Abstract
This study targeted developing a novel Zinc oxide with alpha hematite nanoflowers (NFs)-loaded bee venom (Bv) (Bv-ZnO@αFe2O3 NFs) as a bio-natural product from bees to combine both the advantages of combination magnetic properties and the antimicrobial and anti-biofilm properties on isolated coliform bacteria from the effluent of wastewater treatment plants. About 24 isolates of treated wastewater isolates were multidrug resistant (MDR). The phylogenetic grouping of Escherichia coli (E. coli) and Klebsiella pneumonia (K. pneumonia) showed that the largest group was Group A, followed by Group B2 and Group B1. Fourier transform infrared (FTIR), The X-ray diffraction (XRD), and scanning electron microscopy-energy dispersive X-ray analysis (SEM− EDX) validated the coating operation’s contact with Bv onto ZnO@αFe2O3 NFs. According to high-resolution transmission electron microscopy (TEM) and selected area electron diffraction (SAED), pattern analyses for prepared nanoformulations exhibited a spherical shape of αFe2O3 (~9–15 nm), and floral needle shapes with uniform distribution of size with aggregation of ZnOαFe2O3 and Bv-ZnO@αFe2O3 NFs around (~100–200 nm). The toxicity of Bv-ZnO@αFe2O3 NFs was comparable up to 125 µg mL−1, when it reached 64.79% (IC50, 107.18 µg mL−1). The antibacterial activity showed different zones of inhibition against different isolates. The biofilm inhibitory activity of NPs and NFs showed a highly significant reduction (p < 0.001) in treated biofilms with ZnO@αFe2O3 and Bv-ZnO@αFe2O3. In essence, ZnO@αFe2O3 and Bv-ZnO@αFe2O3 NFs are promising antimicrobials for inhibiting the growth and biofilm of MDR E. coli and K. pneumonia isolates, thereby, biocontrol of wastewater.
1. Introduction
Fecal coliform bacteria can cause illnesses both in the hospital and in the community. One of the essential characteristics is multiple resistance to broad-spectrum antibiotics. Due to the excessive use of antimicrobial medications as a treatment, multidrug-resistant (MDR) bacteria like E. coli and K. pneumonia have become more common in recent years [1]. Antibiotics are an important tool in lowering the sickness or mortality caused by bacterial infections in humans. The unrestricted use of antibiotics was the primary cause of MDR bacterial strains’ establishment and dissemination [2,3]. The requirement for water will surely increase as the world population continues to rise. As a result, there will be a greater need globally for effective and cost-effective water treatment solutions. An important development could result from the industry’s adoption of nanotechnology [4]. Wastewater treatment by using nanotechnology has attracted more attention in recent years owing to its important criteria as small sizes providing large specific surface areas; nanomaterials have great adsorption capacities and reactivity with heavy metals, organic pollutants, inorganic anions, and bacteria [5].
Antimicrobial drug resistance is on the rise, necessitating the quest for fresh candidates with fresh modes of operation [6,7,8]. Venoms and their peptides from a variety of creatures or critters, including bees, snakes, wasps, and scorpions, provide potential antibacterial medicines for use against diverse pathogenic diseases. Bee venom (Bv), one of several bee products rich in bioactive chemicals, is a natural substance that exhibits various effects against several illness causes [9,10]. Their Bv is made of bioactive molecules such as melittin, apamin, phospholipase, histamine, dopamine, norepinephrine, and other components [11,12]. Bee venom therapy has been utilized as a supplemental and alternative treatment. Furthermore, bee venom is an effective antimicrobial [13], anti-inflammatory, anti-apoptosis, anti-fibrosis, and anti-atherosclerosis agent [14,15], as well as an antibacterial agent against a variety of Gram-positive/negative bacteria strains [16,17,18]. Using bee product combinations instead of conventional medication, according to Zambrano et al. [19] and Tanuğur and Kekeçoğlu [20], may have beneficial results.
Drug delivery systems refer to nanotechnological devices that are designed to synthesize and store drug molecules in appropriate configurations. Moreover, they expedite the delivery of medications to the specific targeted site of the multidrug resistance microbial, hence optimizing treatment effectiveness and reducing non-specific accumulation in the body of the drugs [21,22,23]. Among that, the most widely used inorganic nanoparticles are metal-based nanoparticles (MNPs), which provide a potential defense against antibiotic resistance. They display efficacy against bacteria that have already gained resistance and employ modes of action that are entirely different from those reported for conventional antibiotics [24]. However, they target several biomolecules that interfere with the emergence of resistant strains [25]. Particularly, MNPs have proven to offer broad-spectrum antibacterial effects on Gram-positive and negative bacteria. For instance, αFe2O3NPs and ZnO NPs display concentration-dependent antibacterial action against Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus [26,27].
We expected that the combination of metal nanoparticles such as ZnO@ αFe2O3 might exhibit a synergistic effect for an increased antibacterial effect where the reactive oxygen species (ROS) generation on the surfaces of alpha hematite Fe2O3 NPs along with bio-safe material with high catalytic and high photochemical activities of ZnO NPs. So, in this regard, we developed novel αFe2O3 nanoparticles exo situ the surface of ZnO nanoflowers by the coprecipitation of Zn2+ and Fe2+ via one reaction catalase in an easy and inexpensive method and loaded bee venom (Bv) as a bio-natural product from bees to combine both the advantages of combination nontoxic, antimicrobial, anti-biofilm properties and magnetic properties of Bv, ZnO nanoflowers, and αFe2O3 nanoparticles. After the process, the activity of the nanoflowers’ combination was investigated for targeting and eradicating multidrug-resistant fecal coliform biofilm isolates from nine sites throughout the effluent of wastewater treatment plants in Minya Governorate during January 2022 and February 2022.
2. Materials and Methods
2.1. Bee Venom Collection
The honeybee venom was collected from Carniolan hybrid Apis mellifera carnica. The commercial apiary in Motobes region, Kafr El-Sheikh Governorate, Egypt, during the active summer of 2022.
2.2. Bv-Collecting Device
The Bv-collecting electric shock device CJ 401 (Chung-Jin Biotech Ltd., Ansan, Republic of Korea) consists of a digital control board, five bee venom collection frames, wire electrodes, battery, input/output voltage: 12 VDC (timer ON: 3 s and timer-OF: 6 s). Finally, collector frames: 46 cm × 28 cm.
2.3. Bee Venom Collection
Honeybees were subjected to a bee venom-collecting electric shock device; there are five parallel wires of electrode-equipped Bv-collecting frames of the device. Every frame in a hive was put on top of the combs before being wired to an electro-stimulator. Bee workers are stimulated by electrical impulses to sting via a latex sheet that was put on a glass plate of the device frame. Bees that came in touch with the wires were mildly shocked by the electricity and stung the glass surface. The other bees were agitated and mobilized due to the alarm odor released by the venom, and they also began to sting. The fresh Bv frames were painstakingly packaged inside a unique container to ensure safe transmission to the laboratory throughout 24–48 h. A sharp scraper processed the dry Bv scraping under laboratory conditions.
2.4. Isolation of Fecal Coliform Bacteria from Treated Wastewater Samples
In this study, multidrug-resistant, fecal coliform isolates were isolated from nine sites throughout the effluent of wastewater treatment plants in Minya Governorate between January 2022 and February 2022. Seventy-six samples were collected into sterilized bottles and transferred immediately to the laboratory. Analyses were initiated as soon as possible after collection (not more than 24 h). All sample analysis steps were carried out under aseptic conditions. The membrane filter technique was used by placing the membrane filter on an M-FC liquid medium agar (Oxoid, Basingstoke, UK) to detect and enumerate thermo-tolerant (fecal) coliform. Then, typical blue colonies were streaked on a CHROMagar™ ESBL (CHROMagar 4, place du 18 Juin 1940, F-75006 Paris, France) plate to differentiate between different types of pathogens. Independent interpretations were given to each culture plate. The type of coloration or the colorless aspect of each of the colonies growing on CHROMagar™ ESBL was recorded. E. coli were given a dark pink to reddish color and Klebsiella was given a metallic blue (Scheme 1).
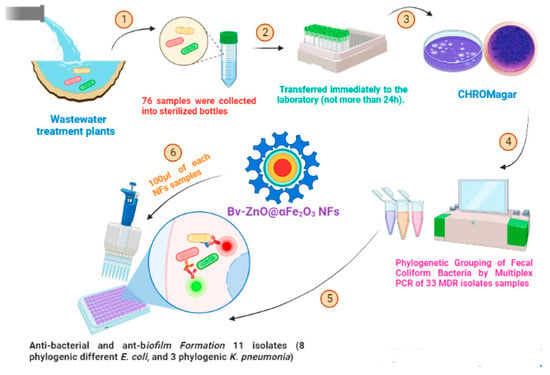
Scheme 1.
Illustration of the structure of Bv-ZnO@αFe2O3 NFs and the process of eradicating isolated bacteria.
2.5. Detection of Isolates’ Multidrug-Resistant Fecal Coliform
In all, 54 fecal coliform isolates were tested for antibiotic susceptibility using Kirby–Bauer Disc Diffusion Method [28]. The antimicrobials used were supplied by Bioanalyse®, Turkey and included Penicillins (Ampicillin AMP10 µg, Piperacillin PRL30 µg), Aminoglycosides (Gentamicin; CN 100 µg, Carbapenems (Meropenem; MEM 30 µg), Cephalosporins (Ceftazidime; CAZ 30 µg, Cefepime; FEP 30 µg, Cefpodoxime; CPD 10 µg, Cefotaxime; CTX 10 µg), Fluoroquinolones (Ciprofloxacin; CIP 5 µg, Ofloxacin; OFX 5 µg), Penicillins+β- lactamase inhibitors (Ampicillin-sulbactam SAM 20 µg), Sulphamethoxazole-trimethoprim; SXT 1.25/23.75 μg, Phenolics (Chloramphenicol; C 30 µg), and Monobactams (Aztreonam; ATM 10 µg) (Oxoid, UK). According to the Clinical and Laboratory Standards Institute, the zones of inhibition were determined, and the findings of the antimicrobial susceptibility test were classified as susceptible (S), intermediate (I), and resistant (R) (CLSI 2019) [29]. When an isolate was resistant to one antimicrobial agent in at least three separate classes, it was deemed MDR. The number of antibiotics the isolate is resistant to was divided by the total number of tested antibiotics to arrive at the multiple antibiotic resistance (MAR) index.
2.6. Phylogenetic Grouping of Fecal Coliform Bacteria by Multiplex PCR
Based on the presence or lack of three genes (chuA, yjaA, and tspE4.C2), triplex polymerase chain reaction (PCR) was used to classify the 33 MDR isolates into four separate phylogenetic groups (A, B1, B2, and D) and seven subgroups (A0, A1, B1, B22, B23, D1, and D2) as shown in Table S1 by using all the available combinations according to the technique of Clermont [30]. Phylogenetic grouping primers were documented in Table S2. In a 25 μL reaction mixture, multiplex PCR was accomplished using the following components: the master solution (12.5 μL, containing Tris-HCl (CAS 1185-53-1 EMPROVE® EXPERT), pH 8.5, (NH4)2SO4, 2 mM MgCl2, 0.2% Tween 20, 0.4 mM dNTPs, 0.2 units/L Ampliqon Taq DNA polymerase, and inert red dye), each primer (1 L from 10 M), nuclease-free water (2.5 μL), and template DNA (4 μL) [31]. The following settings were used for amplification as illustrated in Table S3. The PCR products were examined under UV transillumination by running the PCR products through 1.5% (w/v) agarose in Tris-Borate-EDTA (10X TBE) buffer (Genome Diagnostics Private Limited, Krishna Bhawan, Janak Puri, New Delhi, India) at 132 V for 20 min and seeing the results under UV transillumination.
2.7. Preparation of ZnO@αFe2O3 NFs
The coprecipitation technique utilized to synthesize ZnO@αFe2O3 NFs via a single catalase cross-link reaction may hold the most promise in simplicity and effectiveness. In brief, FeSO4 × 7H2O and ZnSO4 (Aladdin Chemical Reagent Company; Shanghai, China), were dissolved in 100 mL of distilled deionized water (DIH2O) in a 2:1 molar ratio and heated at 80 °C for 15 min. The iron and zinc solution was then quickly added to 100 mL−1 of 1.5 M NaOH and agitated at 1000 rpm for an hour at the same temperature. Eventually, the solution turned a hazy brown color. To eliminate any impurities or absorbed ions, the NFs were separated using a strong magnet and washed four times in DIH2O (Milli-Q water) and once in 0.01 M HCl. After that, the generated NFs were separated from the solution by centrifugation at 15,000 rpm for 5 min, which was subsequently dried in a fan oven at 60 °C overnight. Finally, DIH2O was used to dilute the NF solution to a total volume of 50 mL; unloaded ZnO, and αFe2O3 NPs were prepared according to our previously published papers with some modifications [26].
2.8. Bee Venom-Coated ZnO@αFe2O3 NFs
Following, 2.5% (w/v) of PVA (Shanghai, China) was coated onto 50 mL of ZnO@αFe2O3 NFs under vigorous magnetic stirring at 1000 rpm for 15 h at 25 to 27 °C (pH 7.2) according to the emulsion-coacervation method [32,33] with some modifications. Then, 20 mL−1 of 1.3% (w/v) of pure Bv in DIH2O was stirred for 10 min and added onto PVA-ZnO@αFe2O3 NFs and the mixture was vigorously stirred for 15 min at the same temperature previously. The surplus non-coated PVA polymers and all extra medicines were removed from the Bv-ZnO@αFe2O3 NFs by centrifugation at 7000 rpm for 15 min, many times, with additional ddH2O each time, and were dried in a vacuum for 24 h at 55–60 °C.
2.9. Characterizations of Prepared NFs
Fourier transform infrared (FTIR) spectra were acquired in KBr pellets using a Jasco FT-IR5300 model spectrophotometer. The surface of the material was investigated by scanning electron microscopy (SEM) (JSM 6390, JEOL DATUM Ltd., Tokyo, Japan) with electron energy-dispersive X-ray analysis (Carl Zeiss, Jena, Germany). The particle size and characterization of the samples were carried out by high-resolution transmission electron microscopy (TEM) (JCM-7000 Neo Scope™, Benchtop TEM; JEOL, Tokyo, Japan). The X-ray diffraction (XRD) patterns were obtained at ambient temperature using a Philips (XPERT MPD) X-ray diffractometer. The diffractometer used Cu Kα radiation with a voltage of 40 kV and a current of 45 mA. The scanning speed for the 2θ angle was set at 0.05°/min, spanning the range of 10–80°. The X-ray photoelectron spectroscopy (XPS) experiments were conducted using a VG ESCALAB MkII system equipped with an Al-Kα X-ray source and a 150 mm hemispherical analyzer. The analyzer had a pass energy of 20 eV. The calibration of the XPS system’s binding energy was performed by establishing the Au 4f7/2 peak at 84.0 eV [34], as the reference peak for all subsequent binding energy measurements. Before conducting the XPS experiment, the powder samples were subjected to pressing on an indium foil.
2.10. Cell Viability and Cytotoxicity Study of Prepared NFs
The viability and cytotoxic percentage assessment of Vero-derived kidney epithelia isolated from the green monkey’s cell line were evaluated with MTT. After incubation, the medium was evacuated from the wells. MTT was then added to each well at a concentration of 6 µg/mL−1. A 60 μL solution of dimethylsulfoxide (DMSO, Solarbio Science and Technology, Beijing, China) was used to spread the cells. Finally, via a microplate reader, the specimens’ absorbance spectra were measured at 595 nm:
where OD optical density test means the cells exposed to the αFe2O3 NPs, ZnO NFs, ZnO@αFe2O3 NFs, and Bv-ZnO@αFe2O3 samples; control means the control sample, and blank means the wells without Vero cells [35]. Log concentrations versus (%) cell viabilities were plotted with a logarithmic graph, which was then used to determine the IC50 values.
2.11. Antibacterial Susceptibility Assay of Prepared NFs
The susceptibility of 11 MDR E. coli and K. pneumonia isolates (representative to different phylogenetic groups) to four nanocomposites was assessed by agar well diffusion method [36,37]. Bacterial isolates were grown in BHI broth for an extended period, with the turbidity set at 5 × 105 CFU/mL−1. The nutrient agar medium was covered with the bacterial inoculum (Himedia, India). Using a micropipette, 0.1 mL−1 of a variety of NFs were distributed into wells with a diameter of 5 mm on plates. The wells were then incubated at 37 °C overnight. Each substance’s zone of inhibition was quantified.
2.12. Determination of MIC and MBC of NFs against Tested Isolates of Bacteria
Using the usual dilution approach, a broth microdilution assay was used to estimate the MIC in 96-multi-well microtiter plates (CLSI M07-A8). In order to obtain concentrations ranging from 512 to 2.5 g mL−1, 100 microliters of TSB (Himedia, Mumbai, India) were dispersed evenly across all wells. A volume of 100 microliters from each NF (1024–2.5 µg mL−1) was pipetted into the wells of the first row of the microtiter plate. Finally, 100 µL of freshly made, 0.5 McFarland matching turbid bacterial solution was put into each well. Each plate contained two columns that served as both positive and negative controls. Wrapped plates were incubated for 18–24 h at 37 °C. The plates were visually inspected for the presence or absence of turbidity against a dark background. The MIC was determined as the lowest concentration at which there was no discernible bacterial growth when compared to controls. By sub-culturing 20 µL from the clear wells of the MIC test on MHA, MBC was ascertained. The minimum amount of NFs (MBC) was needed to eradicate 99.9% of the original bacterial inoculum.
2.13. Screening of MDR Isolates for Biofilm Formation by Tissue Culture Plate Method
Eleven isolates (8 phylogenic different E. coli and 3 phylogenic K. pneumonia) isolated from nine sites throughout the effluent of wastewater treatment plants in Minya Governorate were screened for biofilm production by the Tissue Culture Plate Method [38]. Briefly, an overnight culture of each isolate was added to 10 mL−1 TSB supplemented with 1% filter sterile glucose (HiMedia, India), then it was incubated at 37 °C for 24 h. The culture was further diluted 1:100 with fresh medium; 200 µL of diluted culture was added into sterile 96-well flatbottomed microtiter plates. TCPs were incubated at 37 °C for 24 h. The blank wells were only inoculated with sterile TSB with 1% glucose. The experiment was conducted three times using the crystal violet staining method. The biofilms were measured, then content of the wells were removed, and the wells were washed twice with phosphate-buffered saline (PBS) (Solarbio, Beijing, China) to remove what was not attached, then the wells were air-dried for 45 min. The optical density (OD) was measured at 570 nm using an ELISA plate reader (Biorad, Hertfordshire, Watford, UK). The isolates were classified according to the calculated OD into non-biofilm producers (OD ≤ ODc), weak (ODc < OD ≤ 2ODc), moderate (2ODc < OD ≤ 4ODc), and strong producers (4ODc < OD) [39]. Three standard deviations (SD) over the mean OD of the negative control was established as the cut-off value (ODc).
2.14. In Vitro Evaluation of Anti-Biofilm Activity of NFs
The potential effect of 4 NFs on the inhibition of biofilm formation by E. coli isolate (E7) and K. pneumonia (K3) was assessed by a microplate biofilm assay. A microtiter plate with a U-bottom that has 96 wells and 100 µL of bacterial suspensions (108 CFU/mL−1) in each well was used. After incubating the microplates at 37 °C for 24 h, the final concentrations of the NFs were equal to the MIC, and the final volume was 200 µL per well. Three analyses were then carried out. The inoculum volume and NFs were changed to TSB and sterile water for the control wells. The microtiter plates’ contents were discarded, and the sides and bottom of the wells were carefully cleaned with phosphate-buffered saline (PBS, pH 7.2). This removed any free-floating, non-adherent bacterial cells. The microtiter plate wells were afterwards air-dried for 45 min. After drying, adherent “sessile” bacteria were fixed with sodium acetate (2%, w/v) (Merck, Germany), and the wells were then stained with crystal violet (0.1%, w/v), which was allowed to sit in the dark for 30 min. The extra color was then properly removed by washing the wells with sterile deionized water. The plates were then dried by air once more. Thermo Fisher Scientific’s multi-scan plate reader was used to measure absorbance at 570 nm after adding 200 µL of ethanol (95%, v/v) to each well. Inhibition of biofilm was determined from the formula described by [40,41]:
% Inhibition = 1 − (OD570 sample/OD570 control × 100)
2.15. Scanning Electron Microscopy (SEM)
Biofilm inhibition by different NFs was observed by SEM. The coverslips were put in 24-well flat-bottomed microtiter plates, and the NFs adjusted to MIC were added to the E. coli (E7) suspension 108 CFU, then incubated in static condition for 24 h. After a gentle PBS rinse, the coverslips were fixed with 2.5% glutaraldehyde (Sciences, Fort Washington, PA, USA) at 4 °C for two hours. Following glutaraldehyde fixation, the coverslips were cleaned with PBS for one hour and dehydrated using ethanol at increasing concentrations. Before being examined using SEM (JEOLJSM-7600F; JEOL Ltd., Tokyo, Japan), specimens were air-dried and coated with gold (Emmitech K550, Oldham, UK). The fixed cells were ethanol-dehydrated many times, air-dried, coated with gold-palladium, and analyzed under a SEM.
2.16. Statistical Analysis
One-way ANOVA analysis of variance was applied to test differences in biofilm inhibition between treated biofilms with different NFs followed by the Bonferroni test to examine post hoc significance. The differences between means were statistically significant at p < 0.01 and p < 0.05.
3. Results
3.1. Culture and Identification of Fecal Coliform Bacteria from Treated Wastewater Samples
Out of 54 effluents from wastewater treatment plants from nine sites in Minya Governorate, 33 (61.11%) effluent isolates were verified as positive cultures, of which 21 were confirmed to be E. coli (63.64%) and 12 K. pneumoniae (36.36%) (Figure 1).
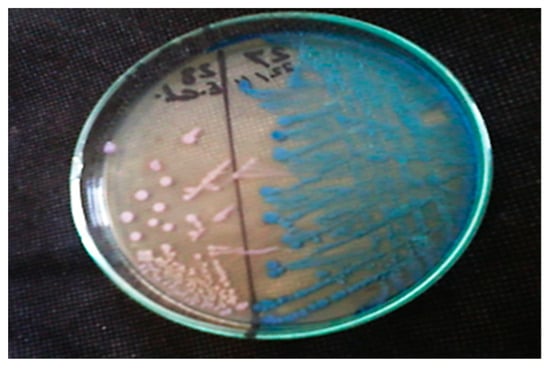
Figure 1.
Growth of two different isolated fecal coliform bacteria on CHROMagar™ ESBL medium; dark pink to reddish colonies represents E. coli and metallic blue colonies represent K. pneumoniae.
3.2. Detection of Antibiotic Susceptibility Pattern of Fecal Isolates
The rates of resistance to different antibiotics in 33 fecal coliform bacterial isolates from treated wastewater samples were 100% resistance for Ampicillin (AMP), Piperacillin (PRL), Ofloxacin (OFX) and Cefotaxime (CTX)), 75.9% for Ceftazidime (CAZ), 79.6% for Cefpodoxime (CPD), 85.2% for Cefepime (FEP). 77.8% for Aztreonam (ATM), 81.5% for Ampicillin/sulbactam (AMC), 64.3% for Sulphamethoxazole-trimethoprim (SXT), 29.6% for Chloramphenicol (C), 33.3% for Gentamicin (CN), 14.8% for Ciprofloxacin (CIP), and 0% for Meropenem (MEM) (Table 1). Of the treated wastewater isolates, 24 (72.73%) were MDR resistant to at least three distinct antimicrobial agents.

Table 1.
Antibiotic susceptibility patterns of fecal isolates.
3.3. Phylogenetic Grouping by Multiplex PCR
The results of the phylogenetic grouping of E. coli and K. pneumoniae were recorded in Table 2. For E. coli, the results showed that Group A compile 66.7%, Group B2, 23.8%, Group B1, 4.8%, and finally, Group D, 4.8%.

Table 2.
Phylogenetic analysis of 33 isolates (21 E. coli + 12 K. pneumoniae).
In our study, the largest group was Group A followed by Group B2, then Group B1 and Group D (Figure 2 and Figure 3). While in the case of K. pneumoniae, it showed that Group A compile 58.3%, Group B2, 33.3%, and finally, Group B1, 8.3%. In this study, the largest group was Group A followed by Group B2 and Group B1 (Figure 2 and Figure 4).
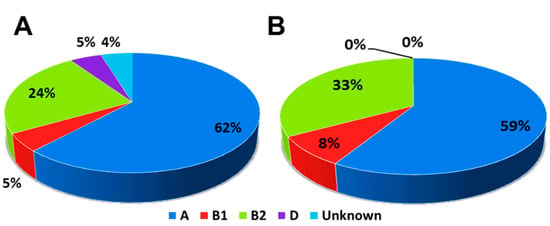
Figure 2.
Pie chart showing percentages of subgroups for (A) E. coli isolates and (B) K. pneumoniae isolates.
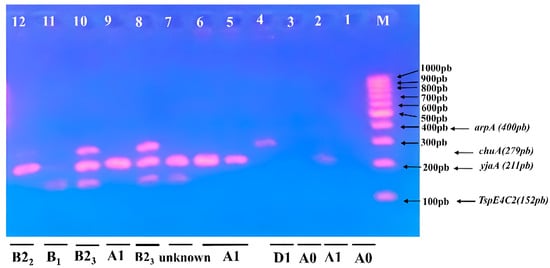
Figure 3.
Triplex PCR-based phylogenetic profile of E. coli isolates. Lane M contained a 100–1000 bp marker showing phylogenetic genes (arpA400 bp—chuA 279 bp—yjaA 211 bp—TspE4C2 152 bp); Lane 8 and 10, subgroup B23; Lane 12, subgroup B22; Lane 11, subgroup B1; Lane 4, subgroup D1; Lane 2, 5, 6, and 9 subgroup A1; Lane 1, 3, subgroup A0, and Lane 7 is unknown.
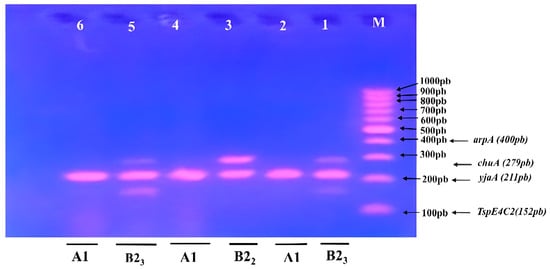
Figure 4.
Triplex PCR-based phylogenetic profile of K. pneumoniae isolates. Lane M contained a 100–1000 bp marker showing phylogenetic genes (arpA400 bp—chuA 279 bp—yjaA 211 bp—TspE4C2 152 bp); Lane 1 and 5, subgroup B23; Lane 3, subgroup B22; Lane 2, 4 and 6, subgroup A1.
3.4. Characterization of Prepared NFs
3.4.1. FTIR Spectra
The inverse spinel structure of the αFe2O3 NPs are shown in Figure 5A. FTIR shows an absorption band at 560 and 680 cm−1, which may be caused by the stretching vibration mode linked to the metal-oxygen absorption band (Fe-O bond). Wide and intense peaks between 1025 and 1406.11 cm−1 in Figure 5B indicate the stretching of the Zn-O bond. The H-C-H and C-O-C stretching vibration caused by PVA adsorbed on the surface of the magnetite NPs was verified by the peaks at 1050 cm−1. All samples showed a large absorption peak at 3420 cm−1, which may be attributable to the typical hydroxyl group (O-H) absorption. This revealed the presence of hydrogen bonding between the PVA polymer and the associated ZnO@αFe2O3 NFs (Figure 5C). Additionally, Figure 5D showed additional peaks at 1620, 1160, 1039, and 1018 cm−1 that correspond to C=C, C-OH, C-O, and C-N stretching bonds and are indicative of the peaks of Bv. The distinctive peaks shown in Figure 5E at 610 and 1360 cm−1 are the vibrational modes of the Fe-O and Zn-O bonds, respectively. The H-C-H vibration mode is shown by the peaks at 869 cm−1, which results from the influence of polymer molecules.
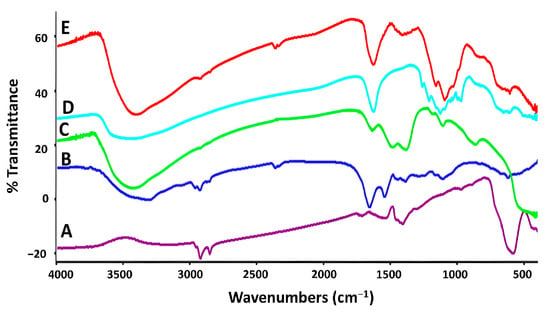
Figure 5.
FTIR spectra of bee venom honey natural product and prepared nanoparticles and nanoflowers: (A) αFe2O3 NPs, (B) ZnO NFs, (C) ZnO@αFe2O3 NFs, (D) Bv, and (E) Bv-ZnO@αFe2O3 NFs.
3.4.2. XRD and XPS
In order to evaluate the crystal phase and degree of crystallinity of the ZnO@α-Fe2O3 NFs in its initial state, the XRD pattern of the catalyst in its as-prepared form was provided. The XRD pattern of the ZnO@α-Fe2O3 NFs is shown in Figure 6A. ZnO@α-Fe2O3 exhibited a strong resemblance to the JCPDS 01-089-0596 pattern of α-Fe2O3 and the JCPDS 36–1451 pattern of ZnO. The ZnO@α-Fe2O3 NFs exhibited distinct and well-defined peaks in its crystallographic profile, indicating the presence of diffraction peaks at angles of 30.8°, 34.8°, 37.9°, 48.2°, 58.4°, 65.8°, 69.3°, 78.6°, 92.7°, and 96.1°. These peaks corresponded to certain crystallographic planes, namely, 100, 002, 101, 102, 110, 103, 112, 004, 202, and 104, respectively. The proposition is made that hematite was evenly distributed among the ZnO particles. The XPS spectra of ZnO@α-Fe2O3 NFs are shown in Figure 3. The ZnO@α-Fe2O3 composite is comprised of the elements zinc (Zn), iron (Fe), and oxygen (O). The resolution spectrum of ZnO@α-Fe2O3 NFs displayed two distinct binding energies at 1022.8 and 1040.7 eV, which were assigned to ZnO and Zn2+-O-Fe3+ species, respectively. Furthermore, the observed peaks corresponding to the Fe 2p energy level at 725.7 eV were ascribed to the presence of Fe3+-O-Zn2+ species. The analysis of the O1s high-resolution spectrum indicated the presence of binding energy around 540.3 eV, which might potentially be associated with α-Fe2O3 (Figure 6B).
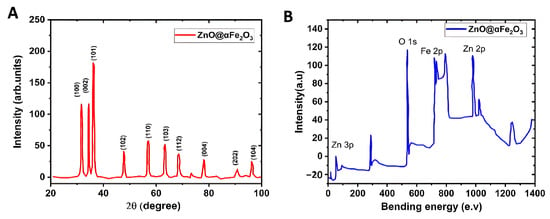
Figure 6.
(A) XRD pattern and (B) XPS spectra of ZnO@α-Fe2O3 NFs.
3.5. Surface Morphology Analysis
3.5.1. TEM and Selected Area Electron Diffraction (SAED)
TEM analysis was used to investigate the surface morphological structure further. The TEM pictures of Fe2O3 NPs, ZnO NFs, ZnO@αFe2O3 NFs, and Bv-ZnO@αFe2O3 NFs are illustrated in Figure 7A–D at a reduced magnification scale of 100 nm, and the SAED patterns are shown in Figure 7E. TEM findings showed the presence of different morphological surfaces of the prepared formulations’ obtained material. Figure 7A shows the largest observed structures of NPs: small particle sizes, spherical-like structures, and particles in an agglomerated condition with an aggregation width of about 9–15 nm of αFe2O3 NPs. However, the morphological structure of ZnO NFs demonstrated flower needle-shaped formations with a width of about 70–80 nm (Figure 7B). Flower-spherical nano-composites of ZnO@αFe2O3 NFs with 100–200 nm size are deposited on the surface of ZnO NPs’ needles (Figure 7C). TEM analysis confirmed that the catalyst reaction of ZnO@αFe2O3-loaded Bv NFs is stable and retains its original shape even after loading the Bv processes carried out with rapid mixing, with an increase in the particle size around 200–300 nm (Figure 7D). The SAED pattern in Figure 7E shows concentric rings of brilliant spots, indicating the preferred orientation and highly crystalline structure of ZnO@αFe2O3 NFs. In Figure 5E, the planes (220) and (311) correspond to the cubic inverse spinel structure of Fe2O3 NFs, whereas the planes (101), (102), (110), and (103) correspond to the wurtzite structure of ZnO. This supports the development of NPs and is consistent with the FTIR data.
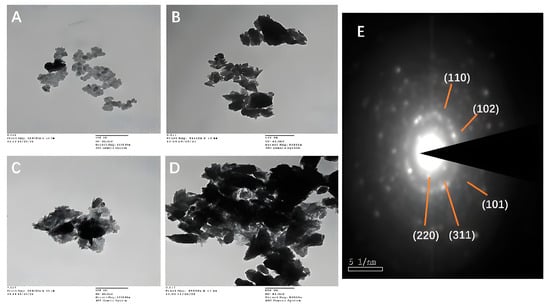
Figure 7.
Average particle size and data regarding size diameter mean of size distributions of prepared samples: (A) αFe2O3, (B) ZnO, (C) ZnO@αFe2O3, and (D) Bv-ZnO@αFe2O3 NFs measured by TEM (scale bar = 100 nm; direct magnifications = 80,000–10,000×), and (E) the SAED patterns of ZnO@αFe2O3 NFs.
3.5.2. SEM-EDX and HRTEM
The structural and morphological characteristics of ZnO@αFe2O3 were examined using SEM and HRTEM. Figure 8A displays the representative SEM image. The composite material consisting of ZnO@αFe2O3 had a layered structure with irregularities and small particles on the surface. These characteristics are believed to be a result of the formation of oxide phases by the calcination of sulphate. Figure 8B shows the findings of an SEM-EDX analysis that revealed the presence of iron at 40.04%, zinc at 10.16%, and oxygen at 49.8%. A prominent peak at 6.54 keV was also seen, indicating the presence of iron (Fe), while peaks at 0.483 keV and 1.283 keV were connected to the presence of oxygen (O) and zinc (Zn), respectively. Several elements identified by SEM-EDX, such as Au, Si, and Cl, serve as capping agents for biogenic ZnO and Fe2O3 NPs, and have previously been demonstrated. The HRTEM image of the ZnO@αFe2O3 sample revealed distinct lattice fringes of ZnO particles. These fringes exhibit an interplanar spacing of 0.247 nm, as shown in Figure 7C. This spacing corresponds to the (110) facet, which is consistent with the findings obtained from XRD analysis. Likewise, the discernible lattice fringes shown by αFe2O3, with an interplanar spacing of 0.492 nm, are indicative of the (102) facet, as confirmed by XRD (Figure 8C).
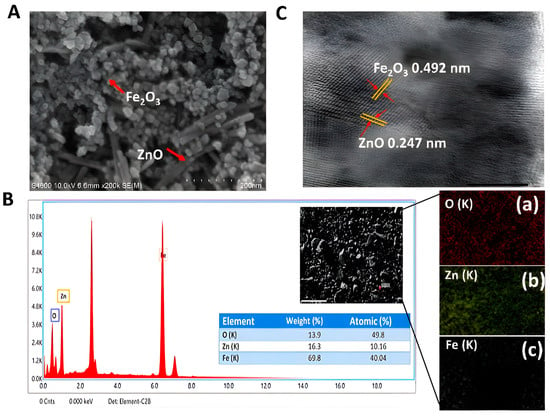
Figure 8.
(A) SEM image (magnification 200 nm), (B) SEM-EDX microphotographs, (C) HRTEM images, and (a–c) the elemental mapping images of ZnO@αFe2O3 composite.
3.6. Cell Viability and Cytotoxicity of NPs and NFs
The capacity of Bv and αFe2O3 NPs, ZnO NFs, and Bv- ZnO@αFe2O3 NFs to block normal Vero cells with concentrations ranging between 31.25 and 1000 µg mL−1 for 24 h at a temperature of 37 °C, at a rate of three replicates for each concentration, as well as the control sample without treatment for comparison. Calculating the percentages of growth inhibition rate compared to the control (100% growth), the amount of the toxicological impact was also assessed, as shown in Figure 8. Toxicity (%) of Bv, αFe2O3 NPs, ZnO NFs, and Bv-ZnO@αFe2O3 against Vero cells was dose-dependent with the concentrations. The toxicity present in Bv was similar up to 62.5 µg mL−1 (IC50, 72.17 µg mL−1). While in αFe2O3 NPs and ZnO NFs, the toxicity present was similar up to 250 and 125 µg mL−1 where the toxicity reached 85.11% (IC50, 185.45 µg mL−1) and 65.24% (IC50, 104.69 µg mL−1), respectively. The toxicity of Bv-ZnO@αFe2O3 NFs was comparable up to 125 µg mL−1, when it reached 64.79% (IC50, 107.18 µg mL−1). Further analysis of Bv > αFe2O3 NPs > ZnO NFs > Bv-ZnO@αFe2O3 NFs revealed that the lowest amount of cytotoxic activity against Vero cells was present at concentrations < 250 µg mL−1, and that the maximum toxicity was present at concentrations > 250 µg mL−1 (Figure 9A). Additionally, the morphological characteristics of Vero cells exposed to various concentrations of Bv, αFe2O3 NPs, ZnO NFs, and Bv-ZnO@αFe2O3 NFs were reported compared to untreated cells. Using the absorbance values revealed after the red dye was captured, and the pertinent concentrations of the Bv, αFe2O3 NPs, ZnO NFs, and Bv-ZnO@αFe2O3 NFs utilized in the cytotoxicity tests, the 3T3 Phototox program was used to determine the IC50 of generated samples in various cell lines. The treated Vero cells displayed notable differences from the control and morphological changes in cells at concentrations of 125 and 500 µg mL−1 of the samples of NPs and NFs, cells’ destructuration were evident and had distinguishable morphological signs of apoptosis (Figure 9B), and morphological alterations were evident in a concentration-dependent manner in the case of αFe2O3 NPs, αFe2O3 NPs, and ZnO NFs’ treatment. Furthermore, Bv-treated cells were more damaged than NPs and NFs-treated cells, which were less homogeneous with a loss of membrane integrity, rounding, and shrinkage but remained intact at 31.25 and 62.50 µg mL−1 (Figure 9B).
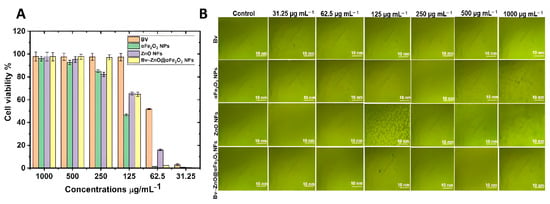
Figure 9.
(A) Cell viability and (B) morphological features of Vero cells treated with different concentrations of Bv, αFe2O3 NPs, ZnO NFs, and Bv-ZnO@αFe2O3 NFs.
3.7. Antibacterial Susceptibility Assay of NPs and NFs
Antibacterial activity analysis of four different NPs and NFs (ZnO, αFe2O3, ZnO@αFe2O3, and Bv-ZnO@αFe2O3 NFs) was performed by the aforementioned agar well diffusion method. As demonstrated in the results in Table 3, the ZnO NFs displayed greater antibacterial activity with the zone of inhibition (2 cm) against E. coli isolate E1 followed by K. pneumonia K1 (1.8 cm). The αFe2O3 NPs showed the zone of inhibition (4 cm) against E7 followed by E5 (3.5 cm). ZnO@αFe2O3 NFs’ zone of inhibition was highest in the E7 isolate (2.6 cm) and then (2.2 cm) in the E3 isolate. Maximum inhibition of Bv-ZnO@αFe2O3 NFs was found in K3 (4.5 cm) followed by E1 (3.8 cm) (Table 3).

Table 3.
Zone of inhibition of different E. coli and K. pneumonia and biofilm production.
3.8. MIC and MBC Determination of NFs
The antibacterial activity of NFs was assessed by measuring MICs and MBCs for two strong biofilm producer isolates E7 and K3. Table 4 showed that E7 MICs’ values were 320 µg/mL−1, 80 µg/mL−1, 160 µg/mL−1, and 10 µg/mL−1 while MBCs’ values were 100 µg/mL−1, 40 µg/mL−1, 97 µg/mL−1, and 20 µg/mL−1 for ZnO, αFe2O3, ZnO@αFe2O3, and Bv-ZnO@αFe2O3, respectively; on the other hand, K3 isolate MICs were 160 µg/mL−1, 40 µg/mL−1, 80 µg/mL−1, and 5 µg/mL−1 while MBCs were 80 µg/mL−1, 10 µg/mL−1, 40 µg/mL−1, and 2.5 µg/mL−1 for ZnO, αFe2O3, ZnO@αFe2O3, and Bv-ZnO@αFe2O3, respectively.

Table 4.
MICs and MBCs of different NPs and NFs against E. coli (E7) and K. pneumonia (K3).
3.9. Biofilm Formation of Isolates and Anti-Biofilm Activity of NPs and NFs
The results clarified that all the MDR examined isolates (E. coli and K. pneumonia) were biofilm producers: 27.3% (3/11) were weak biofilm producers, 27.3% (3/11) were strong biofilm producers, and 45.5% (5/11) were strong biofilm producers. The biofilm inhibitory activity of the four used NFs, ZnO, αFe2O3, ZnO@αFe2O3, and Bv–ZnO@αFe2O3NFs, showed a highly significant reduction (p < 0.001) in treated biofilms with ZnO@αFe2O3 and Bv-ZnO@αFe2O3 (78.16 ± 0.86 and 79.13 ± 1.26), respectively, compared with ZnO and αFe2O3 NFs-treated biofilms (25.13 ± 1.42 and 26.63 ± 2.04), respectively, for the E7 isolate. Also, for K. pneumonia (K3), there was a highly significant reduction (p < 0.001) in treated biofilms with ZnO@αFe2O3 and Bv–ZnO@αFe2O3 NFs (79.06 ± 1.55 and 87.30 ± 1.11), respectively, compared with ZnO and αFe2O3 NFs-treated biofilms (27.70 ± 1.60 and 30.36 ± 1.22) (Figure 10).
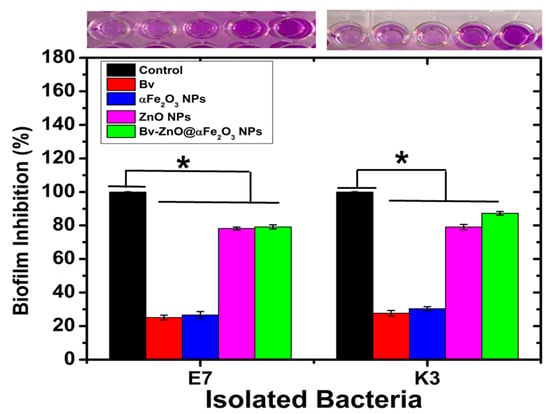
Figure 10.
The biofilm inhibition of MDR E. coli E7 and K. pneumonia K3 treatment with prepared NPs and NFs. Statistical differences using one-way ANOVA (* p < 0.05).
3.10. Biofilm Inhibition for E. coli by SEM
To observe the biofilm inhibition by different NPs and NFs, SEM was applied for E. coli (E7). SEM images indicated that the control group’s bacterial cells had clearly developed biofilms with matrix material (Figure 11A). On the contrary, the presence of MIC of ZnONFs and αFe2O3NPs (Figure 11B,C), respectively, caused the amount of biofilm to decrease, while there was an obvious reduction in the case of treatment with ZnO@αFe2O3 and Bv–ZnO@αFe2O3 NFs (Figure 11D,E), respectively. The results of the anti-biofilm activity were supported by SEM pictures.
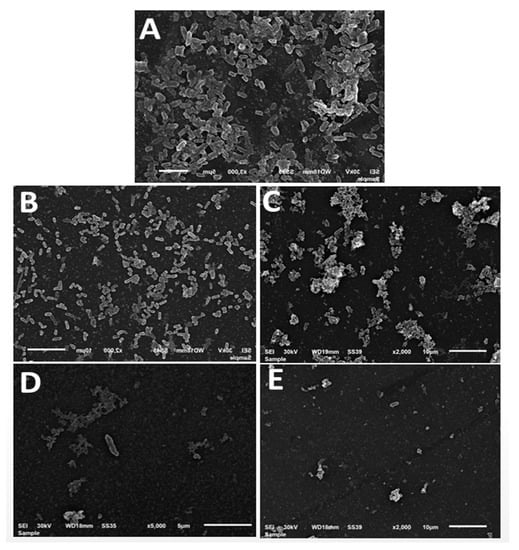
Figure 11.
Scanning electron microscope visualization of E. coli (E7) biofilms treated by differently prepared samples: (A) Control (B) ZnO NFs, (C) αFe2O3, (D) ZnO@αFe2O3NFs NFs, and (E) Bv-ZnO@αFe2O3NFs (scale bar = 5 and 10 nm).
4. Discussion
MDR is becoming more prevalent worldwide, drastically reducing therapy options. Future control of MDR Gram-negative bacilli around the world needs to be highly considered. In this study, about 72.73% isolates of treated wastewater isolates were MDR with resistance to one antimicrobial agent in at least three different groups. Indeed, most of the previous studies focused on E. coli, as a result of using it as an indicator of fecal contamination and to evaluate the spread of antimicrobial resistance (AMR (within the aquatic environment [42]. Studies provided little information on antibiotic-resistant heterotrophic and non-coliform bacteria [43]. These microorganisms can be found in activated sludge and are vital for the emergence and spread of antibiotic resistance (AMR) [44,45]. In this study, about 63.64% were confirmed to be E. coli, which is higher than the previous study by Haberecht et al., who reported that the percentage of total detected bacteria, E. coli, was 37% in wastewater [46]. These elevations in E. coli in wastewater were described in previous studies as being due to the proximity of fecal contamination from humans and animals [47,48,49,50]. However, as reported in previous studies, K. pneumoniae strains were considered to have come from hospitals and receiving wastewater treatment plants [51,52,53]. This theory explains why this study’s treated wastewater contained 36.36% more K. pneumoniae than expected. In agreement with that study, Johnson and Stell presented phylogenetic grouping data that showed isolates in phylogenetic Groups A and B1 which were typically commensal strains, while subgroups D and B2 were related to extra intestinal illnesses [45,54]. In our study, the results of phylogenetic grouping in E. coli isolates showed that the largest group was Group A followed by Group B2, then Group B1 and Group D. In addition, in K. pneumoniae isolates, the largest group was Group A followed by Group B2 and Group B1. The phylogenetic subtype distribution of the MDR strains from treated wastewater, close to the average distribution of total isolates, suggests that the environmental water pathogen might originate from both humans and animals, concurrent with previous studies [55,56].
For simplicity and efficiency, the coprecipitation process used to synthesize ZnO@αFe2O3 NFs via one catalase cross-link reaction could be the most promising [57]. An XRD and XPS confirmed the success of synthesized ZnO@αFe2O3 NFs compared to the previous studies [58,59,60], and FTIR analysis revealed functional groups such as C=C, C-OH, C-O, H-C-H, and C-N. These findings were consistent with prior studies [61,62,63,64]. The existence of a protective organic covering on the NFs’ surface generated by Bv was revealed by TEM, adding to previously published evidence on the involvement of active chemicals and other organic components in preventing aggregate formation [26]. Additionally, TEM, HRTEM, and SEM-EDX analysis demonstrated that the ZnO@αFe2O3- loaded Bv NFs’ catalyst reaction is stable and maintains its original shape even after loading the Bv processes, which are carried out with rapid mixing and result in an increase in particle size of about 200–300 nm; the apparent size increase may be attributable to the success of the Bv-loaded process. Concentric rings of bright spots in the selected area electron diffraction (SAED) pattern attest to the preferred orientation and extremely crystalline structure of ZnO@αFe2O3 NFs [65,66]. The size and shape of NPs affect their reactivity, surface area, rate of absorption into microbial cells, cell surface contact, protein binding, and other features; these factors are crucial for antibacterial applications. The findings show that the proposed nanoparticles of sizes between 100 and 200 nm were sufficiently tiny to fulfill the steric requirement for fast diffusion across the bacterial outer membrane [67]. These results confirmed the successful one-reaction catalase of prepared ZnO@αFe2O3 NFs. It was also found that the bio-coating shell process for ZnO@αFe2O3NPs by Bv honey product did not affect the phase change of formulations. In the current study, the antimicrobial effect of ZnO, Bv-ZnO, ZnO@αFe2O3, and Bv–ZnO@αFe2O3 on 11 isolates (8 phylogenic different E. coli and 3 K. pneumonia) isolated from nine sites throughout the effluent of wastewater treatment plants in Minya Governorate was assessed.
The antibacterial activity was evaluated by observing the inhibition zones as shown in Table 3. This technique makes dispersible nanoparticles (NPs) come into direct contact with the bacteria. The zone’s diameter was shown to rise in opposition to E. coli and K. pneumonia in the case of Bv-ZnO and Bv-ZnO@αFe2O3 as the NC has substantially greater antibacterial efficacy compared to ZnO@αFe2O3 and bare ZnO. The bactericidal effect of ZnO is owed to ROS formation on the surface, damaging the bacterial membrane [68]. The antibacterial activity of Fe2O3 depends on the presence of Fe2+/Fe3+ cations on the particle surface, which has catalytic activity towards the reduction of H2O2 to produce ROS [69], which is an oxidative stress required for bacterial cell damage. Additionally, the interaction between positive and negative charges on Fe2O3 and the bacterial surface plays a crucial role in bacterial death [70]. The results of our study are supported by previous studies [67,71], which examined the higher antibacterial activity of metal oxides on Gram-positive and Gram-negative bacteria, including E. coli and K. pneumonia, as compared to ZnO monoxide. Also, there was a notable synergistic antimicrobial effect of iron oxide nanoparticles loaded with Bv [72].The antibacterial characteristics of Bv, which interact with bacterial cell membrane components and efficiently harm bacterial cells, may be influenced by peptides, enzymes, physiologically active amines, and non-peptide components [73,74].
Thus, the synergistic effect between the three substances Bv, ZnO, and αFe2O3 explains the increased inhibition zones observed after using the NFs (Bv-ZnO and Bv-ZnO@αFe2O3) in all examined samples. Also, that combination helps in reducing toxicity and immunogenicity, allowing their introduction into wastewater as a treatment option. Hence, the widespread application of nanocomposites is mainly determined by their safety in the environment. According to the results in Table 4, all used NPs and NFs proved to have MIC and MBC activities against E. coli isolate (E7) and K. pneumonia isolate (K3); this finding aligns with those attained by [75,76,77], which postulated that a variety of elements, including the technique of manufacture, particle size, and bacterial strains, might affect the bactericidal activity of the employed nanocomposites. Previous investigations [78,79] revealed BV’s potent antibacterial action against K. pneumonia and E. coli supported our work. The bactericidal action of bee venom against Gram-positive and Gram-negative bacteria was also established in earlier research in Korea [80,81].
Biofilms continue to grow and act as a type of crust to protect the microbes underneath. The crust increases bacterial resistance to the effects of the chemicals, physical agents, and disinfectants employed in the wastewater treatment process [82]. The aggregation of Bv peptides can interact with bacterial membranes, producing ion channels and toroidal pores, increasing the permeability of the cytoplasmic membrane of the bacterial cell, and causing its damage. This distinctive intervention system allows antimicrobial peptides to damage the bacteria at different stages [83]. They may also help other substances, such as antibiotics and nanoparticles, get inside the cell, producing significant bactericidal effects with their mechanisms [84,85].
In this work, biofilm inhibitory activity varied between the used NPs and NFs. From the aforementioned results shown in Figure 6, it was found that ZnO@αFe2O3 and Bv-ZnO@αFe2O3 had a solid potential to inhibit biofilm activity by 78.16 ± 0.86 and 79.13 ± 1.26 against E. coli isolate (E7) and by 79.06 ± 1.55 and 87.30 ± 1.11 for K. pneumonia isolate (K3). The increased anti-biofilm effect produced in the case of Bv-ZnO@αFe2O3 may be owed to many reasons, as Bv caused a significant increase in anti-biofilm activity of the used substances; these findings are supported by Dosler et al. [86], who found that Bv melittin caused the inhibition of biofilm formation by sub-MIC concentrations. Picoli et al. found low biofilm production by E. coli isolate (56.3%) due to Bv melittin [87]. The effect of BV is to eliminate biofilm in Gram-positive and Gram-negative bacteria [88,89]. Furthermore, αFe2O3NPs have a less significant antibacterial activity than its significant anti-biofilm potential as it facilitates the delivery of different antimicrobial agents conjugated with it into the biofilm by its magnetic field. [88,90,91]. Bare ZnO NFs against the E7 isolate showed inhibition in biofilm formation by 25.13 ± 1.42, which is similar to the findings of [92]. As biofilm in this study was inhibited by 20% in E. coli isolates after using ZnO NFs, Applerot et al. mentioned a higher inhibitory effect of ZnO NFs in their study than ours. This difference is owed to many factors as different methods are used for the evaluation of biofilm formation, bacterial strain, and NFs’ size as the greater antibacterial action depends on the size of the NFs since the surface area to volume ratio rises with decreasing particle size [93]. ZnO NFs have the ability to inhibit biofilm relay on DapE protein enzymatic activity inhibition that shows protein is vital for the synthesis of peptidoglycans, affecting biofilm formation at its initial stage [94]. Thus, combining the three substances in the Bv-ZnO@αFe2O3 NFs is considered a good candidate for application in controlling fecal coliforms isolated from wastewater compared to other methods.
There are some limitations in this work that need additional investigation. Firstly, it is necessary to examine the in vitro releasing profile of Bv from Bv-ZnO@Fe2O3. Furthermore, due to limited financial resources, we were unable to conduct a comparative investigation of the developed formulations with typical bare ZnO@αFe2O3 NFs using X-ray diffraction (XRD) and X-ray photoelectron spectroscopy (XPS), which are widely recognized as the most effective methods for physiochemical characterization of various components. Furthermore, it is advisable to conduct in vivo experiments in order to validate the effectiveness of the formulations we have developed. Additionally, we recommend comparing the effectiveness of Bv-conjugated nanoflowers in the removal of MDR bacteria with other conventional techniques previously applied in WWTPs, such as chlorination and ozonation, or even ultraviolet radiation in future studies.
5. Conclusions
The obtained results in this study showed high levels of MDR fecal coliform isolated from treated wastewater samples. Meanwhile, using ZnO, αFe2O3, and ZnO@αFe2O3 and especially, Bv-ZnO@αFe2O3 NFs assisted in creating effective anti-biofilm activity and reducing the related biological health risk for isolated K. pneumonia and E. coli isolates. These qualities make them suitable for use as antimicrobials, eliminate the threat of antimicrobial resistance, and serve as a good option for application in controlling fecal coliforms isolated from wastewater compared to other methods. Also, they can be used as a pre-treatment and disinfection technique for wastewater. Additional research based on the bioactivities of such biosynthesized NFs will focus on the antibacterial impact to combat the latest wave of various multi-resistant pathogens.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microbiolres14040102/s1, Table S1. Combination of chuA, yjaA, and TspE4.C2 Genes for Determination of Phylogenetic Subgroups of E. coli isolates; Table S2. Primers of phylogenetic groups used in PCR; Table S3. Multiplex PCR Protocols were described for determination of different Phylogenetic Subgroups of fecal coliform isolates.
Author Contributions
Conceptualization, M.S., E.J.M., M.A. and M.F.S.; methodology, M.S., E.J.M., E.M.F., A.A.A., A.A., A.M.A., A.A.Z., S.A.Z., A.E., M.A. and M.F.S.; software, A.M.A., A.A.Z., S.A.Z., A.E., M.A. and M.F.S.; validation, M.S., E.J.M., E.M.F., A.A.A., A.A., A.M.A., A.A.Z. and S.A.Z.; formal analysis, M.S., E.J.M., E.M.F., A.A.A. and A.A.; resources, A.A., A.M.A., A.A.Z., S.A.Z., A.E., M.A. and M.F.S.; data curation, M.S., E.J.M., E.M.F., A.A.A., A.A., A.M.A., A.A.Z., S.A.Z., A.E., M.A. and M.F.S.; writing—original draft preparation M.S. and E.J.M.; writing—review and editing, M.S., E.J.M., E.M.F., A.A.A., A.A., A.M.A., A.A.Z., S.A.Z., A.E., M.A. and M.F.S.; visualization, M.S., E.J.M., E.M.F., A.A.A., A.A. and M.F.S.; supervision M.S., S.A.Z., A.E., M.A. and M.F.S.; project administration, M.S., A.E., M.A. and M.F.S.; funding acquisition, A.A.Z., S.A.Z., A.E., M.A. and M.F.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We express appreciation and thanks to Mohammed Gaber Taha, Hany Mohamed, and Mohamed Mabrouk, Biochemistry Department, Faculty of Agriculture, Al Azhar University, Egypt, who supported and encouraged us throughout this study. We also extend our gratefulness for the Princess Nourah bint Abdulrahman University Researchers Supporting Project, number PNURSP2023R182, Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Escobar-Páramo, P.; Le Menac’h, A.; Le Gall, T.; Amorin, C.; Gouriou, S.; Picard, B.; Skurnik, D.; Denamur, E. Identification of forces shaping the commensal Escherichia coli genetic structure by comparing animal and human isolates. Environ. Microbiol. 2006, 8, 1975–1984. [Google Scholar] [CrossRef] [PubMed]
- Aarestrup, F.M.; Wegener, H.C.; Collignon, P. Resistance in bacteria of the food chain: Epidemiology and control strategies. Expert Rev. Anti-Infect. Ther. 2008, 6, 733–750. [Google Scholar] [PubMed]
- Alsanie, W.F.; Felemban, E.M.; Farid, M.A.; Hassan, M.M.; Sabry, A.; Gaber, A. Molecular identification and phylogenetic analysis of multidrug-resistant bacteria using 16S rDNA sequencing. J. Pure Appl. Microbiol. 2018, 12, 489–496. [Google Scholar] [CrossRef]
- Tesh, S.J.; Scott, T.B. Iron nanoparticles for water treatment: Is the future free or fixed? Iron oxides: From nature to applications. In Iron Oxides: From Nature to Applications; Wiley-VCH: Weinheim, Germany, 2016; pp. 473–522. [Google Scholar]
- Badshah, I.; Mustafa, N.; Khan, R.; Mashwani, Z.-U.-R.; Raja, N.I.; Almutairi, M.H.; Aleya, L.; Sayed, A.A.; Zaman, S.; Sawati, L.; et al. Biogenic Titanium Dioxide Nanoparticles Ameliorate the Effect of Salinity Stress in Wheat Crop. Agronomy 2023, 13, 352. [Google Scholar]
- Shin, S.-H.; Ye, M.-K.; Choi, S.-Y.; Park, K.-K. The effects of melittin and apamin on airborne fungi-induced chemical mediator and extracellular matrix production from nasal polyp fibroblasts. Toxins 2017, 9, 348. [Google Scholar] [CrossRef] [PubMed]
- Leandro, L.F.; Mendes, C.A.; Casemiro, L.A.; Vinholis, A.H.; Cunha, W.R.; Almeida, R.D.; Martins, C.H. Antimicrobial activity of apitoxin, melittin and phospholipase A 2 of honey bee (Apis mellifera) venom against oral pathogens. An. Acad. Bras. Ciências 2015, 87, 147–155. [Google Scholar] [CrossRef]
- Socarras, K.M.; Theophilus, P.A.; Torres, J.P.; Gupta, K.; Sapi, E. Antimicrobial activity of bee venom and melittin against Borrelia burgdorferi. Antibiotics 2017, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Yacoub, T.; Rima, M.; Karam, M.; Sabatier, J.-M.; Fajloun, Z. Antimicrobials from venomous animals: An overview. Molecules 2020, 25, 2402. [Google Scholar] [CrossRef]
- Neves, R.C.D.; Mortari, M.R.; Schwartz, E.F.; Kipnis, A.; Junqueira-Kipnis, A.P. Antimicrobial and antibiofilm effects of peptides from venom of social Wasp and scorpion on multidrug-resistant Acinetobacter baumannii. Toxins 2019, 11, 216. [Google Scholar] [CrossRef]
- Purushothaman, S.; Nair, R.R.; Harikrishnan, V.; Fernandez, A. Temporal relation of cardiac hypertrophy, oxidative stress, and fatty acid metabolism in spontaneously hypertensive rat. Mol. Cell. Biochem. 2011, 351, 59–64. [Google Scholar] [CrossRef]
- Weis, W.A.; Ripari, N.; Conte, F.L.; da Silva Honorio, M.; Sartori, A.A.; Matucci, R.H.; Sforcin, J.M. An overview about apitherapy and its clinical applications. Phytomed. Plus 2022, 2, 100239. [Google Scholar]
- Pereira, A.F.M.; Albano, M.; Alves, F.C.B.; Andrade, B.F.M.T.; Furlanetto, A.; Rall, V.L.M.; Santos, L.D.D.; Orsi, R.D.O.; Junior, A.F. Influence of apitoxin and melittin from Apis mellifera bee on Staphylococcus aureus strains. Microb. Pathog. 2020, 141, 104011. [Google Scholar] [CrossRef] [PubMed]
- Carpena, M.; Nuñez-Estevez, B.; Soria-Lopez, A.; Simal-Gandara, J. Bee venom: An updating review of its bioactive molecules and its health applications. Nutrients 2020, 12, 3360. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Liu, Y.; Ye, Y.; Wang, X.-R.; Lin, L.-T.; Xiao, L.-Y.; Zhou, P.; Shi, G.-X.; Liu, C.-Z. Bee venom therapy: Potential mechanisms and therapeutic applications. Toxicon 2018, 148, 64–73. [Google Scholar]
- Kocyigit, A.; Guler, E.M.; Kaleli, S. Anti-inflammatory and antioxidative properties of honey bee venom on Freund’s Complete Adjuvant-induced arthritis model in rats. Toxicon 2019, 161, 4–11. [Google Scholar]
- Sobral, F.; Sampaio, A.; Falcão, S.; Queiroz, M.J.R.; Calhelha, R.C.; Vilas-Boas, M.; Ferreira, I.C. Chemical characterization, antioxidant, anti-inflammatory and cytotoxic properties of bee venom collected in Northeast Portugal. Food Chem. Toxicol. 2016, 94, 172–177. [Google Scholar] [CrossRef]
- Han, S.; Yeo, J.; Baek, H.; Lin, S.-M.; Meyer, S.; Molan, P. Postantibiotic effect of purified melittin from honeybee (Apis mellifera) venom against Escherichia coli and Staphylococcus aureus. J. Asian Nat. Prod. Res. 2009, 11, 796–804. [Google Scholar] [CrossRef]
- Debnath, S.; Sharma, D.; Chaudhari, S.Y.; Sharma, R.; Shaikh, A.A.; Buchade, R.S.; Kesari, K.K.; Abdel-Fattah, A.M.; Algahtani, M.; Mheidat, M.; et al. Wheat ergot fungus-derived and modified drug for inhibition of intracranial aneurysm rupture due to dysfunction of TLR-4 receptor in Alzheimer’s disease. PLoS ONE 2023, 18, e0279616. [Google Scholar] [CrossRef]
- Akash, S.; Baeza, J.; Mahmood, S.; Mukerjee, N.; Subramaniyan, V.; Islam, M.R.; Gupta, G.; Rajakumari, V.; Chinni, S.V.; Ramachawolran, G.; et al. Development of a new drug candidate for the inhibition of Lassa virus glycoprotein and nucleoprotein by modification of evodiamine as promising therapeutic agents. Front. Microbiol. 2023, 14, 1206872. [Google Scholar] [CrossRef]
- Rayaprolu, B.M.; Strawser, J.J.; Strawser, G. Anyarambhatla, i. pharmacy, Excipients in parenteral formulations: Selection considerations and effective utilization with small molecules and biologics. Drug Dev. Ind. Pharm. 2018, 44, 1565–1571. [Google Scholar] [CrossRef]
- Vargason, A.M.; Anselmo, A.C.; Mitragotri, S. The evolution of commercial drug delivery technologies. Nat. Biomed. Eng. 2021, 5, 951–967. [Google Scholar] [CrossRef] [PubMed]
- Gong, D.; Celi, N.; Zhang, D.; Cai, J. Magnetic biohybrid microrobot multimers based on chlorella cells for enhanced targeted drug delivery. ACS Appl. Mater. Interfaces 2022, 14, 6320–6330. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Sharaf, M.; Ahmed, I.; Khan, T.U.; Shabana, S.; Arif, M.; Kazmi, S.S.U.H.; Liu, C. Potential utility of nano-based treatment approaches to address the risk of Helicobacter pylori. Expert Rev. Anti-Infect. Ther. 2022, 20, 407–424. [Google Scholar] [CrossRef]
- Slavin, Y.N.; Asnis, J.; Häfeli, U.O.; Bach, H. Metal nanoparticles: Understanding the mechanisms behind antibacterial activity. J. Nanobiotechnol. 2017, 15, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Alshawwa, S.Z.; Mohammed, E.J.; Hashim, N.; Sharaf, M.; Selim, S.; Alhuthali, H.M.; Alzahrani, H.A.; Mekky, A.E.; Elharrif, M.G. In Situ Biosynthesis of Reduced Alpha Hematite (α-Fe2O3) Nanoparticles by Stevia Rebaudiana L. Leaf Extract: Insights into Antioxidant, Antimicrobial, and Anticancer Properties. Antibiotics 2022, 11, 1252. [Google Scholar] [CrossRef]
- Javidi, S.; Nafchi, A.M.; Moghadam, H.H. Synergistic effect of nano-ZnO and Mentha piperita essential oil on the moisture sorption isotherm, antibacterial activity, physicochemical, mechanical, and barrier properties of gelatin film. J. Food Meas. Charact. 2022, 16, 964–974. [Google Scholar] [CrossRef]
- Bauer, A. Antibiotic susceptibility testing by a standardized single disc method. Am. J. Clin. Pathol. 1966, 45, 149–158. [Google Scholar] [CrossRef]
- Kahlmeter, G.; Giske, C.G.; Kirn, T.J.; Sharp, S.E. Point-counterpoint: Differences between the European Committee on Antimicrobial Susceptibility Testing and Clinical and Laboratory Standards Institute recommendations for reporting antimicrobial susceptibility results. J. Clin. Microbiol. 2019, 57, e01129-19. [Google Scholar] [CrossRef]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef]
- Singla, M.; Gupta, S.; Porwal, O.; Binjawhar, D.N.; Sayed, A.A.; Mittal, P.; El-Demerdash, F.M.; Algahtani, M.; Singh, S.K.; Dua, K.; et al. Theoretical design for covering Engeletin with functionalized nanostructure-lipid carriers as neuroprotective agents against Huntington’s disease via the nasal-brain route. Front. Pharmacol. 2023, 14, 1218625. [Google Scholar]
- Sharaf, M.; Sewid, A.H.; Hamouda, H.; Elharrif, M.G.; El-Demerdash, A.S.; Alharthi, A.; Hashim, N.; Hamad, A.A.; Selim, S.; Alkhalifah, D.H.M. Rhamnolipid-Coated Iron Oxide Nanoparticles as a Novel Multitarget Candidate against Major Foodborne E. coli Serotypes and Methicillin-Resistant S. aureus. Microbiol. Spectr. 2022, 10, e00250-22. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Neoh, K.; Kang, E. Synthesis and in vitro anti-cancer evaluation of tamoxifen-loaded magnetite/PLLA composite nanoparticles. Biomaterials 2006, 27, 5725–5733. [Google Scholar] [CrossRef] [PubMed]
- Kant, R.; Pathak, S.; Dutta, V.J.S.E.M.; Cells, S. Design and fabrication of sandwich-structured α-Fe2O3/Au/ZnO photoanode for photoelectrochemical water splitting. Sol. Energy Mater. Sol. Cells 2018, 178, 38–45. [Google Scholar] [CrossRef]
- Mutz, B.; Belimov, M.; Wang, W.; Sprenger, P.; Serrer, M.-A.; Wang, D.; Pfeifer, P.; Kleist, W.; Grunwaldt, J.-D. Potential of an alumina-supported Ni3Fe catalyst in the methanation of CO2: Impact of alloy formation on activity and stability. ACS Catal. 2017, 7, 6802–6814. [Google Scholar] [CrossRef]
- Magaldi, S.; Mata-Essayag, S.; De Capriles, C.H.; Pérez, C.; Colella, M.; Olaizola, C.; Ontiveros, Y. Well diffusion for antifungal susceptibility testing. Int. J. Infect. Dis. 2004, 8, 39–45. [Google Scholar] [CrossRef]
- Valgas, C.; Souza, S.M.D.; Smânia, E.F.; Jr, A.S. Screening methods to determine antibacterial activity of natural products. Braz. J. Microbiol. 2007, 38, 369–380. [Google Scholar] [CrossRef]
- Salim, M.; Ullah, I.; Saljoqi, A.U.R.; Gökçe, A.; Ahmad, S.; Almutairi, M.H.; Sayed, A.A.; Aleya, L.; Abdel-Daim, M.M.; Shah, M. Life table study of Sitotroga cerealella on different cereals and its implications on the performance of the egg parasitoid (Trichogramma chilonis) under laboratory conditions. Sci. Rep. 2023, 13, 10961. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Hola, V.; Bonaventura, G.D.; Djukić, S.; Ćirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. Apmis 2007, 115, 891–899. [Google Scholar] [CrossRef]
- Jadhav, S.; Shah, R.; Bhave, M.; Palombo, E.A. Inhibitory activity of yarrow essential oil on Listeria planktonic cells and biofilms. Food Control 2013, 29, 125–130. [Google Scholar] [CrossRef]
- Abdallah, F.B.; Lagha, R.; Gaber, A. Biofilm inhibition and eradication properties of medicinal plant essential oils against methicillin-resistant Staphylococcus aureus clinical isolates. Pharmaceuticals 2020, 13, 369. [Google Scholar] [CrossRef]
- Watkinson, A.; Micalizzi, G.; Graham, G.; Bates, J.; Costanzo, S. Antibiotic-resistant Escherichia coli in wastewaters, surface waters, and oysters from an urban riverine system. Appl. Environ. Microbiol. 2007, 73, 5667–5670. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Han, B.; Gu, J.; Wang, C.; Wang, P.; Ma, Y.; Cao, J.; He, Z. Fate of antibiotic resistant cultivable heterotrophic bacteria and antibiotic resistance genes in wastewater treatment processes. Chemosphere 2015, 135, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Zanotto, C.; Bissa, M.; Illiano, E.; Mezzanotte, V.; Marazzi, F.; Turolla, A.; Antonelli, M.; Morghen, C.D.G.; Radaelli, A. Identification of antibiotic-resistant Escherichia coli isolated from a municipal wastewater treatment plant. Chemosphere 2016, 164, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Selim, S.; Faried, O.A.; Almuhayawi, M.S.; Saleh, F.M.; Sharaf, M.; El Nahhas, N.; Warrad, M. Incidence of Vancomycin-Resistant Staphylococcus aureus Strains among Patients with Urinary Tract Infections. Antibiotics 2022, 11, 408. [Google Scholar] [CrossRef]
- Salim, M.; Kamran, M.; Khan, I.; Saljoqi, A.U.R.; Ahmad, S.; Almutairi, M.H.; Sayed, A.A.; Aleya, L.; Abdel-Daim, M.M.; Shah, M. Effect of larval diets on the life table parameters of dengue mosquito, Aedes aegypti (L.) (Diptera: Culicidae) using age-stage two sex life table theory. Sci. Rep. 2023, 13, 11969. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Vishwas, S.; Kaur, A.; Kaur, H.; Kakoty, V.; Khursheed, R.; Chaitanya, M.; Babu, M.; Awasthi, A.; Corrie, L.; et al. Harnessing role of sesamol and its nanoformulations against neurodegenerative diseases. Biomed. Pharmacother. 2023, 1, 115512. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, A.S.; Layton, A.C.; Mailloux, B.J.; Culligan, P.J.; Williams, D.E.; Smartt, A.E.; Sayler, G.S.; Feighery, J.; McKay, L.D.; Knappett, P.S. Comparison of fecal indicators with pathogenic bacteria and rotavirus in groundwater. Sci. Total Environ. 2012, 431, 314–322. [Google Scholar] [CrossRef]
- Gilbert, R.; De Louvois, J.; Donovan, T.; Little, C.; Nye, K.; Ribeiro, C.; Richards, J.; Roberts, D.; Bolton, F. Guidelines for the microbiological quality of some ready-to-eat foods sampled at the point of sale. PHLS Advisory Committee for Food and Dairy Products. Commun. Dis. Public Health 2000, 3, 163–167. [Google Scholar]
- Gruber, J.S.; Ercumen, A.; Colford, J.M., Jr. Coliform bacteria as indicators of diarrheal risk in household drinking water: Systematic review and meta-analysis. PLoS ONE 2014, 9, e107429. [Google Scholar] [CrossRef]
- Alouache, S.; Estepa, V.; Messai, Y.; Ruiz, E.; Torres, C.; Bakour, R. Characterization of ESBLs and associated quinolone resistance in Escherichia coli and Klebsiella pneumoniae isolates from an urban wastewater treatment plant in Algeria. Microb. Drug Resist. 2014, 20, 30–38. [Google Scholar] [CrossRef]
- Surleac, M.; Barbu, I.C.; Paraschiv, S.; Popa, L.I.; Gheorghe, I.; Marutescu, L.; Popa, M.; Sarbu, I.; Talapan, D.; Nita, M. Whole genome sequencing snapshot of multi-drug resistant Klebsiella pneumoniae strains from hospitals and receiving wastewater treatment plants in Southern Romania. PLoS ONE 2020, 15, e0228079. [Google Scholar] [CrossRef] [PubMed]
- Loudermilk, E.M.; Kotay, S.M.; Barry, K.E.; Parikh, H.I.; Colosi, L.M.; Mathers, A.J. Tracking Klebsiella pneumoniae carbapenemase gene as an indicator of antimicrobial resistance dissemination from a hospital to surface water via a municipal wastewater treatment plant. Water Res. 2022, 213, 118151. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Stell, A.L. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 2000, 181, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.-Y.; Cai, J.-C.; Zhou, H.-W.; Chi, D.; Zhang, X.-F.; Chen, W.-L.; Zhang, R.; Chen, G.-X. Molecular typing of CTX-M-producing Escherichia coli isolates from environmental water, swine feces, specimens from healthy humans, and human patients. Appl. Environ. Microbiol. 2013, 79, 5988–5996. [Google Scholar] [CrossRef]
- Zhi, S.; Stothard, P.; Banting, G.; Scott, C.; Huntley, K.; Ryu, K.; Otto, S.; Ashbolt, N.; Checkley, S.; Dong, T. Characterization of water treatment-resistant and multidrug-resistant urinary pathogenic Escherichia coli in treated wastewater. Water Res. 2020, 182, 115827. [Google Scholar] [CrossRef]
- Długosz, O.; Szostak, K.; Krupiński, M.; Banach, M. Synthesis of Fe3O4/ZnO nanoparticles and their application for the photodegradation of anionic and cationic dyes. Int. J. Environ. Sci. Technol. 2021, 18, 561–574. [Google Scholar] [CrossRef]
- Smith, R.D.; Prévot, M.S.; Fagan, R.D.; Zhang, Z.; Sedach, P.A.; Siu, M.K.J.; Trudel, S.; Berlinguette, C.P.J.S. Photochemical route for accessing amorphous metal oxide materials for water oxidation catalysis. Science 2013, 340, 60–63. [Google Scholar] [CrossRef]
- Thirunavukkarasu, K.; Sankaranarayanan, T.; Pandurangan, A.; Shanthi, R.V.; Sivasanker, S. The role of surface Zn 2+ ions in the transesterification of vegetable oils over ZnO supported on Al2O3 and Fe2O3. Catal. Sci. Technol. 2014, 4, 851–860. [Google Scholar] [CrossRef]
- Mao, N.J.S.R. Investigating the heteronjunction between ZnO/Fe2O3 and g-C3N4 for an enhanced photocatalytic H2 production under visible-light irradiation. Sci. Rep. 2019, 9, 12383. [Google Scholar] [CrossRef]
- Fakhr, M.A.; Mazrou, Y.S.; Ellmouni, F.Y.; ElSaied, A.; Elhady, M.; Elkelish, A.; Nour, I.H. Investigating the Phenotypic Plasticity of the Invasive Weed Trianthema portulacastrum L. Plants 2022, 11, 77. [Google Scholar] [CrossRef]
- Sawant, V.; Bamane, S. PEG-beta-cyclodextrin functionalized zinc oxide nanoparticles show cell imaging with high drug payload and sustained pH responsive delivery of curcumin in to MCF-7 cells. J. Drug Deliv. Sci. Technol. 2018, 43, 397–408. [Google Scholar] [CrossRef]
- Kumar, S.R.; Paulpandi, M.; ManivelRaja, M.; Mangalaraj, D.; Viswanathan, C.; Kannan, S.; Ponpandian, N. An in vitro analysis of H1N1 viral inhibition using polymer coated superparamagnetic Fe 3 O 4 nanoparticles. RSC Adv. 2014, 4, 13409–13418. [Google Scholar] [CrossRef]
- Alsamadany, H.; Mansour, H.; Elkelish, A.; Ibrahim, M.F.M. Folic Acid Confers Tolerance against Salt Stress-Induced Oxidative Damages in Snap Beans through Regulation Growth, Metabolites, Antioxidant Machinery and Gene Expression. Plants 2022, 30, 1459. [Google Scholar] [CrossRef]
- Rossi, T.; Penfold, T.; Rittmann-Frank, M.; Reinhard, M.; Rittmann, J.; Borca, C.; Grolimund, D.; Milne, C.; Chergui, M. Characterizing the structure and defect concentration of ZnO nanoparticles in a colloidal solution. J. Phys. Chem. C 2014, 118, 19422–19430. [Google Scholar] [CrossRef]
- Rajan, A.S.; Khan, A.; Asrar, S.; Raza, H.; Das, R.K.; Sahu, N.K. Synthesis of ZnO/Fe3 O4/rGO nanocomposites and evaluation of antibacterial activities towards E. coli and S. aureus. IET Nanobiotechnol. 2019, 13, 682–687. [Google Scholar] [CrossRef]
- Bahari, A.; Roeinfard, M.; Ramzannezhad, A.; Khodabakhshi, M.; Mohseni, M. Nanostructured features and antimicrobial properties of Fe3O4/ZnO Nanocomposites. Natl. Acad. Sci. Lett. 2019, 42, 9–12. [Google Scholar] [CrossRef]
- Oprea, O.; Andronescu, E.; Ficai, D.; Ficai, A.; Oktar, F.N.; Yetmez, M. ZnO applications and challenges. Curr. Org. Chem. 2014, 18, 192–203. [Google Scholar] [CrossRef]
- Pradhan, L.; Srivastava, R.; Bahadur, D. pH-and thermosensitive thin lipid layer coated mesoporous magnetic nanoassemblies as a dual drug delivery system towards thermochemotherapy of cancer. Acta Biomater. 2014, 10, 2976–2987. [Google Scholar] [CrossRef]
- Gudkov, S.V.; Burmistrov, D.E.; Serov, D.A.; Rebezov, M.B.; Semenova, A.A.; Lisitsyn, A.B. Do iron oxide nanoparticles have significant antibacterial properties? Antibiotics 2021, 10, 884. [Google Scholar] [CrossRef]
- Sharma, N.; Jandaik, S.; Kumar, S.; Chitkara, M.; Sandhu, I.S. Synthesis, characterisation and antimicrobial activity of manganese-and iron-doped zinc oxide nanoparticles. J. Exp. Nanosci. 2016, 11, 54–71. [Google Scholar] [CrossRef]
- Neupane, B.P.; Chaudhary, D.; Paudel, S.; Timsina, S.; Chapagain, B.; Jamarkattel, N.; Tiwari, B.R. Himalayan honey loaded iron oxide nanoparticles: Synthesis, characterization and study of antioxidant and antimicrobial activities. Int. J. Nanomed. 2019, 14, 3533. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, V.; Feio, M.J.; Bastos, M. Role of lipids in the interaction of antimicrobial peptides with membranes. Prog. Lipid Res. 2012, 51, 149–177. [Google Scholar] [CrossRef] [PubMed]
- Čujová, S.; Slaninová, J.; Monincová, L.; Fučík, V.; Bednárová, L.; Štokrová, J.; Hovorka, O.; Voburka, Z.; Straka, J.; Čeřovský, V. Panurgines, novel antimicrobial peptides from the venom of communal bee Panurgus calcaratus (Hymenoptera: Andrenidae). Amino Acids 2013, 45, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, L.-L.; Zhang, A.-T.; Xie, P.; Lu, J.-J.; Zou, X.-T. Antibacterial effects of zinc oxide nanoparticles on Escherichia coli K88. Afr. J. Biotechnol. 2012, 11, 10248–10254. [Google Scholar]
- Yousef, J.M.; Danial, E.N. In vitro antibacterial activity and minimum inhibitory concentration of zinc oxide and nano-particle zinc oxide against pathogenic strains. J. Health Sci. 2012, 2, 38–42. [Google Scholar] [CrossRef]
- Elmi, F.; Alinezhad, H.; Moulana, Z.; Salehian, F.; Tavakkoli, S.M.; Asgharpour, F.; Fallah, H.; Elmi, M.M. The use of antibacterial activity of ZnO nanoparticles in the treatment of municipal wastewater. Water Sci. Technol. 2014, 70, 763–770. [Google Scholar] [CrossRef]
- Samy, R.P.; Gopalakrishnakone, P.; Thwin, M.; Chow, T.; Bow, H.; Yap, E.; Thong, T. Antibacterial activity of snake, scorpion and bee venoms: A comparison with purified venom phospholipase A2 enzymes. J. Appl. Microbiol. 2007, 102, 650–659. [Google Scholar] [CrossRef]
- Hegazi, A.G.; El-Feel, M.; Abdel-Rahman, E.; Al-Fattah, A. Antibacterial activity of bee venom collected from Apis mellifera carniolan pure and hybrid races by two collection methods. Int. J. Curr. Microbiol. App Sci. 2015, 4, 141–149. [Google Scholar]
- Park, S.; Park, B.; Yun, S.; Kang, H.; So, B. Antimicrobial activities of honey bee venom against pathogens isolated from clinical bovine mastitis in Korea. Planta Medica 2013, 79, PL16. [Google Scholar] [CrossRef]
- Fadl, A.E.-W. Antibacterial and antibiofilm effects of bee venom from (Apis mellifera) on multidrug-resistant bacteria (MDRB). Al-Azhar J. Pharm. Sci. 2018, 58, 60–80. [Google Scholar] [CrossRef]
- Marques, S.C.; Rezende, J.D.G.O.S.; Alves, L.A.D.F.; Silva, B.C.; Alves, E.; Abreu, L.R.D.; Piccoli, R.H. Formação de biofilme por Staphylococcus aureus na superfície de aço inoxidável e vidro e sua resistência a alguns sanificantes químicos. Braz. J. Microbiol. 2007, 38, 538–543. [Google Scholar] [CrossRef]
- Batoni, G.; Maisetta, G.; Esin, S. Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochim. Biophys. Acta BBA-Biomembr. 2016, 1858, 1044–1060. [Google Scholar] [CrossRef] [PubMed]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef] [PubMed]
- Tossi, A.; Sandri, L. Giangaspero, Amphipathic, α-helical antimicrobial peptides. Pept. Sci. 2000, 55, 4–30. [Google Scholar] [CrossRef]
- Dosler, S.; Karaaslan, E.; Gerceker, A.A. Antibacterial and anti-biofilm activities of melittin and colistin, alone and in combination with antibiotics against Gram-negative bacteria. J. Chemother. 2016, 28, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Picoli, T.; Peter, C.M.; Zani, J.L.; Waller, S.B.; Lopes, M.G.; Boesche, K.N.; de Oliveira Hübner, S.; Fischer, G. Melittin and its potential in the destruction and inhibition of the biofilm formation by Staphylococcus aureus, Escherichia coli and Pseudomonas aeruginosa isolated from bovine milk. Microb. Pathog. 2017, 112, 57–62. [Google Scholar] [CrossRef]
- Bandara, H.; Nguyen, D.; Mogarala, S.; Osiñski, M.; Smyth, H. Magnetic fields suppress Pseudomonas aeruginosa biofilms and enhance ciprofloxacin activity. Biofouling 2015, 31, 443–457. [Google Scholar] [CrossRef]
- Estevão-Costa, M.-I.; Sanz-Soler, R.; Johanningmeier, B.; Eble, J.A. Snake venom components in medicine: From the symbolic rod of Asclepius to tangible medical research and application. Int. J. Biochem. Cell Biol. 2018, 104, 94–113. [Google Scholar] [CrossRef]
- Ranmadugala, D.; Ebrahiminezhad, A.; Manley-Harris, M.; Ghasemi, Y.; Berenjian, A. The effect of iron oxide nanoparticles on Bacillus subtilis biofilm, growth and viability. Process Biochem. 2017, 62, 231–240. [Google Scholar] [CrossRef]
- Rumyantceva, V.; Rumyantceva, V.; Koshel, E.; Vinogradov, V. Biocide-conjugated magnetite nanoparticles as an advanced platform for biofilm treatment. Ther. Deliv. 2019, 10, 241–250. [Google Scholar] [CrossRef]
- Shakerimoghaddam, A.; Ghaemi, E.A.; Jamalli, A. Zinc oxide nanoparticle reduced biofilm formation and antigen 43 expressions in uropathogenic Escherichia coli. Iran. J. Basic Med. Sci. 2017, 20, 451. [Google Scholar] [PubMed]
- Applerot, G.; Lellouche, J.; Perkas, N.; Nitzan, Y.; Gedanken, A.; Banin, E. ZnO nanoparticle-coated surfaces inhibit bacterial biofilm formation and increase antibiotic susceptibility. Rsc. Adv. 2012, 2, 2314–2321. [Google Scholar] [CrossRef]
- Khan, S.T.; Ahamed, M.; Musarrat, J.; Al-Khedhairy, A.A. Anti-biofilm and antibacterial activities of zinc oxide nanoparticles against the oral opportunistic pathogens R othia dentocariosa and R othia mucilaginosa. Eur. J. Oral Sci. 2014, 122, 397–403. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).