Whole-Genome Sequence of Aeromonas spp. Isolated from a Dairy Farm in Central Texas
Abstract
1. Introduction
2. Materials and Methods
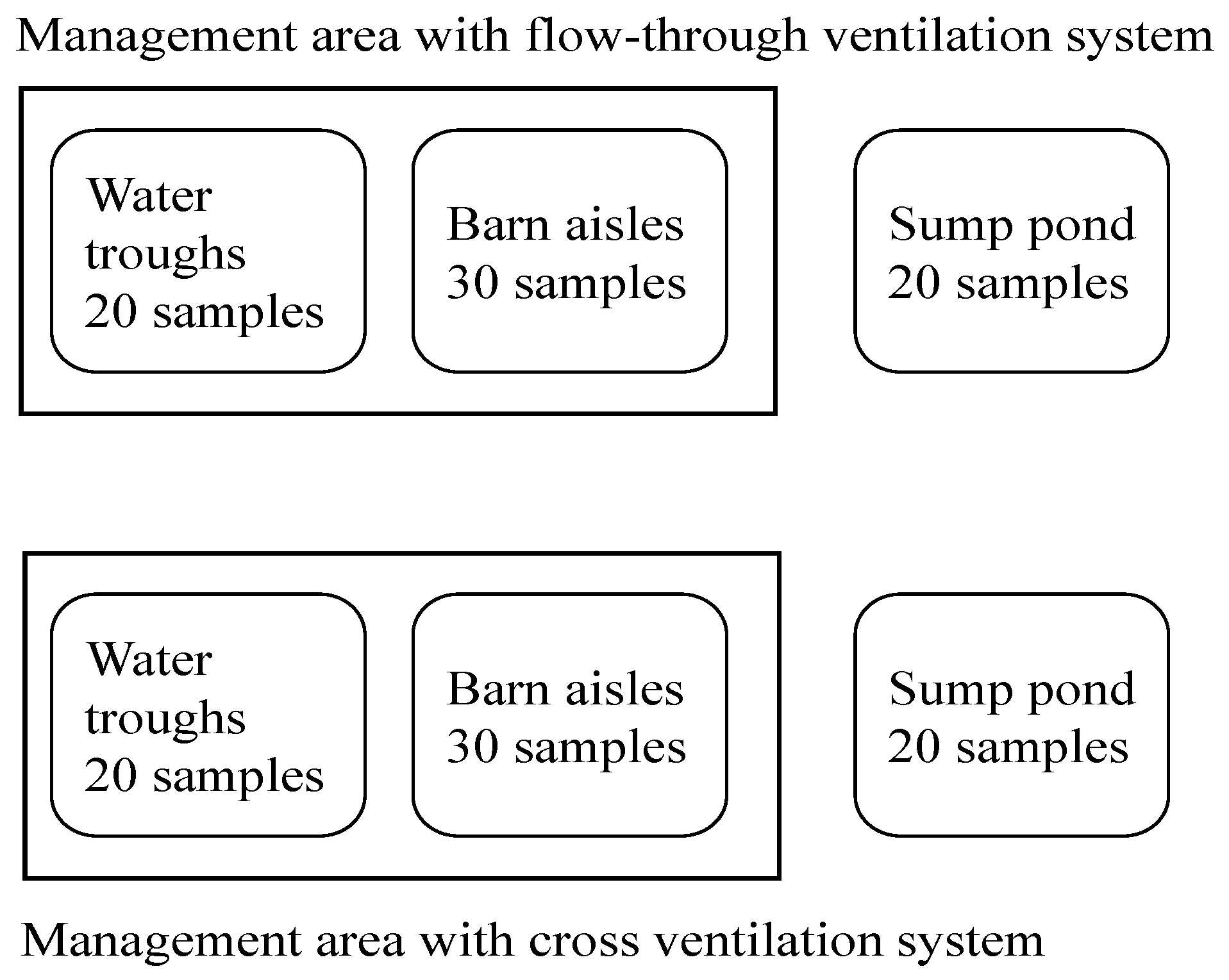
2.1. Sample Collection
2.2. Aeromonas Isolation and Identification
2.3. Statistical Testing
2.4. Antimicrobial Susceptibility Testing
2.5. WGS and Assembly
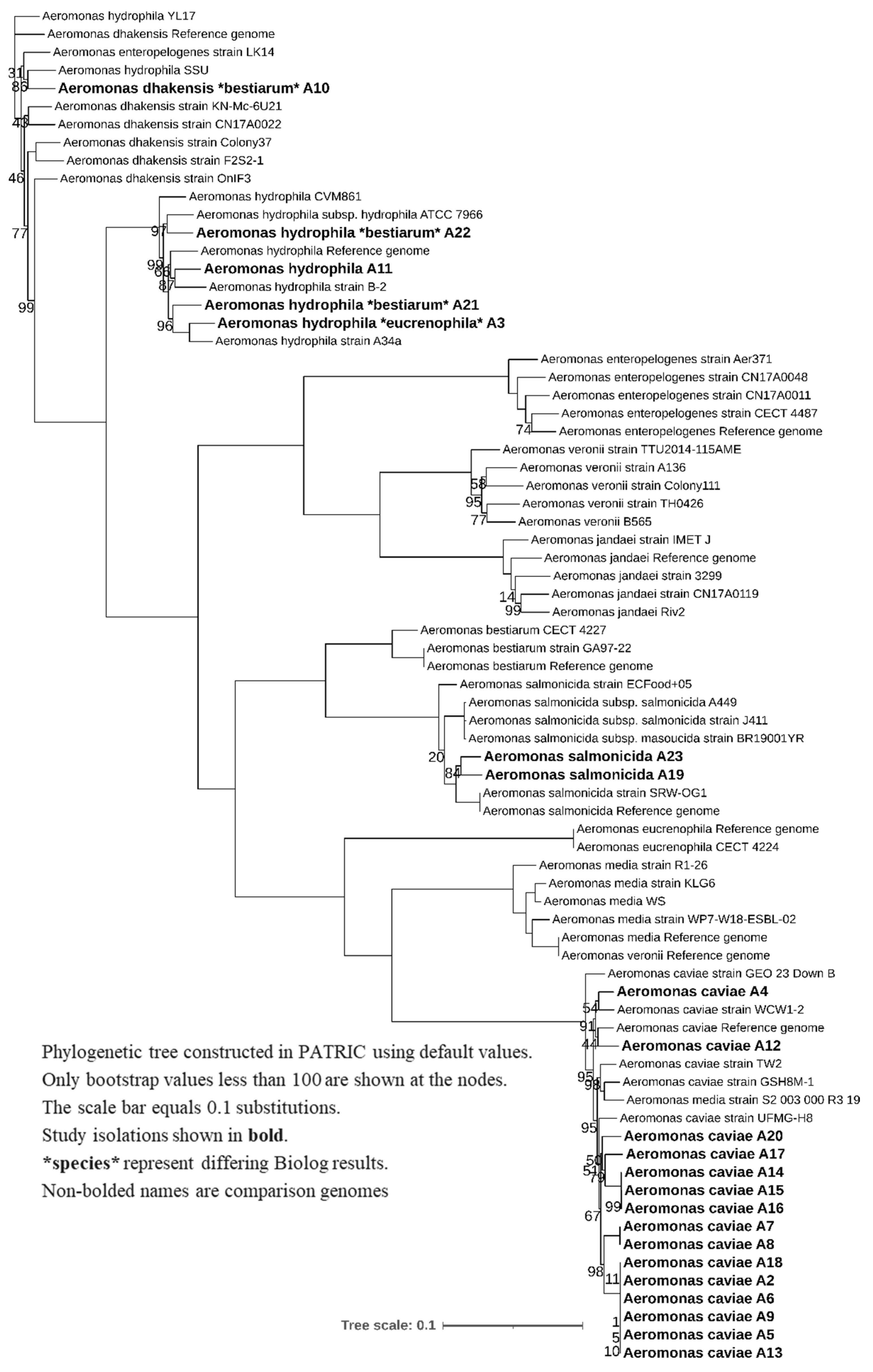
2.6. Phylogenetic Tree Construction
3. Results
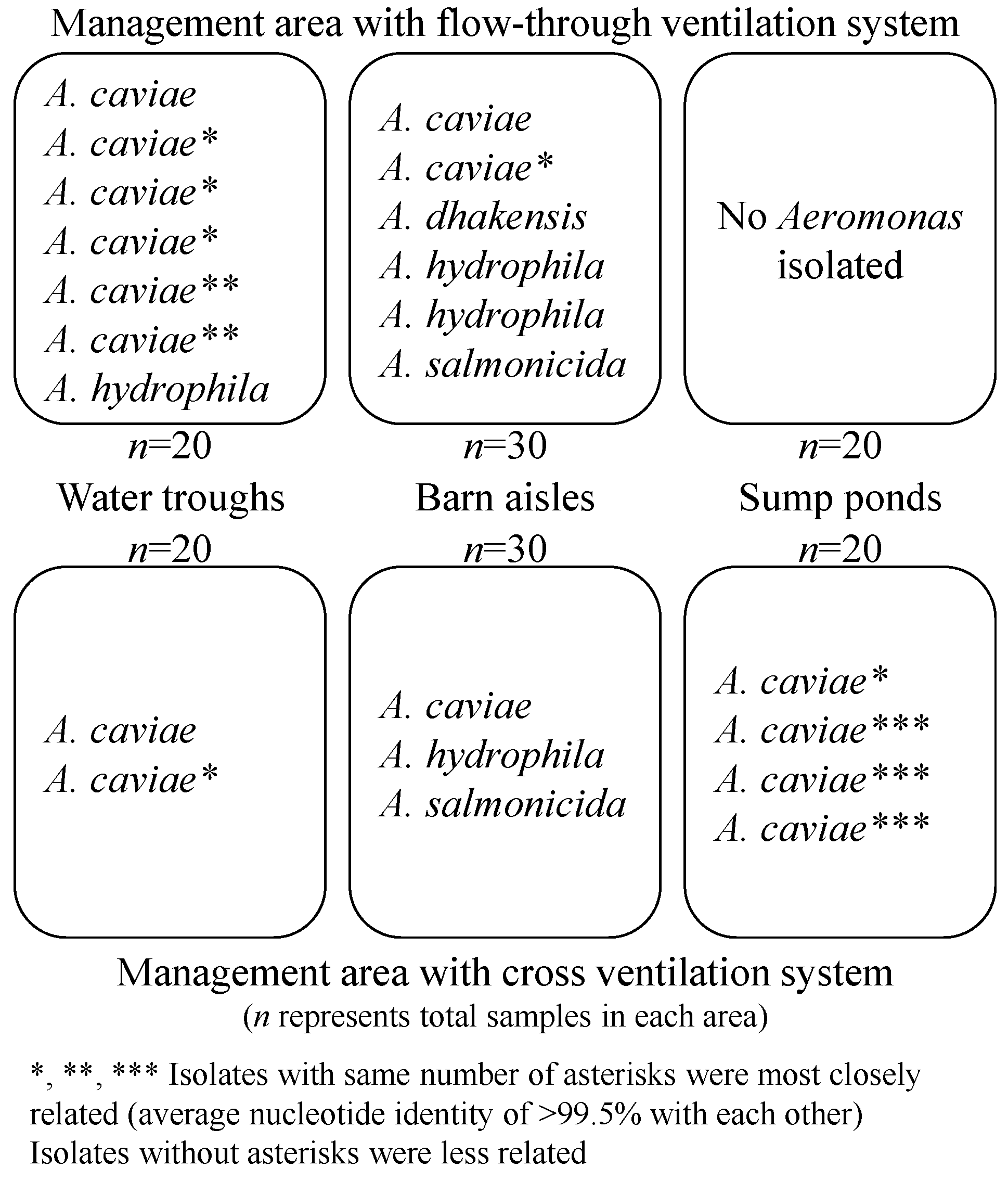
3.1. Aeromonas Collection and Presumptive Speciation
3.2. WGS Phylogeny
3.3. Antimicrobial Resistance
3.4. Mobility Elements
3.5. Aeromonas Virulence Genes
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Bertran, X.; Rubio, M.; Gomez, L.; Llovet, T.; Munoz, C.; Navarro, F.; Miro, E. Taxonomic Identification of Different Species of the Genus Aeromonas by Whole-Genome Sequencing and Use of Their Species-Specific beta-Lactamases as Phylogenetic Markers. Antibiotics 2021, 10, 354. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Bravo, A.; Figueras, M.J. An Update on the Genus Aeromonas: Taxonomy, Epidemiology, and Pathogenicity. Microorganisms 2020, 8, 129. [Google Scholar] [CrossRef] [PubMed]
- Goncalves Pessoa, R.B.; de Oliveira, W.F.; Marques, D.S.C.; Dos Santos Correia, M.T.; de Carvalho, E.; Coelho, L. The genus Aeromonas: A general approach. Microb. Pathog. 2019, 130, 81–94. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. Evolving concepts regarding the genus Aeromonas: An expanding Panorama of species, disease presentations, and unanswered questions. Clin. Infect. Dis. 1998, 27, 332–344. [Google Scholar] [CrossRef]
- Monfort, P.; Baleux, B. Distribution and survival of motile Aeromonas spp. in brackish water receiving sewage treatment effluent. Appl. Environ. Microbiol. 1991, 57, 2459–2467. [Google Scholar] [CrossRef]
- Mackenzie, J.S.; Jeggo, M. The One Health Approach-Why Is It So Important? Trop. Med. Infect. Dis. 2019, 4, 88. [Google Scholar] [CrossRef] [PubMed]
- McEwen, S.A.; Collignon, P.J. Antimicrobial Resistance: A One Health Perspective. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Gray, S.J.; Stickler, D.J. Some observations on the faecal carriage of mesophilic Aeromonas species in cows and pigs. Epidemiol. Infect. 1989, 103, 523–537. [Google Scholar] [CrossRef]
- Gray, S.J.; Stickler, D.J.; Bryant, T.N. The incidence of virulence factors in mesophilic Aeromonas species isolated from farm animals and their environment. Epidemiol. Infect. 1990, 105, 277–294. [Google Scholar] [CrossRef]
- Igbinosa, I.H.; Igumbor, E.U.; Aghdasi, F.; Tom, M.; Okoh, A.I. Emerging Aeromonas species infections and their significance in public health. Sci. World J. 2012, 2012, 625023. [Google Scholar] [CrossRef]
- Neyts, K.; Huys, G.; Uyttendaele, M.; Swings, J.; Debevere, J. Incidence and identification of mesophilic Aeromonas spp. from retail foods. Lett. Appl. Microbiol. 2000, 31, 359–363. [Google Scholar] [CrossRef]
- Palumbo, S.A.; Bencivengo, M.M.; Del Corral, F.; Williams, A.C.; Buchanan, R.L. Characterization of the Aeromonas hydrophila group isolated from retail foods of animal origin. J. Clin. Microbiol. 1989, 27, 854–859. [Google Scholar] [CrossRef]
- Ling, Z.; Yin, W.; Li, H.; Zhang, Q.; Wang, X.; Wang, Z.; Ke, Y.; Wang, Y.; Shen, J. Chromosome-Mediated mcr-3 Variants in Aeromonas veronii from Chicken Meat. Antimicrob. Agents Chemother. 2017, 61, e01272-17. [Google Scholar] [CrossRef]
- Chen, P.L.; Lamy, B.; Ko, W.C. Aeromonas dhakensis, an Increasingly Recognized Human Pathogen. Front. Microbiol. 2016, 7, 793. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. The genus Aeromonas: Taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 2010, 23, 35–73. [Google Scholar] [CrossRef]
- Grim, C.J.; Kozlova, E.V.; Ponnusamy, D.; Fitts, E.C.; Sha, J.; Kirtley, M.L.; van Lier, C.J.; Tiner, B.L.; Erova, T.E.; Joseph, S.J.; et al. Functional genomic characterization of virulence factors from necrotizing fasciitis-causing strains of Aeromonas hydrophila. Appl. Environ. Microbiol. 2014, 80, 4162–4183. [Google Scholar] [CrossRef] [PubMed]
- Grim, C.J.; Kozlova, E.V.; Sha, J.; Fitts, E.C.; van Lier, C.J.; Kirtley, M.L.; Joseph, S.J.; Read, T.D.; Burd, E.M.; Tall, B.D.; et al. Characterization of Aeromonas hydrophila wound pathotypes by comparative genomic and functional analyses of virulence genes. Mbio 2013, 4, e00064-13. [Google Scholar] [CrossRef]
- Nawaz, M.; Khan, S.A.; Khan, A.A.; Sung, K.; Tran, Q.; Kerdahi, K.; Steele, R. Detection and characterization of virulence genes and integrons in Aeromonas veronii isolated from catfish. Food Microbiol. 2010, 27, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Figueras, M.J.; Aldea, M.J.; Fernandez, N.; Aspiroz, C.; Alperi, A.; Guarro, J. Aeromonas hemolytic uremic syndrome. A case and a review of the literature. Diagn. Microbiol. Infect. Dis. 2007, 58, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Pablos, M.; Remacha, M.A.; Rodriguez-Calleja, J.M.; Santos, J.A.; Otero, A.; Garcia-Lopez, M.L. Identity, virulence genes, and clonal relatedness of Aeromonas isolates from patients with diarrhea and drinking water. Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 1163–1172. [Google Scholar] [CrossRef]
- Avison, M.B.; Niumsup, P.; Walsh, T.R.; Bennett, P.M. Aeromonas hydrophila AmpH and CepH beta-lactamases: Derepressed expression in mutants of Escherichia coli lacking creB. J. Antimicrob. Chemother. 2000, 46, 695–702. [Google Scholar] [CrossRef]
- Walsh, T.R.; Hall, L.; MacGowan, A.P.; Bennett, P.M. Sequence analysis of two chromosomally mediated inducible β-lactamases from Aeromonas sobria, strain 163a, one a class D penicillinase, the other an AmpC cephalosporinase. J. Antimicrob. Chemother. 1995, 36, 41–52. [Google Scholar] [CrossRef]
- Walsh, T.R.; Stunt, R.A.; Nabi, J.A.; MacGowan, A.P.; Bennett, P.M. Distribution and expression of beta-lactamase genes among Aeromonas spp. J. Antimicrob. Chemother. 1997, 40, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, G.A. AmpC beta-lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182. [Google Scholar] [CrossRef]
- Piotrowska, M.; Przygodzinska, D.; Matyjewicz, K.; Popowska, M. Occurrence and Variety of beta-Lactamase Genes among Aeromonas spp. Isolated from Urban Wastewater Treatment Plant. Front. Microbiol. 2017, 8, 863. [Google Scholar] [CrossRef] [PubMed]
- Fosse, T.; Giraud-Morin, C.; Madinier, I.; Labia, R. Sequence analysis and biochemical characterisation of chromosomal CAV-1 (Aeromonas caviae), the parental cephalosporinase of plasmid-mediated AmpC 'FOX' cluster. FEMS Microbiol. Lett. 2003, 222, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Di Pilato, V.; Arena, F.; Giani, T.; Conte, V.; Cresti, S.; Rossolini, G.M. Characterization of pFOX-7a, a conjugative IncL/M plasmid encoding the FOX-7 AmpC-type beta-lactamase, involved in a large outbreak in a neonatal intensive care unit. J. Antimicrob. Chemother. 2014, 69, 2620–2624. [Google Scholar] [CrossRef]
- Ebmeyer, S.; Kristiansson, E.; Larsson, D.G.J. CMY-1/MOX-family AmpC beta-lactamases MOX-1, MOX-2 and MOX-9 were mobilized independently from three Aeromonas species. J. Antimicrob. Chemother. 2019, 74, 1202–1206. [Google Scholar] [CrossRef]
- Piotrowska, M.; Popowska, M. Insight into the mobilome of Aeromonas strains. Front. Microbiol. 2015, 6, 494. [Google Scholar] [CrossRef]
- Rasmussen-Ivey, C.R.; Figueras, M.J.; McGarey, D.; Liles, M.R. Virulence Factors of Aeromonas hydrophila: In the Wake of Reclassification. Front. Microbiol. 2016, 7, 1337. [Google Scholar] [CrossRef]
- Parker, J.L.; Shaw, J.G. Aeromonas spp. clinical microbiology and disease. J. Infect. 2011, 62, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef] [PubMed]
- Bello-Lopez, J.M.; Cabrero-Martinez, O.A.; Ibanez-Cervantes, G.; Hernandez-Cortez, C.; Pelcastre-Rodriguez, L.I.; Gonzalez-Avila, L.U.; Castro-Escarpulli, G. Horizontal Gene Transfer and Its Association with Antibiotic Resistance in the Genus Aeromonas spp. Microorganisms 2019, 7, 363. [Google Scholar] [CrossRef] [PubMed]
- Poole, T.L.; Schlosser, W.D.; Anderson, R.C.; Norman, K.N.; Beier, R.C.; Nisbet, D.J. Whole-Genome Sequence of Aeromonas hydrophila CVM861 Isolated from Diarrhetic Neonatal Swine. Microorganisms 2020, 8, 1648. [Google Scholar] [CrossRef]
- Anandan, S.; Gopi, R.; Devanga Ragupathi, N.K.; Muthuirulandi Sethuvel, D.P.; Gunasekaran, P.; Walia, K.; Veeraraghavan, B. First report of bla(OXA-181)-mediated carbapenem resistance in Aeromonas caviae in association with pKP3-A: Threat for rapid dissemination. J. Glob. Antimicrob. Resist. 2017, 10, 310–314. [Google Scholar] [CrossRef] [PubMed]
- Poole, T.L.; Callaway, T.R.; Bischoff, K.M.; Warnes, C.E.; Nisbet, D.J. Macrolide inactivation gene cluster mphA-mrx-mphR adjacent to a class 1 integron in Aeromonas hydrophila isolated from a diarrhoeic pig in Oklahoma. J. Antimicrob. Chemother. 2006, 57, 31–38. [Google Scholar] [CrossRef]
- Ceylan, E.; Berktas, M.; Agaoglu, Z. The occurrence and antibiotic resistance of motile Aeromonas in livestock. Trop. Anim. Health Prod. 2009, 41, 199–204. [Google Scholar] [CrossRef]
- EPA. Method 1605: Aeromonas in Finished Water by Membrane Filtration; EPA: Washington, DC, USA, 2000. [Google Scholar]
- CLSI. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. In Approved Standard M07, 11th ed.; Clinical Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- CLSI. Performance standards for antimicrobial susceptibility testing. In M100, 31st ed.; Clinical Laboratory Standards Institute: Wayne, PA, USA, 2021. [Google Scholar]
- Stratev, D.; Vashin, I.; Daskalov, H. Determination of Beta-haemolytic activity and Minimum Inhibitory Concentrations of antimicrobial drugs against Aeromonas hydrophila strains isolated from fish products. Bulg. J. Vet. Med. 2015, 18, 239–247. [Google Scholar] [CrossRef]
- Davis, J.J.; Wattam, A.R.; Aziz, R.K.; Brettin, T.; Butler, R.; Butler, R.M.; Chlenski, P.; Conrad, N.; Dickerman, A.; Dietrich, E.M.; et al. The PATRIC Bioinformatics Resource Center: Expanding data and analysis capabilities. Nucleic Acids Res. 2020, 48, D606–D612. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Moller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Boratyn, G.M.; Thierry-Mieg, J.; Thierry-Mieg, D.; Busby, B.; Madden, T.L. Magic-BLAST, an accurate RNA-seq aligner for long and short reads. BMC Bioinform. 2019, 20, 405. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- BV-BRC. Phylogenetic Tree Service. Available online: https://www.bv-brc.org/docs/tutorial/phylogenetic_tree/phylogenetic_tree.html (accessed on 11 August 2022).
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Seshadri, R.; Joseph, S.W.; Chopra, A.K.; Sha, J.; Shaw, J.; Graf, J.; Haft, D.; Wu, M.; Ren, Q.; Rosovitz, M.J.; et al. Genome sequence of Aeromonas hydrophila ATCC 7966T: Jack of all trades. J. Bacteriol. 2006, 188, 8272–8282. [Google Scholar] [CrossRef]
- Blackwell, G.A.; Holt, K.E.; Bentley, S.D.; Hsu, L.Y.; Hall, R.M. Variants of AbGRI3 carrying the armA gene in extensively antibiotic-resistant Acinetobacter baumannii from Singapore. J. Antimicrob. Chemother. 2017, 72, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Karah, N.; Dwibedi, C.K.; Sjostrom, K.; Edquist, P.; Johansson, A.; Wai, S.N.; Uhlin, B.E. Novel Aminoglycoside Resistance Transposons and Transposon-Derived Circular Forms Detected in Carbapenem-Resistant Acinetobacter baumannii Clinical Isolates. Antimicrob. Agents Chemother. 2016, 60, 1801–1818. [Google Scholar] [CrossRef]
- Talagrand-Reboul, E.; Jumas-Bilak, E.; Lamy, B. The Social Life of Aeromonas through Biofilm and Quorum Sensing Systems. Front. Microbiol. 2017, 8, 37. [Google Scholar] [CrossRef]
- Jiang, B.; Howard, S.P. The Aeromonas hydrophila exeE gene, required both for protein secretion and normal outer membrane biogenesis, is a member of a general secretion pathway. Mol. Microbiol. 1992, 6, 1351–1361. [Google Scholar] [CrossRef]
- Webb, H.E.; Bugarel, M.; den Bakker, H.C.; Nightingale, K.K.; Granier, S.A.; Scott, H.M.; Loneragan, G.H. Carbapenem-Resistant Bacteria Recovered from Faeces of Dairy Cattle in the High Plains Region of the USA. PLoS ONE 2016, 11, e0147363. [Google Scholar] [CrossRef] [PubMed]
| Sample ID | Farm Sample # | Site | Biolog ID | WGS ID | Resistance Profile (MIC) µg/uL | SRA Accession Number | Biosample Accession Number |
|---|---|---|---|---|---|---|---|
| Samples collected from a management area that used flow-through ventilation | |||||||
| A2 | AFT2 | WT | A. caviae | A. caviae | Am(32)Ap(>32) | SRR15274966 | SAMN20441443 |
| A3 | AFT3 | WT | A. eucrenophilia | A. hydrophila | PS | SRR15274965 | SAMN20441444 |
| A4 | AFT1 | WT | A. caviae | A. caviae | Ap(>32) | SRR14289137 | SAMN18813779 |
| A5 | AFT5 | WT | A. caviae | A. caviae | Ap(>32) | SRR14289136 | SAMN18813781 |
| A6 | AFT6 | WT | A. caviae | A. caviae | Ap(>32) | SRR14289145 | SAMN18813782 |
| A7 | AFT9 | WT | A. caviae | A. caviae | Ap(>32) | SRR14289144 | SAMN18813783 |
| A8 | AFT9-5 | WT | A. caviae | A. caviae | Ap(>32) | SRR14289143 | SAMN18813784 |
| A9 | AFW1-3 | BA | A. caviae | A. caviae | Ap(>32) | SRR14289142 | SAMN18813785 |
| A10 | AFW3-2 | BA | A. bestiarum | A. dhakensis | Ap(>32)Am(>32)F(>32)T(8)Ax(2) | SRR15274964 | SAMN20441445 |
| A11 | AFW4-2 | BA | A. hydrophila | A. hydrophila | Ap(>32)Am(>32)F(>32)T(>8) | SRR14289141 | SAMN18813786 |
| A12 | AFW5-2 | BA | A. caviae | A. caviae | Ap(>32)Te(16) | SRR14289140 | SAMN18813787 |
| A21 | AFW7-3 | BA | A. bestiarum | A. hydrophila | Ap(>32) | SRR15274963 | SAMN20441446 |
| A23 | AFW10-2 | BA | A. salmonicida | A. salmonicida | Ap(>32) | SRR14289146 | SAMN18813796 |
| Samples collected from a management area that used cross-ventilation | |||||||
| A18 | AXT6 | WT | A. caviae | A. caviae | Ap(>32) | SRR14289149 | SAMN18813793 |
| A20 | AXT9 | WT | A. caviae | A. caviae | Ap(>32) | SRR14289147 | SAMN18813795 |
| A13 | AXS1-1 | SP | A. caviae | A. caviae | Ap(>32) | SRR14289139 | SAMN18813788 |
| A14 | AXS1-4 | SP | A. caviae | A. caviae | Ap(>32) | SRR14289138 | SAMN18813789 |
| A15 | AXS1-8-1 | SP | A. caviae | A. caviae | Ap(>32) | SRR14289135 | SAMN18813790 |
| A16 | AXS1-8-2 | SP | A. caviae | A. caviae | Ap(>32) | SRR14289134 | SAMN18813791 |
| A17 | AXW6-3w | BA | A. caviae | A. caviae | Ap(>32) | SRR14289133 | SAMN18813792 |
| A19 | AXW6-3y | BA | A. salmonicida ss pectinolytica | A. salmonicida | PS | SRR14289148 | SAMN18813794 |
| A22 | AXW8-2 | BA | A. bestiarum | A. hydrophila | Am(>32)Ap(>32)F(32)T(>8) | SRR15274962 | SAMN20441447 |
| Gene | A3—A. hydrophila | A10—A. dhakensis | A11—A. hydrophila | A21—A. hydrophila | A22—A. hydrophila | A19—A. salmonicida | A23—A. salmonicida | A2—A. caviae | A5—A. caviae | A6—A. caviae | A9—A. caviae | A13—A. caviae | A18—A. caviae | A7—A. caviae | A8—A. caviae | A4—A. caviae | A12—A. caviae | A14—A. caviae | A15—A. caviae | A16—A. caviae | A17—A. caviae | A20—A. caviae |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ampH_1 | 97.7 | 95.3 | 97.9 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ampH_2 | 97.7 | 95.3 | - | 97.1 | 98.9 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| blaMOX-6 | - | - | - | - | - | - | - | 96.6 | 96.6 | 96.6 | 96.6 | 96.6 | 96.6 | 97.6 | 97.6 | 97.2 | 97.2 | - | - | - | 97.3 | 97.6 |
| blaMOX-7 | - | - | - | - | - | - | - | 96.6 | 96.6 | 96.6 | 96.6 | 96.6 | 96.6 | - | - | - | - | 97.5 | 97.5 | 97.5 | - | - |
| cphA1 | - | 95.1 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| cphA1 | - | - | - | - | - | - | 96.0 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| cphA5 | - | - | - | - | - | 95.9 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| imiH | - | - | - | 96.6 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| tet(E) | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 99.9 | - | - | - | - | - |
| Gene | A3—A. hydrophila | A10—A. dhakensis | A11—A. hydrophila | A21—A. hydrophila | A22—A. hydrophila | A19—A. salmonicida | A23—A. salmonicida | A2—A. caviae | A5—A. caviae | A6—A. caviae | A9—A. caviae | A13—A. caviae | A18—A. caviae | A7—A. caviae | A8—A. caviae | A4—A. caviae | A12—A. caviae | A14—A. caviae | A15—A. caviae | A16—A. caviae | A17—A. caviae | A20—A. caviae |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ICEEcoUMN026-1 | - | - | - | - | - | 99.3 | - | - | - | - | - | - | - | 99.2 | 99.2 | 99.6 | 99.2 | - | - | 99.3 | - | - |
| IS5 | - | - | - | - | - | 99.3 | - | - | - | - | - | - | - | 99.2 | 99.2 | 99.7 | 99.4 | - | - | - | - | - |
| ISAeme15 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 100.0 | 99.7 | - | - | - | - | - |
| ISAeme16 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 99.8 | 99.8 | - | - | - | - | - |
| ISAeme19 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 99.5 | 99.6 | - | - | - | - | - |
| ISAeme21 | - | - | - | - | - | - | - | 96.6 | 96.6 | 96.6 | 96.6 | 96.6 | 96.6 | - | - | - | - | - | - | - | - | - |
| ISAeme3 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 96.8 | - | - | - | - |
| ISAhy1 | - | - | 95.5 | 96.1 | 97.1 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ISAhy3 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 99.0 | - | - | - | - |
| ISAs1 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 99.3 | - | - | - | - | - | - |
| ISAs15 | - | - | - | - | - | - | 99.9 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ISAs16 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 99.3 | - | - | - | - | - | - |
| ISAs17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 95.6 | - | 99.6 | - | - | - | - |
| ISAs18 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 95.3 | - | - | - | - | - | - |
| ISAs19 | - | - | - | - | - | - | - | - | - | - | 95.3 | 96.1 | - | - | - | 97.8 | - | - | - | - | - | - |
| ISAs20 | - | - | - | - | - | - | - | - | - | - | - | - | - | 98.2 | 98.2 | - | 99.6 | - | - | - | - | - |
| ISAs22 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 100.0 | - | - | - | - | - | - |
| ISAs23 | - | - | - | - | - | 98.4 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ISAs24 | - | - | - | - | - | 99.0 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ISAs26 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 99.6 | 100.0 | 99.7 | 99.4 | 99.3 | - | - |
| ISAs31 | - | - | - | - | - | 97.2 | 97.2 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ISAs32 | - | - | - | - | - | - | 96.5 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ISAs34 | - | - | - | - | - | 98.1 | 98.2 | - | - | - | - | - | - | 99.7 | 99.7 | - | - | - | 99.0 | 99.0 | - | - |
| ISAs5 | - | - | - | - | - | - | 95.5 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ISAs7 | - | - | - | - | - | - | 95.5 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ISAve3 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 96.6 | 96.5 | 96.5 | 96.5 | - | - |
| ISEc28 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 100.0 | 100.0 | - | - | - | - | - |
| MITEAeme1 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 100.0 | - | - | - | - | - | - |
| Tn6180 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 100.0 | 100.0 | - | - | - | - | - |
| Tn6234 | - | - | - | - | - | 98.7 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Tn6240 | - | - | - | - | - | 98.7 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Tn6279 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 100.0 | 100.0 | - | - | - | - | - |
| VFDB Identifier | A3—A. hydrophila | A10—A. dhakensis | A11—A. hydrophila | A21—A. hydrophila | A22—A. hydrophila | A19—A. salmonicida | A23—A. salmonicida | A2—A. caviae | A5—A. caviae | A6—A. caviae | A9—A. caviae | A13—A. caviae | A18—A. caviae | A7—A. caviae | A8—A. caviae | A4—A. caviae | A12—A. caviae | A14—A. caviae | A15—A. caviae | A16—A. caviae | A17—A. caviae | A20—A. caviae |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YP_856360 | - | - | 96.5 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_008043135 | 95.3 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_858602 | - | - | 96.8 | 96.3 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_008045131 | 97.4 | 95.7 | - | 98.7 | 97.6 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_008043134 | - | - | - | 97.9 | 96.7 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 145297253 | - | - | - | - | - | 97.3 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 145299407 | - | - | - | - | - | 96.2 | 96.4 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_855102 | 96.9 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_857337 | - | - | 96.8 | 96.8 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 111143381 | - | 98.2 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 145297589 | - | - | - | - | - | 97.5 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 145300643 | - | - | - | - | - | 98.9 | 95.7 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_855103 | - | - | - | - | - | - | - | 95.1 | 95.1 | 95.1 | 95.1 | 95.1 | 95.1 | 95.3 | 95.3 | - | 95.0 | - | - | - | - | 95.3 |
| YP_856379 | 97.0 | 95.9 | 97.4 | 96.5 | 97.4 | - | - | 96.3 | 96.3 | 96.3 | 96.3 | 96.3 | 96.3 | - | - | - | - | - | - | - | - | 97.0 |
| 145298366 | - | - | - | - | - | - | 98.3 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 145299432 | - | - | - | - | - | - | 97.1 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_857806 | - | - | - | - | 98.4 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_857151 | - | - | - | - | 98.5 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 145300802 | - | - | - | - | - | 97.8 | 97.9 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 145297730 | - | - | - | - | - | 97.5 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 507519905 | - | - | - | - | 97.7 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_858122 | - | - | - | 97.6 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_856293 | - | - | - | - | 99.2 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 145299468 | - | - | - | - | - | 96.0 | 96.8 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_857189 | 95.7 | - | 97.9 | 97.7 | 97.5 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_008041840 | - | - | 97.9 | 97.3 | 98.0 | - | - | - | - | - | - | - | - | - | - | 97.8 | - | - | - | - | - | - |
| 145297762 | - | - | - | - | - | 96.7 | 96.3 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 145297254 | - | - | - | - | - | - | 95.3 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_008043465 | 97.8 | - | 99.0 | 99.0 | 98.8 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 145299828 | - | - | - | - | - | 98.6 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_856993 | - | - | - | 96.2 | 95.6 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 145300215 | - | - | - | - | - | 98.5 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 145300516 | - | - | - | - | - | 97.5 | 97.6 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_855918 | - | - | - | 98.8 | 98.6 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_008042625 | 98.0 | - | 97.0 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_001140282 | - | - | - | - | - | 97.1 | 97.5 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 145300595 | - | - | - | - | - | 98.7 | 99.3 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_858308 | - | - | 99.5 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_855659 | - | 96.4 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 145300501 | - | - | - | - | - | 98.0 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_857681 | 96.5 | - | 97.0 | 97.2 | 97.0 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 145298152 | - | - | - | - | - | 97.5 | 97.9 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_857247 | - | - | 98.1 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 145298624 | - | - | - | - | - | - | 98.1 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 145297880 | - | - | - | - | - | 98.1 | 98.6 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 507523204 | 97.8 | 96.0 | 97.5 | 97.2 | 97.8 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_855347 | 97.4 | - | - | 97.8 | 99.2 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 145298519 | - | - | - | - | - | 97.9 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 145299774 | - | - | - | - | - | 98.7 | 98.4 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 145300766 | - | - | - | - | - | 98.0 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| YP_858604 | - | 96.5 | - | - | 95.6 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 507520337 | - | - | 97.4 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Identifier | Gene Description | A3—A. hydrophila | A10—A. dhakensis | A11—A. hydrophila | A21—A. hydrophila | A22—A. hydrophila |
|---|---|---|---|---|---|---|
| AB237183 | A. hydrophila ahlip11 gene for lipase | - | 97.4 | - | - | - |
| AF419157 | A. hydrophila isolate SSU enterotoxin (ast) | - | - | 96.1 | 96.5 | - |
| DQ650654 | A. hydrophila strain AH-1 LafK (lafK) | 97.6 | - | 98.3 | 98.9 | 98.8 |
| GQ856318 | A. hydrophila strain BSK-10 phospholipid-cholesterol acyltransferase (GCAT) | - | 97.0 | - | - | 98.7 |
| JN215210 | A. hydrophila strain FSP 384/10 flagellin (fla) | - | - | 95.5 | - | - |
| U81555 | A. hydrophila hemolysin (hlyA) | 96.7 | 95.5 | 96.5 | 96.4 | 96.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poole, T.L.; Schlosser, W.D.; Crippen, T.L.; Swiger, S.L.; Norman, K.N.; Anderson, R.C. Whole-Genome Sequence of Aeromonas spp. Isolated from a Dairy Farm in Central Texas. Microbiol. Res. 2023, 14, 161-176. https://doi.org/10.3390/microbiolres14010014
Poole TL, Schlosser WD, Crippen TL, Swiger SL, Norman KN, Anderson RC. Whole-Genome Sequence of Aeromonas spp. Isolated from a Dairy Farm in Central Texas. Microbiology Research. 2023; 14(1):161-176. https://doi.org/10.3390/microbiolres14010014
Chicago/Turabian StylePoole, Toni L., Wayne D. Schlosser, Tawni L. Crippen, Sonja L. Swiger, Keri N. Norman, and Robin C. Anderson. 2023. "Whole-Genome Sequence of Aeromonas spp. Isolated from a Dairy Farm in Central Texas" Microbiology Research 14, no. 1: 161-176. https://doi.org/10.3390/microbiolres14010014
APA StylePoole, T. L., Schlosser, W. D., Crippen, T. L., Swiger, S. L., Norman, K. N., & Anderson, R. C. (2023). Whole-Genome Sequence of Aeromonas spp. Isolated from a Dairy Farm in Central Texas. Microbiology Research, 14(1), 161-176. https://doi.org/10.3390/microbiolres14010014





