination of galactose-decorated ɣ-poly(glutamic acid) nanoparticles and low-intensity ultrasound. Langmuir 2014, 30, 5510–5517. [Google Scholar] [CrossRef] [PubMed]



| 1st C and N Source | 2nd C and N Source | 3rd C and N Source | 4th C and N Source |
|---|---|---|---|
| Glutamic acid | Glucose | Citric acid | Glutamic acid |
| Glucose | Ammonium Sulfate | Ammonium Sulfate | Glucose |
| Ammonium sulfate | KH2PO4 | KH2PO4 | KH2PO4 |
| KH2PO4 | Na2HPO4.12H2O | Na2HPO4.12H2O | Na2HPO4.12H2O |
| Na2HPO4.12H2O | MgSO4.7H2O | MgSO4.7H2O | MgSO4.7H2O |
| MgSO4.7H2O | MnSO4.nH2O | MnSO4.nH2O | MnSO4.nH2O |
| MnSO4.nH2O | FeCl3.6H2O | FeCl3.6H2O | FeCl3.6H2O |
| FeCl3.6H2O | CaCl2 | CaCl2 | CaCl2 |
| CaCl2 | Biotin | ||
| Biotin |
| Microbial Isolates | ɣ-PGA NPs Size (nm) |
|---|---|
| B1 | 340 |
| B2 | 342 |
| B3 | 615 |
| B4 | 218 |
| B5 | 190 |
| B6 | 220 |
| B7 | 615 |
| ɣ-PGA NPs Concentration (μg/mL) | Hemolysis % | |||
|---|---|---|---|---|
| 8 h | 12 h | 24 h | 48 h | |
| 1 | 1 ± 0.024 | 1.8 ± 0.031 | 2.2 ± 0.023 | 3 ± 0.047 |
| 2 | 1.3 ± 0.028 | 1.8 ± 0.036 | 2.3 ± 0.026 | 3.4 ± 0.04 |
| 4 | 1.4 ± 0.035 | 1.9 ± 0.019 | 2.3 ± 0.021 | 3.5 ± 0.042 |
| 8 | 1.5 ± 0.032 | 1.9 ± 0.025 | 2.4 ± 0.014 | 3.5 ± 0.041 |
| 16 | 1.5 ± 0.031 | 1.9 ± 0.042 | 2.5 ± 0.019 | 3.5 ± 0.032 |
| 32 | 1.5 ± 0.03 | 2.0 ± 0.012 | 2.5 ± 0.023 | 3.7 ± 0.029 |
| 64 | 1.6 ± 0.022 | 2.0 ± 0.043 | 2.5 ± 0.027 | 3.7 ± 0.04 |
| 128 | 1.6 ± 0.022 | 2.0 ± 0.04 | 2.6 ± 0.037 | 3.8 ± 0.019 |
| 256 | 1.7 ± 0.024 | 2.1 ± 0.035 | 2.7 ± 0.038 | 3.8 ± 0.034 |
| 512 | 1.7 ± 0.025 | 2.2 ± 0.035 | 3 ± 0.039 | 3.9 ± 0.02 |
| ɣ-PGA NPs Concentration (μg/mL) | Percentage of Treated Cells |
|---|---|
| 1 | 98 ± 0.68 |
| 2 | 96 ± 0.2 |
| 4 | 96 ± 0.48 |
| 8 | 95 ± 0.57 |
| 16 | 94 ± 0.29 |
| 32 | 93 ± 0.63 |
| 64 | 93 ± 0.21 |
| 128 | 92 ± 0.3 |
| 256 | 90 ± 0.73 |
| 512 | 90 ± 0.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elsayed, E.M.; Farghali, A.A.; Zanaty, M.I.; Abdel-Fattah, M.; Alkhalifah, D.H.M.; Hozzein, W.N.; Mahmoud, A.M. In Vitro Cytotoxicity and Hemolysis Effect of Poly-Gamma-Glutamic Acid Nano-Polymer Biosynthesized Using Some Isolates of Bacillus spp. Microbiol. Res. 2023, 14, 1720-1735. https://doi.org/10.3390/microbiolres14040118
Elsayed EM, Farghali AA, Zanaty MI, Abdel-Fattah M, Alkhalifah DHM, Hozzein WN, Mahmoud AM. In Vitro Cytotoxicity and Hemolysis Effect of Poly-Gamma-Glutamic Acid Nano-Polymer Biosynthesized Using Some Isolates of Bacillus spp. Microbiology Research. 2023; 14(4):1720-1735. https://doi.org/10.3390/microbiolres14040118
Chicago/Turabian StyleElsayed, Eman M., Ahmed A. Farghali, Mohamed I. Zanaty, Medhat Abdel-Fattah, Dalal Hussien M. Alkhalifah, Wael N. Hozzein, and Ahmed M. Mahmoud. 2023. "In Vitro Cytotoxicity and Hemolysis Effect of Poly-Gamma-Glutamic Acid Nano-Polymer Biosynthesized Using Some Isolates of Bacillus spp." Microbiology Research 14, no. 4: 1720-1735. https://doi.org/10.3390/microbiolres14040118
APA StyleElsayed, E. M., Farghali, A. A., Zanaty, M. I., Abdel-Fattah, M., Alkhalifah, D. H. M., Hozzein, W. N., & Mahmoud, A. M. (2023). In Vitro Cytotoxicity and Hemolysis Effect of Poly-Gamma-Glutamic Acid Nano-Polymer Biosynthesized Using Some Isolates of Bacillus spp. Microbiology Research, 14(4), 1720-1735. https://doi.org/10.3390/microbiolres14040118






