The Molecular Epidemiology of Enterovirus in a Birth Cohort in Nepal
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Data Collection
2.2. Study Type and Sample Size
2.3. Ethic Approval
2.4. Nucleic Acid Extraction
2.5. Real Time RT PCR of EV
2.6. Enterovirus Typing
2.7. Statistical Analysis
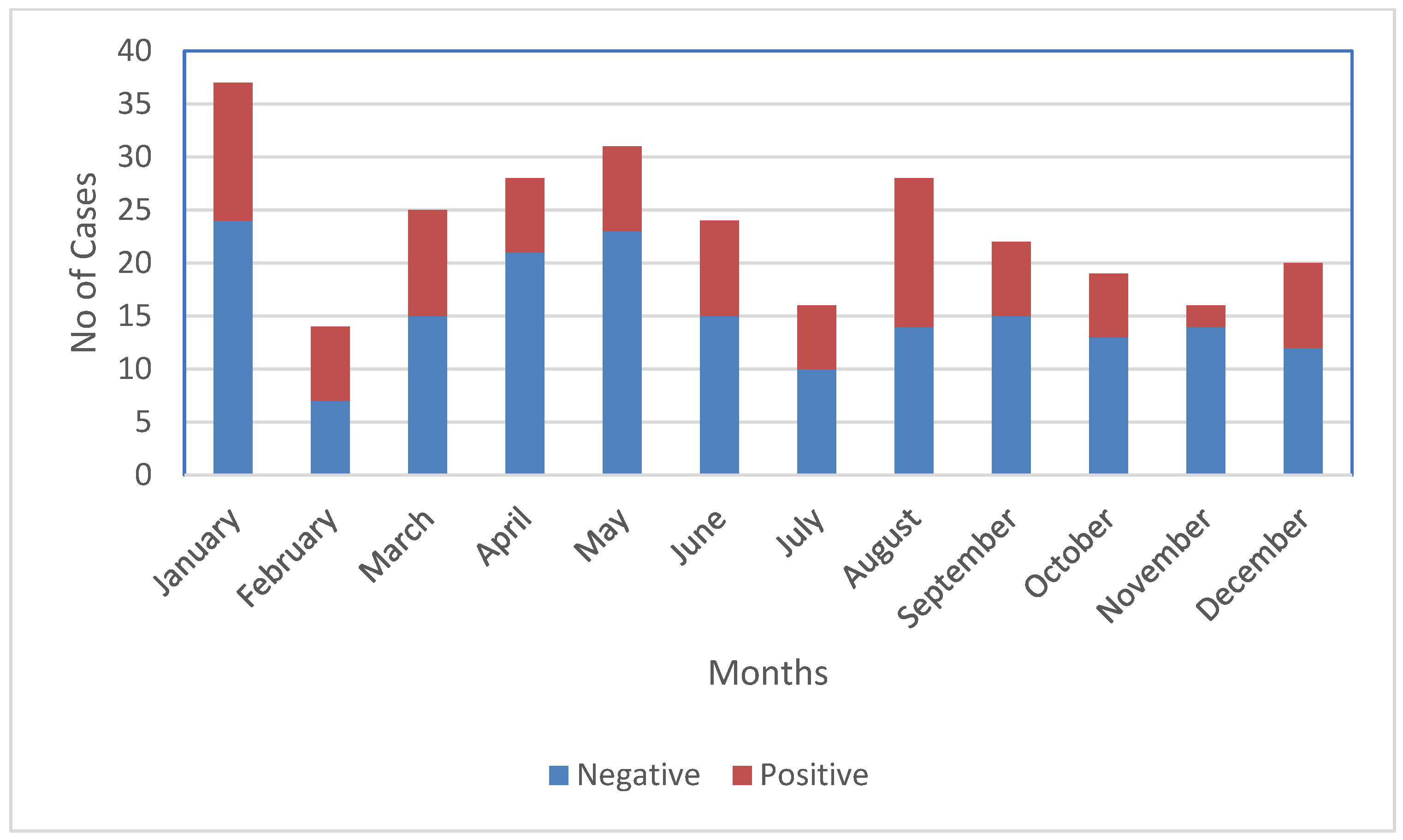
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zell, R.; Delwart, E.; Gorbalenya, A.E.; Hovi, T.; King, A.M.Q.; Knowles, N.J.; Lindberg, A.M.; Pallansch, M.A.; Palmenberg, A.C.; Reuter, G.; et al. ICTV Virus Taxonomy Profile: Picornaviridae. J. Gen. Virol. 2017, 98, 2421–2422. [Google Scholar] [CrossRef] [PubMed]
- Jiao, M.M.A.; Apostol, L.N.; de Quiroz-Castro, M.; Jee, Y.; Roque, V.; Mapue, M.; Navarro, F.M.; Tabada, C.F.; Tandoc, A. Non-polio enteroviruses among healthy children in the Philippines. BMC Public Health 2020, 20, 167. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.M.; Zhang, Y.; Scheuermann, R.H. Epidemiology and Sequence-Based Evolutionary Analysis of Circulating Non-Polio Enteroviruses. Microorganisms 2020, 8, 1856. [Google Scholar] [CrossRef]
- Dhole, T.N.; Ayyagari, A.; Chowdhary, R.; Shakya, A.K.; Shrivastav, N.; Datta, T.; Prakash, V. Non-polio enteroviruses in acute flaccid paralysis children of India: Vital assessment before polio eradication. J. Paediatr. Child Health 2009, 45, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Hassan, F.; Schuster, J.E.; Simenauer, A.; Selvarangan, R.; Halpin, R.A.; Lin, X.; Fedorova, N.; Stockwell, T.B.; Lam, T.T.-Y.; et al. Molecular Evolution and Intraclade Recombination of Enterovirus D68 during the 2014 Outbreak in the United States. J. Virol. 2015, 90, 1997–2007. [Google Scholar] [CrossRef]
- Majer, A.; McGreevy, A.; Booth, T.F. Molecular Pathogenicity of Enteroviruses Causing Neurological Disease. Front. Microbiol. 2020, 11, 540. [Google Scholar] [CrossRef]
- Chansaenroj, J.; Tuanthap, S.; Thanusuwannasak, T.; Duang-In, A.; Klinfueng, S.; Thaneskongtong, N.; Vutithanachot, V.; Vongpunsawad, S.; Poovorawan, Y. Human enteroviruses associated with and without diarrhea in Thailand between 2010 and 2016. PLoS ONE 2017, 12, e0182078. [Google Scholar] [CrossRef]
- Shen, X.-X.; Qiu, F.-Z.; Li, G.-X.; Zhao, M.-C.; Wang, J.; Chen, C.; Zhao, L.; Qi, J.-J.; Liu, H.; Zhang, Y.; et al. A case control study on the prevalence of enterovirus in children samples and its association with diarrhea. Arch. Virol. 2019, 164, 63–68. [Google Scholar] [CrossRef]
- Fazelipour, M.; Makvandi, M.; Samarbafzadeh, A.; Nisi, N.; Azaran, A.; Jalilian, S.; Pirmoradi, R.; Nikfar, R.; Shamsizadeh, A.; Ahmadi Angali, K. Detection of Enteroviruses in Children with Acute Diarrhea. Arch. Clin. Infect. Dis. 2019, 14, e83916. [Google Scholar] [CrossRef]
- Abe, O.; Kimura, H.; Minakami, H.; Akami, M.; Inoue, M.; Saito, A.; Otsuki, K. Outbreak of gastroenteritis caused by echovirus type 6 in an orphanage in Japan. J. Infect. 2000, 41, 285–286. [Google Scholar] [CrossRef]
- Bina Rai, S.; Wan Mansor, H.; Vasantha, T.; Norizah, I.; Chua, K.B. An outbreak of echovirus 11 amongst neonates in a confinement home in Penang, Malaysia. Med. J. Malays. 2007, 62, 223–226. [Google Scholar]
- Patel, J.R.; Daniel, J.; Mathan, V.I. An epidemic of acute diarrhoea in rural southern India associated with echovirus type 11 infection. J. Hyg. 1985, 95, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, R.P.; Kirchner, E.; Moura, R.A.; Murahovschi, J. Acute gastroenteritis associated with coxsackie B-6 virus in infants and children, in São Paulo, Brazil. Rev. Do Inst. De Med. Trop. De Sao Paulo 1977, 19, 73–76. [Google Scholar]
- Shrestha, P.S.; Shrestha, S.K.; Bodhidatta, L.; Strand, T.; Shrestha, B.; Shrestha, R.; Chandyo, R.K.; Ulak, M.; Mason, C.J. Bhaktapur, Nepal: The MAL-ED Birth Cohort Study in Nepal. Clin. Infect. Dis. 2014, 59, S300–S303. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, S.K.; Shrestha, J.; Andreassen, A.K.; Strand, T.A.; Dudman, S.; Dembinski, J.L. Genetic Diversity of Astrovirus in Children From a Birth Cohort in Nepal. Front. Microbiol. 2021, 11, 588707. [Google Scholar] [CrossRef] [PubMed]
- Jokela, P.; Joki-Korpela, P.; Maaronen, M.; Glumoff, V.; Hyypiä, T. Detection of human picornaviruses by multiplex reverse transcription-PCR and liquid hybridization. J. Clin. Microbiol. 2005, 43, 1239–1245. [Google Scholar] [CrossRef]
- Verstrepen, W.; Bruynseels, P.; Mertens, A.H. Evaluation of a rapid real-time RT-PCR assay for detection of enterovirus RNA in cerebrospinal fluid specimens. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2002, 25 (Suppl. S1), S39–S43. [Google Scholar] [CrossRef]
- Watkins-Riedel, T.; Woegerbauer, M.; Hollemann, D.; Hufnagl, P. Rapid diagnosis of Enterovirus infections by real-time PCR on the LightCycler using the TaqMan format. Diagn. Microbiol. Infect. Dis. 2002, 42, 99–105. [Google Scholar] [CrossRef]
- Rotbart, H.A. Enzymatic RNA amplification of the enteroviruses. J. Clin. Microbiol. 1990, 28, 438–442. [Google Scholar] [CrossRef]
- Kares, S.; Lönnrot, M.; Vuorinen, P.; Oikarinen, S.; Taurianen, S.; Hyöty, H. Real-time PCR for rapid diagnosis of entero- and rhinovirus infections using LightCycler. J. Clin. Virol. 2004, 29, 99–104. [Google Scholar] [CrossRef]
- Oberste, M.S.; Pallansch, M.A. Enterovirus molecular detection and typing. Rev. Res. Med. Microbiol. 2005, 16, 163–171. [Google Scholar] [CrossRef]
- Verstrepen, W.A.; Kuhn, S.; Kockx, M.M.; Van De Vyvere, M.E.; Mertens, A.H. Rapid detection of enterovirus RNA in cerebrospinal fluid specimens with a novel single-tube real-time reverse transcription-PCR assay. J. Clin. Microbiol. 2001, 39, 4093–4096. [Google Scholar] [CrossRef] [PubMed]
- Rotbart, H.A.; Sawyer, M.H.; Fast, S.; Lewinski, C.; Murphy, N.; Keyser, E.F.; Spadoro, J.; Kao, S.Y.; Loeffelholz, M. Diagnosis of enteroviral meningitis by using PCR with a colorimetric microwell detection assay. J. Clin. Microbiol. 1994, 32, 2590–2592. [Google Scholar] [CrossRef] [PubMed]
- Nix, W.A.; Oberste, M.S.; Pallansch, M.A. Sensitive, Seminested PCR Amplification of VP1 Sequences for Direct Identification of All Enterovirus Serotypes from Original Clinical Specimens. J. Clin. Microbiol. 2006, 44, 2698–2704. [Google Scholar] [CrossRef] [PubMed]
- Rao, D.C.; Reddy, H.; Sudheendra, K.; Raghavendra, A.; Varadharaj, V.; Edula, S.; Goparaju, R.; Ratnakar, B.; Srinivasa Rao, A.S.; Maiya, P.P.; et al. Non-polio enterovirus association with persistent diarrhea in children as revealed by a follow-up study of an Indian cohort during the first two years of life. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2014, 61, 125–131. [Google Scholar] [CrossRef]
- Pallansch, M. Enteroviruses: Polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses. Fields Virol. 2006, 1, 840–867. [Google Scholar]
- Bubba, L.; Pellegrinelli, L.; Pariani, E.; Primache, V.; Amendola, A.; Binda, S. A novel multiplex one-step real-time RT-PCR assay for the simultaneous identification of enterovirus and parechovirus in clinical fecal samples. J. Prev. Med. Hyg. 2015, 56, E57–E60. [Google Scholar]
- Chitambar, S.; Gopalkrishna, V.; Chhabra, P.; Patil, P.; Verma, H.; Lahon, A.; Arora, R.; Tatte, V.; Ranshing, S.; Dhale, G.; et al. Diversity in the enteric viruses detected in outbreaks of gastroenteritis from Mumbai, Western India. Int. J. Environ. Res. Public Health 2012, 9, 895–915. [Google Scholar] [CrossRef]
- Duan, X.; Chen, Z.; Li, X.; Yuan, P.; Long, L. Virus Shedding in Patients With Hand, Foot and Mouth Disease Induced by EV71, CA16 or CA6: Systematic Review and Meta-analysis. Pediatr. Infect. Dis. J. 2021, 40, 289–294. [Google Scholar] [CrossRef]
- Zhang, Y.; Cao, J.; Zhang, S.; Lee, A.J.; Sun, G.; Larsen, C.N.; Zhao, H.; Gu, Z.; He, S.; Klem, E.B. Genetic changes found in a distinct clade of Enterovirus D68 associated with paralysis during the 2014 outbreak. Virus Evol. 2016, 2, vew015. [Google Scholar] [CrossRef]
- Witsø, E.; Palacios, G.; Cinek, O.; Stene, L.C.; Grinde, B.; Janowitz, D.; Lipkin, W.I.; Rønningen, K.S. High prevalence of human enterovirus a infections in natural circulation of human enteroviruses. J. Clin. Microbiol. 2006, 44, 4095–4100. [Google Scholar] [CrossRef] [PubMed]
- Patil, P.R.; Chitambar, S.D.; Gopalkrishna, V. Molecular surveillance of non-polio enterovirus infections in patients with acute gastroenteritis in Western India: 2004–2009. J. Med. Virol. 2015, 87, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Rao, D.C.; Ananda Babu, M.; Raghavendra, A.; Dhananjaya, D.; Kumar, S.; Maiya, P.P. Non-polio enteroviruses and their association with acute diarrhea in children in India. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2013, 17, 153–161. [Google Scholar] [CrossRef]
- Tapparel, C.; Siegrist, F.; Petty, T.J.; Kaiser, L. Picornavirus and enterovirus diversity with associated human diseases. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2013, 14, 282–293. [Google Scholar] [CrossRef]
- Qiao, X.; Liu, X.; Wang, Y.; Li, Y.; Wang, L.; Yang, Q.; Wang, H.; Shen, H. Analysis of the epidemiological trends of enterovirus A in Asia and Europe. J. Infect. Chemother. Off. J. Jpn. Soc. Chemother. 2023, 29, 316–321. [Google Scholar] [CrossRef]
- Chen, P.; Wang, H.; Tao, Z.; Xu, A.; Lin, X.; Zhou, N.; Wang, P.; Wang, Q. Multiple transmission chains of coxsackievirus A4 co-circulating in China and neighboring countries in recent years: Phylogenetic and spatiotemporal analyses based on virological surveillance. Mol. Phylogenetics Evol. 2018, 118, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Zou, G.; Liao, Q.; Zhou, Y.; Liu, F.; Dai, B.; Liu, J.; Chen, Z.; Xing, W.; Yang, L.; et al. Spectrum of Enterovirus Serotypes Causing Uncomplicated Hand, Foot, and Mouth Disease and Enteroviral Diagnostic Yield of Different Clinical Samples. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2018, 67, 1729–1735. [Google Scholar] [CrossRef]
- Yang, Q.; Zhang, Y.; Yan, D.; Zhu, S.; Wang, D.; Ji, T.; Li, X.; Song, Y.; Gu, X.; Xu, W. Two Genotypes of Coxsackievirus A2 Associated with Hand, Foot, and Mouth Disease Circulating in China since 2008. PLoS ONE 2016, 11, e0169021. [Google Scholar] [CrossRef]
- Chansaenroj, J.; Auphimai, C.; Puenpa, J.; Mauleekoonphairoj, J.; Wanlapakorn, N.; Vuthitanachot, V.; Vongpunsawad, S.; Poovorawan, Y. High prevalence of coxsackievirus A2 in children with herpangina in Thailand in 2015. Virus Dis. 2017, 28, 111–114. [Google Scholar] [CrossRef]
- Chen, S.-P.; Huang, Y.-C.; Li, W.-C.; Chiu, C.-H.; Huang, C.-G.; Tsao, K.-C.; Lin, T.-Y. Comparison of Clinical Features Between Coxsackievirus A2 and Enterovirus 71 During the Enterovirus Outbreak in Taiwan, 2008: A Children’s Hospital Experience. J. Microbiol. Immunol. Infect. 2010, 43, 99–104. [Google Scholar] [CrossRef]
- Lu, H.; Hong, M.; Zhang, Y.; Xiao, J.; Zhang, M.; Zhang, K.; Song, Y.; Han, Z.; Yang, Q.; Wang, D.; et al. A novel interspecies recombinant enterovirus (Enterovirus A120) isolated from a case of acute flaccid paralysis in China. Emerg. Microbes Infect. 2020, 9, 1733–1743. [Google Scholar] [CrossRef] [PubMed]
- Oberste, M.S.; Michele, S.M.; Maher, K.; Schnurr, D.; Cisterna, D.; Junttila, N.; Uddin, M.; Chomel, J.J.; Lau, C.S.; Ridha, W.; et al. Molecular identification and characterization of two proposed new enterovirus serotypes, EV74 and EV75. J. Gen. Virol. 2004, 85, 3205–3212. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Y.; Hong, M.; Li, X.; Zhu, S.; Yan, D.; Wang, D.; An, H.; Tsewang; Han, J.; et al. Isolation and characterization of a Chinese strain of human enterovirus 74 from a healthy child in the Tibet Autonomous Region of China. Arch. Virol. 2012, 157, 1593–1598. [Google Scholar] [CrossRef] [PubMed]
- Bessaud, M.; Pillet, S.; Ibrahim, W.; Joffret, M.L.; Pozzetto, B.; Delpeyroux, F.; Gouandjika-Vasilache, I. Molecular characterization of human enteroviruses in the Central African Republic: Uncovering wide diversity and identification of a new human enterovirus A71 genogroup. J. Clin. Microbiol. 2012, 50, 1650–1658. [Google Scholar] [CrossRef] [PubMed]
- Peacey, M.; Hall, R.J.; Wang, J.; Todd, A.K.; Yen, S.; Chan-Hyams, J.; Rand, C.J.; Stanton, J.A.; Huang, Q.S. Enterovirus 74 infection in children. PLoS ONE 2013, 8, e76492. [Google Scholar] [CrossRef]
- Sousa, I.P., Jr.; Burlandy, F.M.; Tavares, F.N.; da Silva, E.E. Enterovirus B74 associated with hand, foot and mouth disease. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2018, 65, 15–17. [Google Scholar] [CrossRef]
- Alhazmi, A.; Sane, F.; Lazrek, M.; Nekoua, M.P.; Badia-Boungou, F.; Engelmann, I.; Alidjinou, E.K.; Hober, D. Enteroviruses and Type 1 Diabetes Mellitus: An Overlooked Relationship in Some Regions. Microorganisms 2020, 8, 1458. [Google Scholar] [CrossRef]
- Nekoua, M.P.; Alidjinou, E.K.; Hober, D. Persistent coxsackievirus B infection and pathogenesis of type 1 diabetes mellitus. Nat. Rev. Endocrinol. 2022, 18, 503–516. [Google Scholar] [CrossRef]

| Characteristics (n) | EV Positive | Percentage | p Value * |
|---|---|---|---|
| Age in Months | 0.17 | ||
| <6 (117) | 48 | 41 | |
| 6–12 (64) | 22 | 34.4 | |
| 12–24 (67) | 20 | 29.8 | |
| 25–36 (32) | 7 | 21.9 | |
| Seasons | 0.15 | ||
| Spring, Mar–May (84) | 25 | 29.8 | |
| Summer, Jun–Aug (68) | 29 | 42.6 | |
| Autumn, Sept–Nov (57) | 15 | 26.3 | |
| Winter, Dec–Feb (71) | 28 | 39.4 |
| EV Species | EV Serotype | No. of Typed Virus (%) |
|---|---|---|
| EV-A | Coxsackievirus (CV-A2) | 3 (13) |
| Coxsackievirus (CV-A4) | 1 (4.3) | |
| Enterovirus (EV-A120) | 1 (4.3) | |
| EV-B | Coxsackievirus (CV-B2) | 1 (4.3) |
| Coxsackievirus (CV-B4) | 1 (4.3) | |
| Coxsackievirus (CV-B6) | 1 (4.3) | |
| Echovirus (E-1) | 1 (4.3) | |
| Echovirus (E-2) | 1 (4.3) | |
| Echovirus (E-5) | 1 (4.3) | |
| Echovirus (E-6) | 2 (8.7) | |
| Echovirus (E-9) | 1 (4.3) | |
| Echovirus (E-11) | 1 (4.3) | |
| Echovirus (E-17) | 1 (4.3) | |
| Echovirus (E-19) | 3 (13) | |
| Echovirus (E-20) | 1 (4.3) | |
| Echovirus (E-30) | 2 (8.7) | |
| Enterovirus (EV-B74) | 1 (4.3) | |
| Total | 23 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shrestha, S.K.; Shrestha, J.; Strand, T.A.; Numanovic, S.; Andreassen, A.K.; Dembinski, J.L.; Vikse, R.; Dudman, S. The Molecular Epidemiology of Enterovirus in a Birth Cohort in Nepal. Microbiol. Res. 2023, 14, 909-917. https://doi.org/10.3390/microbiolres14030063
Shrestha SK, Shrestha J, Strand TA, Numanovic S, Andreassen AK, Dembinski JL, Vikse R, Dudman S. The Molecular Epidemiology of Enterovirus in a Birth Cohort in Nepal. Microbiology Research. 2023; 14(3):909-917. https://doi.org/10.3390/microbiolres14030063
Chicago/Turabian StyleShrestha, Sanjaya K., Jasmin Shrestha, Tor A. Strand, Sanela Numanovic, Ashild K. Andreassen, Jennifer L. Dembinski, Rose Vikse, and Susanne Dudman. 2023. "The Molecular Epidemiology of Enterovirus in a Birth Cohort in Nepal" Microbiology Research 14, no. 3: 909-917. https://doi.org/10.3390/microbiolres14030063
APA StyleShrestha, S. K., Shrestha, J., Strand, T. A., Numanovic, S., Andreassen, A. K., Dembinski, J. L., Vikse, R., & Dudman, S. (2023). The Molecular Epidemiology of Enterovirus in a Birth Cohort in Nepal. Microbiology Research, 14(3), 909-917. https://doi.org/10.3390/microbiolres14030063






