Abstract
Lactic acid bacteria are widely studied microorganisms and are one of the prevalent groups of bacteria in the oral cavity microbiome. This work aimed to isolate new lactic acid bacterial strains from the human oral cavity and evaluate their characteristics and probiotic potential. Twelve strains were isolated and identified as belonging to several genera in the family Lactobacillaceae. Screening for antimicrobial activity was held, where two of the strains showed antagonistic activity against Streptococcus mutans and most of the strains expressed inhibition against Escherichia coli, Bacillus subtilis, and Bacillus cereus. The ability of the studied strains to autoaggregate and bind to mucin was assessed, showing autoaggregative properties and mucin binding at 5 logs CFU/mL. The survival ability in simulated oral and gastrointestinal conditions and growth dynamics with different gastrointestinal stress factors was studied. Most of the strains showed a good growth potential in the presence of oral and gastrointestinal stress factors. All tested strains exhibited high survival rates in the simulated oral environment, thus having the potential for colonizing the oral cavity and their beneficial properties to be applied. These results are a good basis for continuing the research into these strains so they can be included in new functional products for oral health.
1. Introduction
The oral cavity has a number of different niches, each of them colonized with microorganisms with a different microbial composition. The synergism and interactions between the oral microorganism populations assist the human body against unwanted invasions of exogenous microflora, but if an imbalance occurs, the microbial flora can cause different diseases in the oral cavity, as well as systemic diseases. Therefore, the oral microbiome plays an important role in the human microbial community and health [1].
The Food and Agriculture Organization of the United Nations (FAO) and the World Health Organization (WHO) defines probiotics as live microorganisms, which provide health benefits to the host when administered in adequate amounts [2,3]. Moreover, when consumed as a monoculture or mixed culture, probiotics affect the host by enhancing the quality of the indigenous microflora in the gastrointestinal tract (GIT) [4]. New bacterial isolates with the potential to be used as probiotics need to be fully characterized [5]. To be considered as probiotic candidates, the genera and species of the microorganisms must be identified as per the internationally accepted methods and nomenclature [6]. Additionally, antibiotic susceptibility is one of the important standards for the range of use of lactic acid bacteria (LAB). Investigating the antibiotic susceptibility profiles is based on three main criteria: the possible horizontal transfer of antibiotic resistance genes to pathogenic microorganisms, the connection of lactobacilli in some disease cases, and the possibility of combining antibiotics with probiotics for a therapeutic application [7].
The probiotic properties of LAB are widely studied, so they can be used as probiotic supplements for human healthcare. A number of strains belonging to Lactobacillus and Bifidobacterium genera and some yeast representatives have a significant history in medical and food industry use and have been subject to comprehensive and thorough monitoring and evaluation [3]. Different species of the Lactobacillus genus, including Lacticaseibacillus rhamnosus and Limosilactobacillus fermentum, have been identified as effective probiotic microorganisms for decades by fulfilling the established criteria for probiotics [8]. Despite the established characteristics, many of the current research focuses on new isolates regarding a different range of use [9]. The Weissella genus, however, has begun to increase interest only in the past few years owing to its observed probiotic potential and many emerging applications [10].
Probiotics targeting oral healthcare can successfully compete with pathogenic species and support the growth of beneficial bacteria, so they can positively contribute mainly to the prevention of oral diseases as well as their therapy [11,12,13,14,15]. Several bacterial and yeast species can exhibit pathogenic effects and colonize different organs in the human body, including the oral cavity [16]. Since pathogenic microorganisms can survive even in unfavorable conditions and spread relatively easily, it is important to study the possibility of LAB to inhibit their growth. The use of probiotic products against infections of the soft and hard oral tissues by pathogenic species such as Streptococcus mutans, Streptococcus sobrinus, Porphyromonas gingivalis, and Candida albicans has been relatively well studied [17].
To exhibit their effectiveness, oral probiotics must be able to survive, adhere, and colonize the oral tissues. The oral cavity is the entry point of the GIT and the received probiotic microorganisms are initially exposed to the saliva. During this first contact with the host, the bacterial survival ability of the oral environment is of utmost importance. The protein components of the saliva, including lysozyme, can affect the viability or activity of the probiotic strains, such as the adhesion and metabolic activity. The influence of saliva on the microbial establishment can, on the one hand, inhibit microbial colonization, but on the other, it can promote it [18,19]. Saliva is naturally high in nutrients, which makes a suitable environment for the growth of microorganisms. However, the naturally present lysozyme acts as an antimicrobial agent [20]. Lysozyme has the property of hydrolyzing the polysaccharide layer of the cell wall of Gram-positive bacteria, which results in the disruption of the cellular membrane integrity [21]. Another property is its role in the formation of the acquired enamel and mucosal pellicle, alongside other salivary proteins, such as mucin. Lysozyme can promote the adhesion of different microorganisms but it can also inhibit an adhesion and colonization of potentially pathogenic species [22]. The transfer of a probiotic from the oral cavity to the next parts of the GIT is very likely, so it is important that their ability to survive in this transition is studied as well. Microorganisms that enter the upper and lower parts of the GIT are influenced by the presence of digestive enzymes, pancreatic enzymes, and bile salts [23,24]. For the selection of probiotic LAB, it is required that they can resist the stressful factors of the GIT to exhibit their probiotic properties [25,26].
Microbial aggregation is one of the important properties of probiotic bacteria and is a prerequisite for a successful adhesion and preventing pathogens from surface space. Autoaggregation leads to the formation of structures that facilitate the interactions between cells, which are important for the formation of the dental biofilm and its characteristics toward pathogen elimination. When autoaggregating, microorganisms from the same species are able to form aggregates, which is associated with binding to the mucosal tissue [27]. The ability to adhere to the mucus is an important prerequisite for potential probiotics in order to successfully colonize the GIT and significant for the competition with different pathogenic species. The mucosal layer plays an important role in the formation of endogenous microbiota in individuals, which provides a suitable environment for microorganisms. Therefore, the adhesion to the mucosa and formation of balanced microbial community, probiotic strains included, contributes to the competitive advantage in the gastrointestinal ecosystem [28,29]. Many clinical studies involving the colonization and persistence of probiotic microorganisms show that they do not permanently colonize the GIT and provide benefits to the host for a brief time after their last application. In that case, it is important to evaluate their capability to adhere as an adhesion is a widely used criterion for the selection of probiotic strains. It has been assumed that probiotic effectiveness is promoted by an adhesion to the mucus and increases the residence time. This extends the presence of probiotic microorganisms and the administration of their beneficial effects [30].
In this context, the aim of this work was to assess newly isolated LAB strains, as a part of the human oral cavity microbiome, for their probiotic potential in regard of basic properties as: the enzyme activity profile; antimicrobial activity against pathogens, which were commonly associated with oral diseases or transited through the oral cavity; an evaluation of their properties, related with maintaining viability in the simulated conditions of the saliva and stomach enzymes, and growth expression in the presence of the different stressful factors of the lower parts of GIT; and an assessment of the adhesive potential by their ability to autoaggregate and adhere on the mucosal tissue by a mucin binding assay. The species identification of all the isolates was also part of our present work, carried out by a polyphasic taxonomic characterization. The establishment of probiotic characteristics in the studied strains is a prerequisite for the inclusion of these new oral isolates in the category of already established probiotic LAB strains.
2. Materials and Methods
2.1. Isolation and Characterization of Oral Samples
Oral samples were taken with dry swabs from the cervical area of the teeth of volunteers. Informed consent was obtained from all volunteers who provided oral samples for this study. The samples were spread on Man, Rogosa, Sharpe (MRS) (Merck, Darmstadt, Germany) agar plates and cultivated at 37 °C for 48 h. Single colonies were isolated, inoculated in MRS broth and cultivated for 24 h at 37 °C. Initial screening was performed to determine the affiliation of the newly isolated strains to LAB. The Gram staining technique was used to determine the morphological characteristics of the strains with the 77730 Gram Staining Kit (Sigma-Aldrich, St. Louis, MO, USA) according to the classic methodology [31]. The peroxidase activity was assayed, using Quantofix® Peroxide 100 test-strips (MACHEREY-NAGEL, Düren, Germany) and the catalase activity was tested [32] with 3% hydrogen peroxide solution.
2.2. Identification of Lactic Acid Bacteria
The newly isolated strains were identified based on the 16S rDNA gene sequencing analysis. The genomic DNA was extracted using a Quick-DNA Fungal/Bacterial Miniprep Kit (Zymo Research, Irvine, CA, USA) following the manufacturer’s protocol. A PCR reaction was performed, with the obtained genomic DNA as a template, with the primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) for the 16S rDNA gene amplification, using the PuReTaq Ready-To-Go PCR Beads (Cytiva, Amersham, UK). The PCR products were visualized electrophoretically and sent for sequencing service at a sequencing services provider (Macrogen Europe B.V.; Amsterdam, the Netherlands). The obtained 16S rDNA gene sequences were processed and compared with the closest matching sequences in the National Center for Biotechnology Information (NCBI) GenBank, using the basic local alignment search tool (BLAST) algorithm.
Furthermore, the strains were identified from their protein profile, acquired by the matrix-assisted laser desorption ionizing time-of-flight mass spectrometer (MALDI-TOF MS) QuanTOF (IntelliBio, Qingdao, China) [33] and compared by peptide mass fingerprint (PMF) matching for authenticity of identification of the newly isolated strains. The results were acquired by the QuanID Microbial Test application and interpreted with an identification score. According to the application database, an identification score above >1000 is considered highly confident.
2.3. Enzymatic Activity
For assaying the enzymatic activity, the twelve tested strains were cultivated in MRS broth at 37 °C for 24 h and the cultures were concentrated by centrifugation at 6000× g for 10 min. The strains were analyzed using API® ZYM strips (BioMérieux, Marcy-l`Etoile, France) according to the manufacturer’s manual. The presence of enzyme activity was determined according to the coloration intensity.
2.4. Antimicrobial Activity against Test Pathogens
The isolated oral LAB strains were analyzed for the antimicrobial activity using cell-free supernatant (CFS) against several test pathogens, including Streptococcus mutans ATCC 25175, Candida albicans ATCC 10231, Bacillus subtilis ATCC 6633, Bacillus cereus ATCC 11778, Escherichia coli ATCC 25922, Staphylococcus aureus subsp. aureus ATCC 25923, Staphylococcus epidermidis ATCC 12228, Pseudomonas aeruginosa ATCC 27853, and Propionibacterium acnes (isolate) by the agar well diffusion method [34]. Fresh 24 h cultures of the test pathogens were used and standardized to a concentration of 0.5 McFarland. Petri dishes with Mueller–Hinton (MH) agar, brain heart infusion (BHI) agar, and malt extract (ME) agar were inoculated with their corresponding test pathogen by spreading and wells were made with a cork borer. Native CFS and neutralized CFS (neutralized with 1 N NaOH to pH 7) were obtained from the culture broth of the LAB strains and inoculated into the wells then incubated at the test pathogens’ optimal growth temperatures for 24 h. Growth inhibition was observed by measuring the diameter of the appeared halo zones.
2.5. Survival Ability in Simulated Oral Conditions
An oral stress test was held to determine the survivability of the newly isolated LAB strains in simulated oral conditions [35,36,37]. A saliva-resembling electrolyte saline solution with added lysozyme was made firsthand, comprised of NaCl (6.2 g/L), KCl (2.2 g/L), CaCl2 (0.22 g/L), NaHCO3 (1.2 g/L) + 150 mg/L lysozyme (HiMedia, Mumbai, India) with 30,000 U/mg activity. The biomass from 24 h LAB cultures was resuspended in the solution and incubated at 37 °C for 10 min and control samples without solution treatment were prepared. Adequate dilutions were made, and the controls and the treated samples were then cultivated in MRS agar plates at 37 °C for 48 h. The results were reported as colony-forming units per milliliter (CFU/mL) from the viable cell count.
2.6. Survival Ability in the Presence of Pepsin
The stress test in the presence of pepsin was held to determine the survivability of the newly isolated LAB strains [38]. LAB cultures for 24 h were inoculated in MRS broth with corrected pH (pH 2) and supplemented with 0.3% pepsin (HiMedia, Mumbai, India) in 24-well flat bottom plates. LAB cultures inoculated in plain MRS broth were used as a positive control. The plates were incubated in the SPECTROstar® Nano Microplate Reader (BMG LABTECH, Ortenberg, Germany) at 37 °C for 3 h and measurements were set to be automatically taken every hour. The samples were then cultivated in MRS agar for 48 h and the results were reported as CFU/mL from the viable cell count.
2.7. Growth Dynamics in the Presence of Stress Factors
An in vitro growth dynamics analysis was held to evaluate the stability of the newly isolated LAB strains in the presence of the different stress factors in the GIT [39] with some modifications. The analysis was performed with a SPECTROstar® Nano Microplate Reader in a 24-well flat bottom plate. LAB cultures for 24 h were inoculated in wells with MRS broth as a positive control and with MRS broth with different stress factors added: lysozyme 150 mg/L (Fluka, Sygma-Aldrich, Steinheim, Germany) with 78,643 U/mg activity; pancreatin 0.1% (HiMedia, India) and pH brought to pH 7; ox-bile 0.3%, 0.5%, and 1.0% (Sigma-Aldrich, USA). Blank wells with uninoculated MRS broth were used as a negative control. The prepared plate was then incubated in the microplate reader at 37 °C for 24 h and measurements were set to be automatically taken every two hours by the OD at 600 nm.
2.8. Autoaggregation and Mucin Binding Capability
An autoaggregation assay was held based on Zhang et al. [40] and Unban et al. [41] with some modifications. The bacterial cultures were centrifuged at 6000× g for 5 min, washed twice with PBS, and brought to 108 CFU/mL. After vortexing, aliquots were measured with a SPECTROstar® Nano Microplate Reader at 600 nm (Ainitial). The cell cultures were then incubated at 37 °C statically for 4 h and the upper most fractions were measured (Afinal). The autoaggregation percentage was calculated with the equation:
where Ainitial and Afinal are the absorbance measured at 0 and 4 h, respectively.
Autoaggregation (%) = (Ainitial − Afinal)/(Ainitial) × 100
The newly isolated LAB strains were evaluated for their ability to bind to mucin as described by Monteiro et al. [42] with some modifications. Volumes of 100 µL of PBS mucin solution in a concentration of 10 mg/mL were added in a 96-well flat bottom plate and incubated overnight at 4 °C. The wells were washed twice with PBS and 2% (w/v) bovine serum albumin (BSA) solution (SERVA Electrophoresis, Heidelberg, Germany) was added for incubation at 4 °C for 4 h then washed again with PBS. Then, 24 h cultures from the tested LAB strains were washed with PBS and standardized to a concentration of 0.5 McFarland. Bacterial suspensions were added as 100 µL aliquots to individual wells and incubated at 37 °C for 1 h. Then, the wells were washed 10 times with PBS for the removal of the non-adherent cells, treated with 0.5% Triton X-100 (Sigma-Aldrich, USA), and incubated at room temperature in an orbital shaker at 150 rpm to release the adhered bacteria. The cells were scraped from the wells and adequate dilutions were cultivated in MRS agar plates at 37 °C for 48 h. The results were reported as CFU/mL from the viable cell count.
2.9. Data Analyses
All experiments were performed in triplicate. The obtained data were analyzed by Microsoft Excel built-in functions and the results were expressed as the mean, along with standard deviation when possible. Two-tailed Student’s t-test was carried out as a statistical evaluation to detect differences between the controls and samples. A p-value below 0.05 was considered as a statistically significant difference.
3. Results and Discussion
3.1. Isolation and Identification of Lactic Acid Bacteria
In total, 76 samples were taken from 16 volunteers; 64 isolates were isolated from the samples and assessed with a classic sample processing approach. All isolates were evaluated for peroxidase and catalase enzymatic activities, cell morphology, and Gram staining. According to these criteria, 12 strains were preselected, showing negative peroxidase and catalase activities and were observed as Gram-positive rod-shaped cells (Table 1). From these results, all of the selected strains were defined as belonging to the group of LAB.

Table 1.
Characterization and identification of the newly isolated strains.
Comparing the 16S rDNA gene sequences in the NCBI database showed a high percent of identity for all twelve strains. This identification was verified by the PMF matching of the MALDI-TOF MS, measured by the score number of matching protein profiles (Table 1). According to the QuanID Microbial Test application database, an interpretation of the identification score resulting in a score above >1000 is highly confident.
The twelve strains were identified as representatives from different LAB species. According to the newly proposed nomenclature for the Lactobacillus genera [43], the strains were identified as Limosilactobacillus fermentum, Weissella confusa, Latilactobacillus curvatus, Lactobacillus delbrueckii subsp. lactis, Lactobacillus delbrueckii subsp. sunkii, Lacticaseibacillus rhamnosus, and Lacticaseibacillus paracasei (Table 1).
Determining the nucleotide sequence of the 16S rDNA gene allows the phylogenetic analysis and identification of significant clinical and industrial strains. Although identifying microorganisms using the 16S rDNA is the most used method, in recent years the MALDI-TOF MS has proved to be a very promising tool for identification, classification, and diagnosis [33] and is being widely used [44]. The obtained results from the MALDI-TOF MS analyses provided a highly accurate verification of the 16S rDNA identification of the twelve newly isolated LAB strains.
3.2. Enzymatic Activity
It is important for potential probiotics to be evaluated for the production of certain enzymes to confirm that they do not produce toxic substances. The production of useful enzymes and the inhibition of harmful enzymes exhibit different characteristics, which is dependent on the species of the tested microorganism. Therefore, species that produce potentially toxic substances need to be excluded. The API® ZYM system was used to obtain the enzymatic profiles of the studied strains (Table 2).

Table 2.
Enzymatic activity of the newly isolated strains.
All tested LAB strains exhibited high enzymatic activity for leucine arylamidase (5 according to API® ZYM scale) and acid phosphatase (3–5). Valine arylamidase, α-chymotrypsin, α- and β-galactosidase, and α-glucosidase activity (3–5) was reported for most of the strains. Low lipase (C14) activity (1–2) was exhibited by all of the strains and no enzymatic activity (0–1) was observed for the production of trypsin, β-glucuronidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase.
In a study by Kim et al. [45], the L. paracasei and L. rhamnosus strains included in their work showed high leucine arylamidase activity and no β-glucuronidase or α-mannosidase activity. The expression of arylamidase activities determines the proteolytic activity of the bacterial strains [46]. The high leucine arylamidase activity of the tested LAB strains and the well-expressed strain-specific valine and cystine arylamidase activities qualifies their proteolytic activity. It must be noted that β-glucuronidase is a bacterial enzyme associated with the production of certain harmful carcinogens and negative effects in the colon and liver [47] and the tested strains did not produce this enzymatic activity.
3.3. Antimicrobial Activity against Test Pathogens
All of the isolated strains were analyzed for an antimicrobial capability against the selected oral test pathogens S. mutans and C. albicans, and against gastrointestinal and skin test pathogens that can also colonize the oral cavity, including B. subtilis, B. cereus, E. coli, S. aureus, S. epidermidis, Ps. aeruginosa, and P. acnes by the agar well diffusion method. The results show the presence of antimicrobial activity with the native CFS. Halo zones against E. coli were observed by all of the strains, except L. curvatus KG 12-1, and most of the strains expressed inhibitory activity against B. subtilis and B. cereus. W. confusa AG 2-6 and NN 1 and L. curvatus KG 12-1 did not show an inhibition against the two Bacillus test pathogens, also no inhibition was observed from L. paracasei AV 2-1 against B. subtilis, and from L. rhamnosus NA 1-8 or L. fermentum NA 2-2 against B. cereus. Against Ps. Aeruginosa the strains L. fermentum TC 3-11, L. delbrueckii subsp. Sunkii VG 1, L. delbrueckii subsp. lactis VG 2 and MK 13-1, and L. fermentum NA 2-2 exhibited an inhibition. Three of the studied strains L. delbrueckii subsp. sunkii VG 1, L. delbrueckii subsp. lactis VG 2, and MK 13-1 showed an inhibition against S. aureus. Against S. epidermidis, P. acnes, and S. mutans, W. confusa AG 2-6 and NN 1 expressed inhibition activity. No antimicrobial activity was observed from the native CFS from the studied strains against C. albicans (Table 3). In contrast, the neutralized CFS from all of the newly isolated LAB strains did not show activity against the used test pathogens. It can be assumed that the exhibited antimicrobial activity of the native CFS is mainly due to the production of organic acids from LAB–lactic acid or lactic and acetic acids [48] (Table 3). Further analysis for the antagonistic activity of the tested strains against important oral pathogens will be a part of our work.

Table 3.
Antimicrobial activity of the newly isolated lactic acid bacterial strains (native cell free supernatant) against test pathogens.
The results from the antimicrobial activity of the different tested strains against different test pathogens show specificity, but it was only exhibited from the native CFSs. The production of organic acids is one of the most important mechanisms of the antimicrobial activity of LAB, by which they act as antagonists of many pathogenic species [48]. A study by Ren et al. [49] with Lactobacillus pentosus, Lactiplantibacillus plantarum, and L. paracasei showed antibacterial activity against E. coli, B. cereus, S. aureus, and Salmonella enterica. Another study by Matevosyan et al. [50] evaluated the antibacterial characteristics of LAB strains, including L. rhamnosus, resulting in the inhibition of the pathogens as E. coli, Ps. aeruginosa, S. aureus, Salmonella typhimurium, Bacillus mesentericus, and Micrococcus luteus. Against S. mutans, antibacterial activity is widely reported [51]. According to Sookkhee et al. [52], L. paracasei subsp. paracasei and L. rhamnosus expressed a high antagonizing capacity against some of the significant oral pathogens S. mutans and P. gingivalis. The strains Ligilactobacillus salivarius WB21 and L. fermentum, studied by Strahinic et al. [53], expressed antagonistic activity against S. mutans and Streptococcus pneumoniae.
For opportunistic oral pathogens such as C. albicans, an antagonistic effect is rarely observed and research shows that the inhibitory capability of LAB against Candida representatives is not a species-specific trait and depends on the origin of the LAB isolates [54]. Many studies relate to the inhibitory activity to LAB isolates of a human origin. Other studies report that many used LAB did not exhibit an inhibitory activity against Candida: strains of Lactobacillus gasseri did not have or had partial inhibitory effects, and Lactobacillus crispatus, Limosilactobacillus vaginalis, and L. delbrueckii only expressed a partial inhibition [54]; Bifidobacterium bifidum and L. delbrueckii did not lower the cell density of Candida in a co-cultivation experiment [55]; a study by Liao et al. [56] reported that a non-mutant strain of L. casei did not exhibit inhibition; and Ariningsih et al. [57] reported that 7 of 46 used LAB isolates expressed antagonistic activity against Candida. Many researchers continue to study microorganisms with activity against C. albicans and identify and isolate metabolites with inhibitory effects, since this pathogen causes diseases not only in the oral cavity, but in other organs in the human body.
3.4. Survival Ability in Simulated Oral Conditions
To evaluate the survivability of LAB in the human body, in vitro analyses for their resistance to the conditions of the GIT were made. The purpose of our study was to determine the effect of the saliva-resembling electrolyte saline solution with added lysozyme on the survival of the studied LAB strains when they enter the oral cavity. The results are shown on Figure 1. It can be observed that after treatment with the electrolyte saline solution with added lysozyme, all tested strains maintain the same log CFU/mL as their untreated samples.

Figure 1.
Survival ability of the newly isolated lactic acid bacterial strains under simulated oral stress conditions. Means: a—nonsignificant (p > 0.05); b and c—significant (p < 0.05 and p < 0.01, respectively).
A study by Haukioja et al. [58] evaluated the survivability in saliva and an adhesion to the oral surfaces of Lactobacillus and Bifidobacterium probiotics. All tested strains showed a high survivability, but their adhesion varied, as the Lactobacillus species expressed high adhesion properties. In vitro studies report that Lactobacillus and Bifidobacterium cannot grow in saliva but they can stay viable for 24 h after incubation. The preliminary treatment with lysozyme significantly decreased the adhesion of L. rhamnosus GG, L. rhamnosus Lc705, and L. casei Shirota, but the adhesion properties of Lactobacillus johnsonii La1 and Bifidobacterium lactis Bb12 remained unchanged [59].
Since all tested LAB strains are oral isolates and their natural environment is the oral cavity, it is expected that they can sustain the effect of the saliva. The recorded data show that when treated with the saliva-resembling solution, all of the tested strains show no significant change in CFU/mL compared with the control samples. This points a high survivability potential in a saliva environment, which is a prerequisite for their ability to compete with pathogens and to be a part of the dental biofilm and exhibit their probiotic properties under the conditions of the oral cavity.
3.5. Survival Ability in Low pH and the Presence of Pepsin
The resistance to pepsin in pH 2 was held for 3 h at 37 °C and the log values of CFU/mL from the viable cell count was reported (Figure 2). The results show that most of the tested strains exhibit a decreased cell viability by 2–3 logs. For L. fermentum N 2, N 4-5, TC 3-11, NA 2-2 and L. delbrueckii subsp. lactis MK 13-1, and L. rhamnosus NA 1-8, a decrease of about 2 logs is observed. For W. confusa AG 2-6 and NN 1, and L. delbrueckii subsp. lactis VG 2, a decrease of up to 3 logs is observed for the duration of the experiment. The strains L. curvatus KG 12-1, L. delbrueckii subsp. sunkii VG 1, and L. paracasei AV 2-1 show no survivability in a simulated harsh environment of the stomach in the presence of pepsin in pH 2.
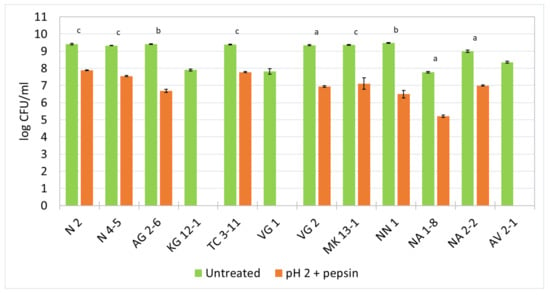
Figure 2.
Survival ability of the newly isolated lactic acid bacterial strains in pepsin in pH 2. Means: a—nonsignificant (p > 0.05); b and c—significant (p < 0.05 and p < 0.01, respectively).
The low pH values in the stomach and the antimicrobial effect of the digestive enzyme pepsin act as a barrier for LAB to survive in the GIT. Because their natural origin is the oral cavity, it is possible that some strains might not withstand very low pH values and the presence of pepsin. The results from this experiment indicate that nine of all the tested strains show a considerable survival ability, which highly favors their possibility to withstand the harsh environment of the stomach and transit to the next sections of the GIT.
Many authors report an expressed sensitivity of many LAB strains under the conditions of the enzymatic activity and high acidity of the stomach environment. Mantzourani et al. [60] reported a decreased viability of 2–5 logs observed within the range of the tested LAB isolates. A study by Tokatlı et al. [61] evaluated the survivability of several LAB strains and reported a decrease in the cell viability of up to 4 logs.
3.6. Growth Dynamics in the Presence of GIT Stress Factors
The growth dynamics of potentially probiotic strains in the presence of various stress factor characteristics of the upper and lower compartments of the GIT were studied. The growth dynamics in the presence of lysozyme were assessed for 24 h at 37 °C and the OD was measured every 2 h at 600 nm. The results are shown in Figure 3 as growth curves. Eight of all the tested strains exhibit similar growth rates as their control samples, except the strains L. curvatus KG 12-1, L. delbrueckii subsp. sunkii VG 1, and L. delbrueckii subsp. lactis VG 2 and MK 13-1. Although the media with added lysozyme influences their growth, they show a high survival ability in these conditions, according to the results in 3.4. Being of an oral origin, the newly isolated LAB strains show high adaptive properties to the presence of this enzyme and have the potential to persist in the environment of the oral cavity and their beneficial properties to be applied.
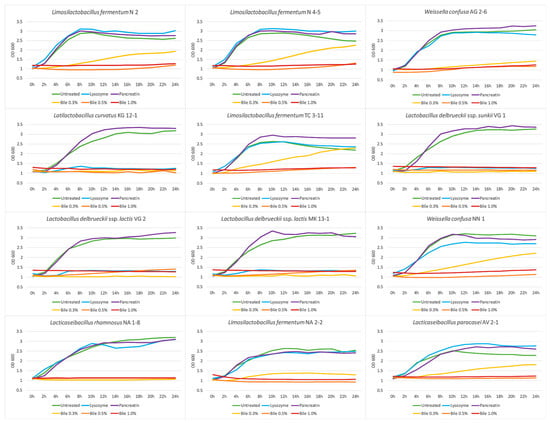
Figure 3.
Growth dynamics of the newly isolated lactic acid bacterial strains in the presence of gastrointestinal stress factors.
Studies by Fang et al. [62] and Jia et al. [39] reported high tolerance levels when cultivating Lactobacillus strains in the presence of different concentrations of added lysozyme. Bosch et al. [63] evaluated the effect of different concentrations of lysozyme on the growth of LAB strains and reported growth ranges of 95.86–49.12%.
The growth dynamic of the studied strains in the presence of pancreatin in pH 7 were assessed. Figure 3 shows that the pancreatic enzymes in neutral pH do not decrease the growth of the tested strains. The observed growth properties in the presence of pancreatin makes the adaptivity of the tested strains highly possible in these conditions. Khagwal et al. [64] reported a resistance to the presence of pancreatin in a concentration of 0.5% for all tested LAB strains in their study.
The growth dynamic in the presence of bile salts were assessed to evaluate the effect of different concentrations of bile salts on the studied strains (Figure 3). The presence of 0.3% bile salts decreases the cell growth for the strains L. fermentum N 2, N 4-5, TC 3-11, and NA 2-2, W. confusa AG 2-6 and NN 1, and L. paracasei AV 2-1 and a lower cell density is reported at the end of the experiment in comparison with the control variants. While for the strains L. curvatus KG 12-1, L. delbrueckii subsp. sunkii VG 1, L. delbrueckii subsp. lactis VG 2 and MK 13-1, and L. rhamnosus NA 1-8, growth inhibition is observed. Bile salts in higher concentrations of 0.5% and 1.0% significantly inhibit the growth of all LAB strains. It is necessary to specify that in physiological conditions, the bile salts concentration of a healthy person is no more than 0.3% [65], so these results indicate that seven of the twelve tested strains can overcome the effect of bile salts in a concentration of 0.3% and exhibit growth in this environment, which makes these strains able to withstand the normal conditions of the colon.
The effect of the bile concentration is reported to be species- and strain-dependent [66]. A study by Alameri et al. [67] reported a 50–60% growth rate in ox-bile medium after 6 h, compared with the control samples, for most of the LAB strains used in their work. A Lactobacillus strain evaluated by Aarti et al. [68] showed a high tolerance for 36 h of cultivating in a bile salts medium.
3.7. Autoaggregation and Mucin Binding Capability
The autoaggregative potential of the tested LAB strains was assessed and presented in Figure 4a. The results show that all the tested strains can autoaggregate and exhibited these properties in the range of 10.3–26.6% for 4 h of incubation. The highest autoaggregation was reported for the trains L. delbrueckii subsp. sunkii VG 1, L. rhamnosus NA 1-8 and L. paracasei AV 2-1. All other LAB strains exhibited below 20% autoaggregative properties. It is important to note that the conditions of the experiment, such as the culture age, time, and temperature, have a significance for the determination of the autoaggregation ability.
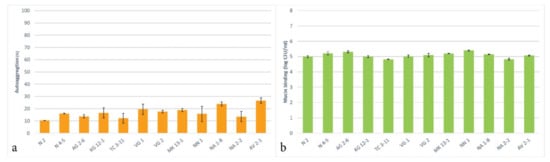
Figure 4.
Autoaggregation (a) and mucin binding (b) of the newly isolated lactic acid bacterial strains.
The ranging autoggregation levels that are observed amongst the LAB strains result from a complex interaction between different bacterial cell surface molecules, such as proteins and polysaccharides [69], and this property is considered to be strain-specific [27]. In other studies, 22 Lactobacillus strains were tested that showed autoaggregative properties in the range of 24 to 41% [70] and several Lactobacillus strains that exhibited autoaggregation in the range of 15 to 21% [71].
For evaluation of the adhesion properties of the tested LAB strains to the mucosal tissue, a mucin binding in vitro analysis was assessed. The results show that all of the tested LAB strains exhibit an expressed ability to bind to mucin, measured at 5 logs CFU/mL (Figure 4b). Exhibiting an adhesion to mucin makes probiotic LAB able to adapt to the environments of GIT, colonize inside the host, and exert their beneficial properties. The assessed adhesion of the tested LAB strains suggests that they possess a capacity for an in vivo colonization and the obtained results have an important relevance for the probiotic use of these strains. A study by Monteiro et al. [42] reported similar mucin binding properties for L. plantarum and L. fermentum strains. Being of oral origin, all tested strains possess the potential ability to adhere to oral mucosa, be included in the oral microbiome, and exhibit their probiotic properties.
4. Conclusions
This work provides relevant information regarding evaluating the characteristics and properties of new LAB isolates of a human origin, specifically from the oral cavity. A complex identification method was applied to acquire the species affiliation, which provided accurate data for all of the twelve new isolates. From the obtained results in this work, a few of the tested LAB strains show a better probiotic potential than the others.
All tested L. fermentum strains exhibit the highest probiotic potential in regard of survival and growth potential in the environment of the oral cavity and in the transition to the next parts of the GIT. Showing viability in the presence of the digestive enzyme pepsin and growth properties in the presence of stress factors, typical for the lower parts of the GIT, places a good basis for continuing research. The observed autoaggregation and mucin binding as adhesive properties puts forward these strains as candidates for a further evaluation of the direct interactions against important oral pathogens, such as S. mutans and C. albicans.
The two tested W. confusa strains exhibited a similar probiotic potential, plus possessing antimicrobial activity with their native CFS against S. mutans and S. epidermidis and P. acnes as well. The probiotic potential of other W. confusa strains regarding GIT tolerance and adhesion properties [72] and observing antimicrobial and antioxidant properties [73] has also been established by other researchers. While wide antimicrobial activity was not being observed by L. rhamnosus N 1-8, the strain showed a promising adhesion potential and survival ability in oral conditions. Many studies have described probiotic strains of this species, such as L. rhamnosus GG being the most well-documented [74]. This makes the two newly isolated W. confusa strains and L. rhanmosus N 1-8 promising for researching other probiotic characteristics.
In the tested L. delbrueckii strains, wider antimicrobial activity was observed against the pathogens E. coli, B. cereus, B. subtilis, S. aureus, and Ps. aeruginosa. Although these strains survive under simulated conditions in the oral cavity and exhibit a mucin binding ability, they do not show growth in the presence of various GIT stress factors. However, it is important to note that all three strains as oral isolates exhibit antagonistic activity against various Gr+ and Gr− pathogens and this is a significant prerequisite for subsequent studies of their interactions with microorganisms in the oral microbiome.
Different properties regarding the probiotic potential were determined for each of the newly isolated LAB strains. The strains L. fermentum N 2 and TC 3-11 and L. delbrueckii subsp. lactis VG 2 showed the most expressed combination of probiotic characteristics. As Weissella is a genus with less rich probiotic knowledge as Lactobacillus, the observed properties of W. confusa AG 2-6 and NN 1 strains makes them interesting for continuing the evaluation. Overall, these tested strains possess characteristics for expanding research to evaluate them as potential probiotics to be included in products for oral health.
Author Contributions
Conceptualization, N.A. and D.N.; methodology, N.A., Y.E. and D.N.; formal analysis, N.A., Y.E. and D.N.; investigation, N.A. and D.N.; writing—original draft preparation, N.A.; writing—review and editing, Y.E. and D.N.; supervision, D.N. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Bulgarian Ministry of Education and Science under the NRP “Healthy Foods for a Strong Bio-Economy and Quality of Life” approved by DCM # 577/17.08.2018 and Scientific Fund of Sofia University “St. Kliment Ohridski” under Project No. 80-10-78/11.05.2022.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Not applicable.
Acknowledgments
This work is partially supported by the Operational Program ‘‘Science and Education for Smart Growth’’, Bulgaria, grant number BG05M2OP001-1.002-0012.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gao, L.; Xu, T.; Huang, G.; Jian, S.; Gu, Y.; Chen, F. Oral microbiomes: More and more importance in oral cavity and whole body. Protein Cell 2018, 9, 488–500. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations/World Health Organization FAO/WHO. Guidelines for the Evaluation of Probiotics in Food; Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food: London, ON, Canada, 2002. [Google Scholar]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Ayivi, R.D.; Ibrahim, S.A. Lactic acid bacteria: An essential probiotic and starter culture for the production of yoghurt. Int. J. Food Sci. Technol. 2022, 57, 7008–7025. [Google Scholar] [CrossRef]
- Sanders, M.E. Probiotics: Definition, sources, selection, and uses. Clin. Infect. Dis. 2008, 46, 58–61. [Google Scholar] [CrossRef]
- Parker, C.T.; Tindall, B.J.; Garrity, G.M. International Code of Nomenclature of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2019, 69, 1–111. [Google Scholar] [CrossRef]
- Gad, G.F.; Abdel-Hamid, A.M.; Farag, Z.S. Antibiotic resistance in lactic acid bacteria isolated from some pharmaceutical and dairy products. Braz. J. Microbiol. 2014, 45, 25–33. [Google Scholar] [CrossRef]
- Pimentel, T.C.; da Costa, W.K.A.; Delfino, T.P.C.; de Oliveira, S.M.L.; Sivieri, K.; Magnani, M. Chapter 5—Foods and supplements as probiotic delivery vehicles. In Probiotics for Human Nutrition in Health and Disease; de Souza, E.L., de Brito Alves, J.L., Fusco, V., Eds.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 115–142. [Google Scholar] [CrossRef]
- Un-Nisa, A.; Khan, A.; Zakria, M.; Siraj, S.; Ullah, S.; Tipu, M.K.; Ikram, M.; Kim, M.O. Updates on the Role of Probiotics against Different Health Issues: Focus on Lactobacillus. Int. J. Mol. Sci. 2023, 24, 142. [Google Scholar] [CrossRef]
- Ahmed, S.; Singh, S.; Singh, V.; Roberts, K.D.; Zaidi, A.; Rodriguez-Palacios, A. The Weissella Genus: Clinically Treatable Bacteria with Antimicrobial/Probiotic Effects on Inflammation and Cancer. Microorganisms 2022, 10, 2427. [Google Scholar] [CrossRef]
- Tekce, M.; Ince, G.; Gursoy, H.; Dirikan Ipci, S.; Cakar, G.; Kadir, T.; Yilmaz, S. Clinical and microbiological effects of probiotic lozenges in the treatment of chronic periodontitis: A 1-year follow-up study. J. Clin. Periodontol. 2015, 42, 363–372. [Google Scholar] [CrossRef]
- Sabatini, S.; Lauritano, D.; Candotto, V.; Silvestre, F.J.; Nardi, G.M. Oral probiotics in the management of gingivitis in diabetic patients: A double blinded randomized controlled study. J. Biol. Regul. Homeost. Agents 2017, 31, 197–202. [Google Scholar]
- Keller, M.K.; Brandsborg, E.; Holmstrom, K.; Twetman, S. Effect of tablets containing probiotic candidate strains on gingival inflammation and composition of the salivary microbiome: A randomised controlled trial. Benef. Microbes 2018, 9, 487–494. [Google Scholar] [CrossRef]
- Nadkerny, P.V.; Ravishankar, P.L.; Pramod, V.; Agarwal, L.A.; Bhandari, S. A comparative evaluation of the efficacy of probiotic and chlorhexidine mouthrinses on clinical inflammatory parameters of gingivitis: A randomized controlled clinical study. J. Indian Soc. Periodontol. 2015, 19, 633–639. [Google Scholar] [CrossRef]
- Grusovin, M.G.; Bossini, S.; Calza, S.; Cappa, V.; Garzetti, G.; Scotti, E.; Gherlone, E.F.; Mensi, M. Clinical efficacy of Lactobacillus reuteri-containing lozenges in the supportive therapy of generalized periodontitis stage III and IV, grade C: 1-year results of a double-blind randomized placebo-controlled pilot study. Clin. Oral Investig. 2020, 24, 2015–2024. [Google Scholar] [CrossRef]
- Parahitiyawa, N.; Scully, C.; Leung, W.; Yam, W.; Jin, L.; Samaranayake, L. Exploring the oral bacterial flora: Current status and future directions. Oral Dis. 2010, 16, 136–145. [Google Scholar] [CrossRef]
- Meurman, J.H.; Stamatova, I. Probiotics: Evidence of Oral Health Implications. Folia Med. 2018, 60, 21–29. [Google Scholar] [CrossRef]
- Bosch, J.A.; Turkenburg, M.; Nazmi, K.; Veerman, C.I.; de Geus, J.C.; Nieuw Amerongen, A.V. Stress as a determinant of saliva-mediated adherence and coadherence of oral and nonoral microorganisms. Psychosom. Med. 2003, 65, 604–612. [Google Scholar] [CrossRef]
- Stamatova, I.; Kari, K.; Vladimirov, S.; Meurman, J.H. In vitro evaluation of yoghurt starter lactobacilli and Lactobacillus rhamnosus GG adhesion to saliva-coated surfaces. Oral Microbiol. Immunol. 2009, 24, 218–223. [Google Scholar] [CrossRef]
- Kilian, M.; Chapple, I.; Hannig, M.; Marsh, P.D.; Meuric, V.; Pedersen, A.M.L.; Tonetti, M.S.; Wade, W.G.; Zaura, E. The oral microbiome—An update for oral healthcare professionals. Br. Dent. J. 2016, 221, 657–666. [Google Scholar] [CrossRef]
- Ellison, R.T., III; Giehl, T.J. Killing of gram-negative bacteria by lactoferrin and lysozyme. J. Clin. Investig. 1991, 88, 1080–1091. [Google Scholar] [CrossRef]
- Lynge Pedersen, A.M.; Belstrøm, D. The role of natural salivary defences in maintaining a healthy oral microbiota. J. Dent. 2019, 80, 3–12. [Google Scholar] [CrossRef]
- Shah, N.P. Probiotic bacteria: Selective enumeration and survival in dairy foods. J. Dairy Sci. 2000, 83, 894–907. [Google Scholar] [CrossRef]
- Pitino, I.; Randazzo, C.L.; Mandalari, G.; Lo Curto, A.; Faulks, R.M.; Le Marc, Y.; Bisignano, C.; Caggia, C.; Wickham, M.S.J. Survival of Lactobacillus rhamnosus strains in the upper gastrointestinal tract. Food Microbiol. 2010, 27, 1121–1127. [Google Scholar] [CrossRef]
- Monteagudo-Mera, A.; Rodríguez-Aparicio, L.B.; Rúa, J.; Martínez-Blanco, H.; Navasa, N.; García-Armesto, M.R.; Ferrero, M.Á. In vitro evaluation of physiological probiotic properties of different lactic acid bacteria strains of dairy and human origin. J. Funct. Foods 2012, 4, 531–541. [Google Scholar] [CrossRef]
- Quinto, E.J.; Jiménez, P.; Caro, I.; Tejero, J.; Mateo, J.; Girbés, T. Probiotic lactic acid bacteria: A review. Food Nutr. Sci. 2014, 5, 1765. [Google Scholar] [CrossRef]
- Trunk, T.; Khalil, H.S.; Leo, J.C. Bacterial autoaggregation. AIMS Microbiol. 2018, 4, 140–164. [Google Scholar] [CrossRef]
- Nishiyama, K.; Sugiyama, M.; Mukai, T. Adhesion Properties of Lactic Acid Bacteria on Intestinal Mucin. Microorganisms 2016, 4, 34. [Google Scholar] [CrossRef]
- Ayivi, R.D.; Gyawali, R.; Krastanov, A.; Aljaloud, S.O.; Worku, M.; Tahergorabi, R.; Silva, R.C.d.; Ibrahim, S.A. Lactic Acid Bacteria: Food Safety and Human Health Applications. Dairy 2020, 1, 202–232. [Google Scholar] [CrossRef]
- Van Tassell, M.L.; Miller, M.J. Lactobacillus adhesion to mucus. Nutrients 2011, 3, 613–636. [Google Scholar] [CrossRef]
- Bartholomew, J.W.; Mittwer, T. The Gram stain. Bacteriol. Rev. 1952, 16, 1–29. [Google Scholar] [CrossRef]
- Duke, P.B.; Jarvis, J.D. The catalase test—A cautionary tale. J. Med. Lab. Technol. 1972, 29, 203–204. [Google Scholar]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Marteau, P.; Minekus, M.; Havenaar, R.; Huis, J.H.J. Survival of Lactic Acid Bacteria in a Dynamic Model of the Stomach and Small Intestine: Validation and the Effects of Bile. J. Dairy Sci. 1997, 80, 1031–1037. [Google Scholar] [CrossRef]
- Bove, P.; Gallone, A.; Russo, P.; Capozzi, V.; Albenzio, M.; Spano, G.; Fiocco, D. Probiotic features of Lactobacillus plantarum mutant strains. Appl. Microbiol. Biotechnol. 2012, 96, 431–441. [Google Scholar] [CrossRef]
- Damodharan, K.; Lee, Y.; Palaniyandi, S.A.; Yang, S.; Suh, J.W. Preliminary probiotic and technological characterization of Pediococcus pentosaceus strain KID7 and in vivo assessment of its cholesterol-lowering activity. Front. Microbiol. 2015, 6, 768. [Google Scholar] [CrossRef]
- Plessas, S.; Kiousi, D.E.; Rathosi, M.; Alexopoulos, A.; Kourkoutas, Y.; Mantzourani, I.; Galanis, A.; Bezirtzoglou, E. Isolation of a Lactobacillus paracasei Strain with Probiotic Attributes from Kefir Grains. Biomedicines 2020, 8, 594. [Google Scholar] [CrossRef]
- Jia, G.C.; Che, N.; Xia, Y.J.; Lai, P.F.; Xiong, Z.Q.; Wang, G.Q.; Zhang, H.; Ai, L.Z. Adhesion to pharyngeal epithelium and modulation of immune response: Lactobacillus salivarius AR809, a potential probiotic strain isolated from the human oral cavity. J. Dairy Sci. 2019, 102, 6738–6749. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, L.; Du, M.; Yi, H.; Guo, C.; Tuo, Y.; Han, X.; Li, J.; Zhang, L.; Yang, L. Antimicrobial activity against Shigella sonnei and probiotic properties of wild lactobacilli from fermented food. Microbiol. Res. 2011, 167, 27–31. [Google Scholar] [CrossRef]
- Unban, K.; Chaichana, W.; Baipong, S.; Abdullahi, A.D.; Kanpiengjai, A.; Shetty, K.; Khanongnuch, C. Probiotic and Antioxidant Properties of Lactic Acid Bacteria Isolated from Indigenous Fermented Tea Leaves (Miang) of North Thailand and Promising Application in Synbiotic Formulation. Fermentation 2021, 7, 195. [Google Scholar] [CrossRef]
- Monteiro, C.R.A.V.; do Carmo, M.S.; Melo, B.O.; Alves, M.S.; dos Santos, C.I.; Monteiro, S.G.; Bomfim, M.R.Q.; Fernandes, E.S.; Monteiro-Neto, V. In Vitro Antimicrobial Activity and Probiotic Potential of Bifidobacterium and Lactobacillus against Species of Clostridium. Nutrients 2019, 11, 448. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Jussiaux, F.; Miot-Sertier, C.; Nguyen-Lopez, D.; Badet, C.; Samot, J. Reliability of MALDI-TOF mass spectrometry to identify oral isolates of Streptococcus salivarius and Lactobacillus spp. Arch. Oral Biol. 2021, 121, 104983. [Google Scholar] [CrossRef]
- Kim, H.; Jeong, Y.; Kim, J.E.; Kim, Y.; Paek, N.S.; Kang, C.H. Anti-obesity Potential of Lactobacillus spp. Isolated from Infant Feces. Biotechnol. Bioprocess E 2021, 26, 575–585. [Google Scholar] [CrossRef]
- Mudryk, Z.J.; Podgórska, B. Enzymatic Activity of Bacterial Strains Isolated from Marine Beach Sediments. Pol. J. Environ. Stud. 2006, 15, 441–448. [Google Scholar]
- Song, M.W.; Chung, Y.; Kim, K.T.; Hong, W.S.; Chang, H.J.; Paik, H.D. Probiotic characteristics of Lactobacillus brevis B13-2 isolated from kimchi and investigation of antioxidant and immune-modulating abilities of its heat-killed cells. LWT 2020, 128, 109452. [Google Scholar] [CrossRef]
- Gao, Z.; Daliri, E.B.M.; Wang, J.; Liu, D.; Chen, S.; Ye, X.; Ding, T. Inhibitory effect of lactic acid bacteria on foodborne pathogens: A review. J. Food Prot. 2019, 82, 441–453. [Google Scholar] [CrossRef]
- Ren, D.; Zhu, J.; Gong, S.; Liu, H.; Yu, H. Antimicrobial Characteristics of Lactic Acid Bacteria Isolated from Homemade Fermented Foods. Biomed. Res. Int. 2018, 2018, 5416725. [Google Scholar] [CrossRef]
- Matevosyan, L.; Bazukyan, I.; Trchounian, A. Antifungal and antibacterial effects of newly created lactic acid bacteria associations depending on cultivation media and duration of cultivation. BMC Microbiol. 2019, 19, 102. [Google Scholar] [CrossRef]
- Vuotto, C.; Longo, F.; Donelli, G. Probiotics to counteract biofilm-associated infections: Promising and conflicting data. Int. J. Oral Sci. 2014, 6, 189–194. [Google Scholar] [CrossRef]
- Sookkhee, S.; Chulasiri, M.; Prachyabrued, W. Lactic acid bacteria from healthy oral cavity of Thai volunteers: Inhibition of oral pathogens. J. Appl. Microbiol. 2001, 90, 172–179. [Google Scholar] [CrossRef]
- Strahinic, I.; Busarcevic, M.; Pavlica, D.; Milasin, J.; Golic, N.; Topisirovic, L. Molecular and biochemical characterizations of human oral lactobacilli as putative probiotic candidates. Oral Microbiol. Immunol. 2007, 22, 111–117. [Google Scholar] [CrossRef]
- Itapary dos Santos, C.; Ramos França, Y.; Duarte Lima Campos, C.; Quaresma Bomfim, M.R.; Oliveira Melo, B.; Assunção Holanda, R.; Santos, V.L.; Gomes Monteiro, S.; Buozzi Moffa, E.; Souza Monteiro, A.; et al. Antifungal and Antivirulence Activity of Vaginal Lactobacillus Spp. Products against Candida Vaginal Isolates. Pathogens 2019, 8, 150. [Google Scholar] [CrossRef]
- Denkova, R.; Yanakieva, V.; Denkova, Z.; Nikolova, V.; Radeva, V. In vitro inhibitory activity of Bifidobacterium and Lactobacillus strains against Candida albicans. Bulg. J. Vet. Med. 2013, 16, 186–197. [Google Scholar]
- Liao, H.; Liu, S.; Wang, H.; Su, H.; Liu, Z. Enhanced antifungal activity of bovine lactoferrin-producing probiotic Lactobacillus casei in the murine model of vulvovaginal candidiasis. BMC Microbiol. 2019, 19, 7. [Google Scholar] [CrossRef]
- Ariningsih, I.; Ramona, Y.; Antara, N. Isolation, Screening, and Characterization of Probiotics (Lactic Acid Bacteria) Antagonistic Against Candida albicans. Metamorf. J. Biol. Sci. 2017, 4, 263. [Google Scholar] [CrossRef]
- Haukioja, A.; Yli-Knuuttila, H.; Loimaranta, V.; Kari, K.; Ouwehand, A.C.; Meurman, J.H.; Tenovuo, J. Oral adhesion and survival of probiotic and other lactobacilli and bifidobacteria in vitro. Oral Microbiol. Immunol. 2006, 21, 326–332. [Google Scholar] [CrossRef]
- Haukioja, A.; Loimaranta, V.; Tenovuo, J. Probiotic bacteria affect the composition of salivary pellicle and streptococcal adhesion in vitro. Oral Microbiol. Immunol. 2008, 23, 336–343. [Google Scholar] [CrossRef]
- Mantzourani, I.; Chondrou, P.; Bontsidis, C.; Karolidou, K.; Terpou, A.; Alexopoulos, A.; Bezirtzoglou, E.; Galanis, A.; Plessas, S. Assessment of the probiotic potential of lactic acid bacteria isolated from kefir grains: Evaluation of adhesion and antiproliferative properties in in vitro experimental systems. Ann. Microbiol. 2019, 69, 751–763. [Google Scholar] [CrossRef]
- Tokatlı, M.; Gülgör, G.; Bağder Elmacı, S.; Arslankoz İşleyen, N.; Özçelik, F. In Vitro Properties of Potential Probiotic Indigenous Lactic Acid Bacteria Originating from Traditional Pickles. BioMed Res. Int. 2015, 2015, 315819. [Google Scholar] [CrossRef]
- Fang, F.; Xu, J.; Li, Q.; Xia, X.; Du, G. Characterization of a Lactobacillus brevis strain with potential oral probiotic properties. BMC Microbiol. 2018, 18, 221. [Google Scholar] [CrossRef]
- Bosch, M.; Nart, J.; Audivert, S.; Bonachera, M.A.; Alemany, A.S.; Fuentes, M.C.; Cuñé, J. Isolation and characterization of probiotic strains for improving oral health. Arch. Oral Biol. 2012, 57, 539–549. [Google Scholar] [CrossRef]
- Khagwal, N.; Sharma, P.K.; Sharma, D.C. Screening and evaluation of Lactobacillus spp. for the development of potential probiotics. Afr. J. Microbiol. Res. 2014, 8, 1573–1579. [Google Scholar] [CrossRef]
- Jose, N.M.; Bunt, C.R.; Hussain, M.A. Comparison of Microbiological and Probiotic Characteristics of Lactobacilli Isolates from Dairy Food Products and Animal Rumen Contents. Microorganisms 2015, 3, 198–212. [Google Scholar] [CrossRef]
- Montville, T.J.; Matthews, K.R. Physiology, growth, and inhibition of microbes in foods. Food Microbiol. Fundam. Front. 2012, 2013, 3–18. [Google Scholar] [CrossRef]
- Alameri, F.; Tarique, M.; Osaili, T.; Obaid, R.; Abdalla, A.; Masad, R.; Al-Sbiei, A.; Fernandez-Cabezudo, M.; Liu, S.Q.; Al-Ramadi, B.; et al. Lactic Acid Bacteria Isolated from Fresh Vegetable Products: Potential Probiotic and Postbiotic Characteristics Including Immunomodulatory Effects. Microorganisms 2022, 10, 389. [Google Scholar] [CrossRef]
- Aarti, C.; Khusro, A.; Varghese, R.; Arasu, M.V.; Agastian, P.; Al-Dhabi, N.A.; Ilavenil, S.; Choi, K.C. In vitro investigation on probiotic, anti-Candida, and antibiofilm properties of Lactobacillus pentosus strain LAP1. Arch. Oral Biol. 2018, 89, 99–106. [Google Scholar] [CrossRef]
- Li, M.; Wang, Y.; Cui, H.; Li, Y.; Sun, Y.; Qiu, H.J. Characterization of Lactic Acid Bacteria Isolated from the Gastrointestinal Tract of a Wild Boar as Potential Probiotics. Front. Vet. Sci. 2020, 7, 49. [Google Scholar] [CrossRef]
- Tuo, Y.; Yu, H.; Ai, L.; Wu, Z.; Guo, B.; Chen, W. Aggregation and adhesion properties of 22 Lactobacillus strains. J. Dairy Sci. 2013, 96, 4252–4257. [Google Scholar] [CrossRef]
- Campana, R.; van Hemert, S.; Baffone, W. Strain-specific probiotic properties of lactic acid bacteria and their interference with human intestinal pathogens invasion. Gut Pathog. 2017, 9, 12. [Google Scholar] [CrossRef]
- Sharma, S.; Kandasamy, S.; Kavitake, D.; Shetty, P.H. Probiotic characterization and antioxidant properties of Weissella confusa KR780676, isolated from an Indian fermented food. LWT 2018, 97, 53–60. [Google Scholar] [CrossRef]
- Lakra, A.K.; Domdi, L.; Hanjon, G.; Tilwani, Y.M.; Arul, V. Some probiotic potential of Weissella confusa MD1 and Weissella cibaria MD2 isolated from fermented batter. LWT 2020, 125, 125–109261. [Google Scholar] [CrossRef]
- Moslem, P.; Hossein, N.; Mahdi, R.; Seyed, N.H.; Seyed, A.S. Lactobacillus rhamnosus Gorbach-Goldin (GG): A top well-researched probiotic strain. J. Med. Microbiol. 2017, 5, 46–59. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).