The Emergence of Carbapenem-Resistant Gram-Negative Bacteria in Mizoram, Northeast India
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Bacterial Isolates
2.2. Minimal Inhibitory Concentration (MIC)
2.3. DNA Isolation
2.4. Screening of Beta-Lactamase Genes
2.5. Identification of Carbapenemase Producers
2.6. Synergy Testing
3. Results
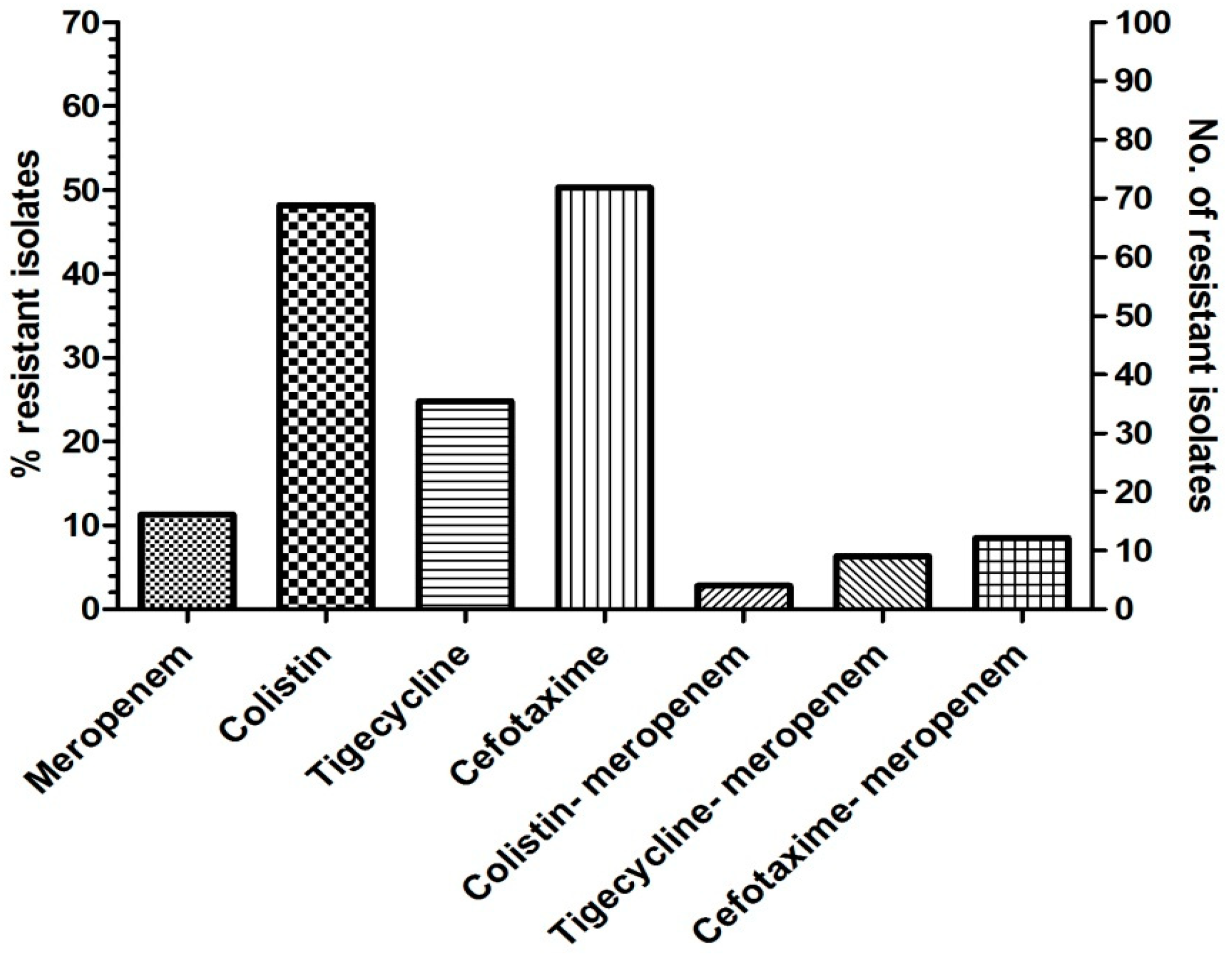
3.1. Bacterial Isolates and Carbapenem Resistance
3.2. Molecular Studies
3.3. Combinational Activity of Colistin-Meropenem
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leptihn, S. Welcome back to the Pre-Penicillin Era. Why we desperately need new strategies in the battle against bacterial pathogens. Infect. Microb. Dis. 2019, 1, 33. [Google Scholar] [CrossRef]
- Georgios, M. Carbapenem Resistance: Overview of the Problem and Future Perspectives. Ther. Adv. Infect. Dis. 2016, 3, 15–21. [Google Scholar] [CrossRef] [Green Version]
- Francis, C.; Donkor, E. Carbapenem Resistance: A Review. Med. Sci. 2017, 6, 1. [Google Scholar] [CrossRef] [Green Version]
- Fatma, E. Carbapenem-Resistant Enterobacteriaceae. Med. J. Islamic World Acad. Sci. 2017, 25, 6–11. [Google Scholar] [CrossRef]
- Aslam, B.; Rasool, M.; Muzammil, S.; Siddique, A.B.; Nawaz, Z.; Shafique, M.; Zahoor, M.A.; Binyamin, R.; Waseem, M.; Khurshid, M.; et al. Carbapenem resistance: Mechanisms and drivers of global menace. In Pathogenic Bacteria; Kırmusaoğlu, S., Bhardwaj, S.B., Eds.; IntechOpen: London, UK, 2020. [Google Scholar] [CrossRef] [Green Version]
- Patrice, N.; Poirel, L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-Negative Bacteria. Clin. Infect. Dis. 2019, 69 (Suppl. 7), S521–S528. [Google Scholar] [CrossRef] [Green Version]
- Vanegas, J.M.; Parra, O.L.; Jiménez, J.N. Molecular Epidemiology of Carbapenem Resistant Gram-Negative Bacilli from Infected Pediatric Population in Tertiary-Care Hospitals in Medellín, Colombia: An Increasing Problem. BMC Infect. Dis. 2016, 16, 463. [Google Scholar] [CrossRef] [Green Version]
- Garg, A.; Garg, J.; Kumar, S.; Bhattacharya, A.; Agarwal, S.; Upadhyay, G.C. Molecular Epidemiology & Therapeutic Options of Carbapenem-Resistant Gram-Negative Bacteria. Indian J. Med. Res. 2019, 149, 285. [Google Scholar] [CrossRef]
- Brink, A.J. Epidemiology of carbapenem-resistant Gram-negative infections globally. Curr. Opin. Infect. Dis. 2019, 32, 609–616. [Google Scholar] [CrossRef]
- Lalhmangaihzuali, F.E.; Varte, Z.; Laldinmawii, G. Antibiotic Resistance Pattern of Uropathogens in Urinary Tract Infections in Children at State Referral Hospital, Falkawn, Mizoram, India. Int. J. Contemp. Pediatrics 2018, 5, 2108–2113. [Google Scholar] [CrossRef] [Green Version]
- Ramesh, N.; Prasanth, M.; Ramkumar, S.; Suresh, M.; Tamhankar, A.J.; Gothandam, K.M.; Karthikeyan, S.; Bozdogan, B. Colistin Susceptibility of Gram-Negative Clinical Isolates from Tamil Nadu, India. Asian Biomed. 2016, 10, 35–39. [Google Scholar] [CrossRef]
- Doyle, D.; Peirano, G.; Lascols, C.; Lloyd, T.; Church, D.L.; Pitout, J.D. Laboratory Detection of Enterobacteriaceae That Produce Carbapenemases. J. Clin. Microbiol. 2012, 50, 3877–3880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nachimuthu, R.; Kannan, V.R.; Bozdogan, B.; Krishnakumar, V.; Pandian, K.S.; Manohar, P. CTX-M-type ESBL-mediated resistance to third-generation cephalosporins and conjugative transfer of resistance in Gram-negative bacteria isolated from hospitals in Tamil Nadu, India. Access Microbiol. 2020, 3, 000142. [Google Scholar] [CrossRef] [PubMed]
- Kargar, M.; Mohammadalipour, Z.; Doosti, A.; Lorzadeh, S.; Japoni-Nejad, A. High Prevalence of Class 1 to 3 Integrons among Multidrug-Resistant Diarrheagenic Escherichia Coli in Southwest of Iran. Osong Public Health Res. Perspect. 2014, 5, 193–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Versalovic, J.; Schneider, M.; De Bruijn, F.J.; Lupski, J.R. Genomic Fingerprinting of Bacteria Using Repetitive Sequence-Based Polymerase Chain Reaction. Methods Mol. Cell. Biol. 1994, 5, 25–40. [Google Scholar]
- Pavel, A.B.; Vasile, C.I. PyElph—A Software Tool for Gel Images Analysis and Phylogenetics. BMC Bioinform. 2012, 13, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Bilung, L.M.; Pui, C.F.; Su’ut, L.; Apun, K. Evaluation of BOX-PCR and ERIC-PCR as Molecular Typing Tools for Pathogenic Leptospira. Dis. Markers 2018, 2018, 1351634. [Google Scholar] [CrossRef] [Green Version]
- Pasteran, F.; Gonzalez, L.J.; Albornoz, E.; Bahr, G.; Vila, A.J.; Corso, A. Triton Hodge Test: Improved Protocol for Modified Hodge Test for Enhanced Detection of NDM and Other Carbapenemase Producers. J. Clin. Microbiol. 2016, 54, 640–649. [Google Scholar] [CrossRef] [Green Version]
- Costa, R.A.; Ortega, D.B.; Fulgêncio, D.L.; Costa, F.S.; Araújo, T.F.; Barreto, C.C. Checkerboard Testing Method Indicates Synergic Effect of Pelgipeptins against Multidrug Resistant Klebsiella Pneumoniae. Biotechnol. Res. Innov. 2019, 3, 187–191. [Google Scholar] [CrossRef]
- Manohar, P.; Shanthini, T.; Ayyanar, R.; Bozdogan, B.; Wilson, A.; Tamhankar, A.J.; Nachimuthu, R.; Lopes, B.S. The Distribution of Carbapenem- And Colistin-Resistance in Gram-Negative Bacteria from the Tamil Nadu Region in India. J. Med. Microbiol. 2017, 66, 874–883. [Google Scholar] [CrossRef]
- Chellapandi, K.; Dutta, T.K.; Sharma, I.; De Mandal, S.; Kumar, N.S.; Ralte, L. Prevalence of Multi Drug Resistant Enteropathogenic and Enteroinvasive Escherichia Coli Isolated from Children with and without Diarrhea in Northeast Indian Population. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 49. [Google Scholar] [CrossRef]
- Mukherjee, S.; Bhattacharjee, A.; Naha, S.; Majumdar, T.; Debbarma, S.K.; Kaur, H.; Dutta, S.; Basu, S. Molecular Characterization of NDM-1-Producing Klebsiella Pneumoniae ST29, ST347, ST1224, and ST2558 Causing Sepsis in Neonates in a Tertiary Care Hospital of North-East India. Infect. Genet. Evol. 2019, 69, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Loh, B.; Chen, J.; Manohar, P.; Yu, Y.; Hua, X.; Leptihn, S. A Biological Inventory of Prophages in A. baumannii Genomes Reveal Distinct Distributions in Classes, Length, and Genomic Positions. Front. Microbiol. 2020, 11, 579802. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Chetia, P.; Puzari, M.; Neog, N.; Borah, A. Menace to the Ultimate Antimicrobials among Common Enterobacteriaceae Clinical Isolates in Part of North-East India. BioRxiv 2019. [Google Scholar] [CrossRef]
- Kar, B.; Sharma, M.; Peter, A.; Chetia, P.; Neog, B.; Borah, A.; Pati, S.; Bhattacharya, D. Prevalence and Molecular Characterization of β-Lactamase Producers and Fluoroquinolone Resistant Clinical Isolates from North East India. J. Infect. Public Health 2021, 14, 628–637. [Google Scholar] [CrossRef] [PubMed]
- Walther-Rasmussen, J.; Høiby, N. OXA-Type Carbapenemases. J. Antimicrob. Chemother. 2006, 57, 373–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.; Kim, C.K.; Yong, D.; Jeong, S.H.; Yum, J.H.; Seo, Y.H.; Docquier, J.D.; Chong, Y. Improved Performance of the Modified Hodge Test with MacConkey Agar for Screening Carbapenemase-Producing Gram-Negative Bacilli. J. Microbiol. Methods 2010, 83, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Manohar, P.; Shanthini, T.; Ekta, P.; Mahesan, J.B.; Gothandam, K.M.; Bozdogan, B.; Ramesh, N. Colistin-Resistant Klebsiella Pneumoniae: Prevalence of Integrons and Synergistic out Turn for Colistin-Meropenem. Arch. Clin. Infect. Dis. 2018, 13, e55099. [Google Scholar] [CrossRef] [Green Version]
| S.no | Isolate ID | Bacteria | MIC | Resistance Gene | Int Gene | |||
|---|---|---|---|---|---|---|---|---|
| Col | Tig | Mer | Cef | |||||
| 1 | EC 2 | E. coli | 1 | 4 | 64 | >128 | blaOXA-48-like | - |
| 2 | EC 20 | E. coli | 2 | <0.12 | 32 | >128 | blaOXA-48-like | - |
| 3 | EC 15 | E. coli | 1 | <0.12 | 128 | >128 | blaOXA-48-like | - |
| 4 | SS 1 | Shigella sonnei | 2 | <0.12 | 128 | >128 | blaOXA-48-like and CTX-M-1 | Intl1 |
| 5 | EC 17 | E. coli | 2 | <0.12 | 32 | >128 | blaOXA-48-like and CTX-M-1 | Intl1 |
| 6 | SF 1 | Shigella flexneri | 2 | 4 | >128 | >128 | CTX-M-1 | Intl1 |
| 7 | EC 4 | E. coli | 2 | 0.5 | 0.5 | >128 | CTX-M-1 | Intl1 |
| 8 | SF 2 | Shigella flexneri | 2 | <0.12 | <0.12 | >128 | CTX-M-1 | Intl1 |
| 9 | EC 8 | E. coli | 1 | <0.12 | <0.12 | >128 | CTX-M-1 | Intl1 |
| 10 | EC 12 | E. coli | 2 | 0.5 | <0.12 | 4 | CTX-M-1 | Intl1 |
| 11 | EC 13 | E. coli | 2 | 0.5 | <0.12 | >128 | CTX-M-1 | Intl1 |
| 12 | KO 1 | K. oxytoca | 16 | 0.5 | <0.12 | >128 | CTX-M-1 | Intl1 |
| 13 | KP 37 | K. pneumoniae | 8 | <0.12 | <0.12 | >128 | CTX-M-1 | Intl1 |
| 14 | EC 47 | E. coli | 4 | <0.12 | <0.12 | >128 | CTX-M-1 | Intl1 |
| 15 | EC 49 | E. coli | 8 | 0.5 | <0.12 | <0.12 | CTX-M-1 | Intl1 |
| 16 | EC 61 | E. coli | 2 | 0.12 | <0.12 | >128 | CTX-M-1 | Intl1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ralte, V.S.C.; Loganathan, A.; Manohar, P.; Sailo, C.V.; Sanga, Z.; Ralte, L.; Zothanzama, J.; Leptihn, S.; Nachimuthu, R.; Kumar, N.S. The Emergence of Carbapenem-Resistant Gram-Negative Bacteria in Mizoram, Northeast India. Microbiol. Res. 2022, 13, 342-349. https://doi.org/10.3390/microbiolres13030027
Ralte VSC, Loganathan A, Manohar P, Sailo CV, Sanga Z, Ralte L, Zothanzama J, Leptihn S, Nachimuthu R, Kumar NS. The Emergence of Carbapenem-Resistant Gram-Negative Bacteria in Mizoram, Northeast India. Microbiology Research. 2022; 13(3):342-349. https://doi.org/10.3390/microbiolres13030027
Chicago/Turabian StyleRalte, Vanlalruati S. C., Archana Loganathan, Prasanth Manohar, Christine Vanlalbiakdiki Sailo, Zothan Sanga, Lalremruata Ralte, John Zothanzama, Sebastian Leptihn, Ramesh Nachimuthu, and Nachimuthu Senthil Kumar. 2022. "The Emergence of Carbapenem-Resistant Gram-Negative Bacteria in Mizoram, Northeast India" Microbiology Research 13, no. 3: 342-349. https://doi.org/10.3390/microbiolres13030027
APA StyleRalte, V. S. C., Loganathan, A., Manohar, P., Sailo, C. V., Sanga, Z., Ralte, L., Zothanzama, J., Leptihn, S., Nachimuthu, R., & Kumar, N. S. (2022). The Emergence of Carbapenem-Resistant Gram-Negative Bacteria in Mizoram, Northeast India. Microbiology Research, 13(3), 342-349. https://doi.org/10.3390/microbiolres13030027









