1. Introduction
Proteus mirabilis is a common cause of urinary tract infections (UTI) [
1]. In humans,
P. mirabilis is frequently associated with complicated, recurrent and catheter-associated UTIs (CAUTI) [
1,
2]. UTIs that are caused by
P. mirabilis are medically demanding because they are prone to persistence and to secondary complications such as urolithiasis, obstruction and pyelonephritis [
2].
P. mirabilis is also relevant in companion animals with UTI since it is the second most common Gram-negative bacteria that is isolated from dogs and cats [
3].
The increasing number of companion animals in modern society has raised concerns regarding the role of dogs and cats as reservoirs of pathogenic and antimicrobial-resistant bacteria [
4,
5]. In a previous study using pulse field gel electrophoresis (PFGE), a high number of clusters (cluster > 80% Dice/UPGMA index) were shown to contain clinical
P. mirabilis strains that were isolated from humans and companion animals (dogs and cats) with UTI [
6].
The closer contact between owners and their pets increases the opportunities for pathogen interchange through direct and indirect contact [
4,
5]. In fact, healthy dogs may share colonizing
Escherichia coli strains with humans who are living in the same household [
7,
8,
9,
10]. The gut has been identified as the major reservoir of uropathogenic bacteria, including
P. mirabilis [
1]. Notably, healthy dogs have been found to become colonized by
E. coli clonal lineages that are known to cause UTI in humans, namely ST131 [
11].
The worldwide
P. mirabilis clonal epidemiology is still poorly understood. This is due, to some extent, to the lack of portable
P. mirabilis molecular typing methods such as multi-locus sequence typing (MLST). Few studies have addressed the gut
P. mirabilis colonization of healthy dogs and cats [
12,
13]. Furthermore, to the best of our knowledge, there is a lack of studies characterizing the sharing of
P. mirabilis between companion animals and humans that are living in close contact.
This study aims to evaluate the P. mirabilis fecal colonization and sharing between companion animals and humans that are living in the same household. Furthermore, the fecal P. mirabilis strains were compared by PFGE with clinical strains from companion animals and humans with UTI.
2. Materials and Methods
2.1. Study Population
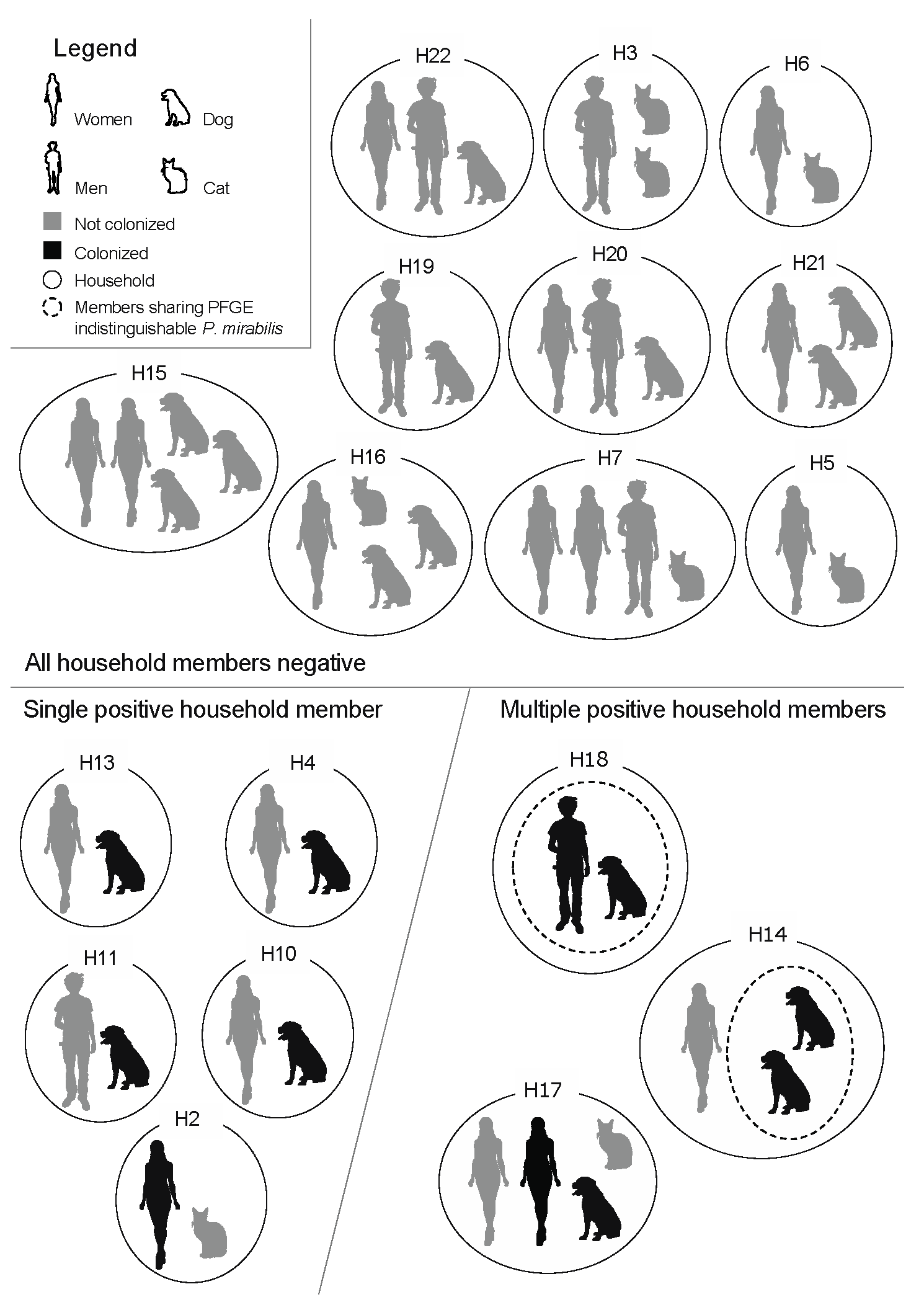
Households that were composed of healthy humans and companion animals (dogs or cats) were enrolled in this study, in 2016. The health status of the participants was accessed through a questionnaire and “healthy” was defined as the absence of known bacterial infections and antimicrobial use in the prior month. The human participants were informed of the main goals of the study and were required to sign a consent form. The studied population included a total of 50 individuals (24 humans, 18 dogs and 8 cats) across 18 households. All human participants had lived with the corresponding companion animals for at least 6 months prior to the study, with the exception of one cat that had been recently adopted to Household 16 (H16). Each family was composed of at least one human and one companion animal (dog or cat), yet the number of humans and companion animals varied among households (
Figure 1). Enrolment was anonymous, therefore the households, humans and animals were numbered and coded with the letters H, Hu and A, respectively.
2.2. Sample Collection and Bacteriological Methods
Ethical approval was obtained from the Comissão de Ética e Bem-Estar Animal (CEBEA) from the Faculty of Veterinary Medicine of the University of Lisbon. The human participants were asked to collect their own fecal samples, as well as the fecal samples from their pets, into sterile containers using non-invasive methods. The fecal samples were immediately stored at 4 °C until processing was undertaken.
After sample homogenization, one gramme of feces was added to 10 mL of sterile 0.85% NaCl (Merk, Darmstadt, Germany) solution and vortexed. Following thorough mixing, 10 μL of the fecal suspension was plated onto MacConkey agar plates (Scharlau, Barcelona, Spain) and Hektoen enteric agar plates (Biokar diagnostics, Beauvais, France). Ten microliters of the fecal suspension was also plated onto MacConkey agar plates and supplemented with 1.5 μg/mL of cefotaxime (Sigma-Aldrich, St. Louis, MO, USA) or meropenem (Sigma-Aldrich, St. Louis, MO, USA).
One gramme of feces was added to 5 mL of tetrathionate broth (tetrathionate broth base [Oxoid, Hampshire, UK] supplemented with iodine [AlliedSignal, Morristown, NJ, USA] and potassium iodide [Scharlau, Barcelona, Spain] according to the manufacturers’ instructions) in order to improve the detection of P. mirabilis. The fecal suspension in tetrathionate broth was homogenized and incubated at 36 ± 1 °C during the 18 h prior to the process of plating the samples in the agar plates, as described above. One tube of tetrathionate broth without a fecal sample and one tube of tetrathionate broth that was inoculated with known P. mirabilis strains were included as negative and positive quality controls, respectively.
After incubation at 36 ± 1 °C for 18h, all of the agar plates were inspected for the presence of suspected Proteus spp. colonies. When present, up to five colonies of each participant were isolated and stored in a 20% glycerol (Sigma-Aldrich, St. Louis, MO, USA) brain heart infusion broth (Biokar diagnostics, Beauvais, France) at −20 °C until further analysis was undertaken. The growth of a high number of colony forming units of Enterobacteriaceae after the direct plating of the samples in the MacConkey agar plates was deemed an appropriate quality control for sample viability.
2.3. Bacteria Identification
The total DNA extraction of each suspected isolate of
Proteus spp. was processed by using the boiling method and the species identification was conducted by PCR using the previously described species-specific primers [
14,
15].
2.4. Population Structure Analysis
The clonal relatedness of all of the
P. mirabilis isolates that were obtained from the households with multiple colonized members was determined by PFGE after overnight
NotI (New England Biolabs, Hertfordshire, UK) restriction. The
P. mirabilis from 70% of the households with a single colonized member were also characterized. Restriction fragments were resolved in a 1% agarose gel (pulse-field grade agarose, NZYtech—Genes and Enzymes, Lisbon, Portugal) using the previously described electrophoresis conditions (1–30 s for 8 h followed by 30–70 s for 16 h at 14 °C, 6 V/cm
2) on a CHEF DR II-apparatus (Bio-rad, Hercules, CA, USA) [
16].
2.5. Clinical Collection of UTI P. mirabilis
The PFGE restriction patterns of the fecal
P. mirabilis strains from the healthy companion animals and humans were compared to a previously characterized collection of clinical
P. mirabilis that were isolated from companion animals (
n = 107) and humans (
n = 76 [
n = 26 hospital patients and
n = 50 community patients]) with UTI [
6].
2.6. Susceptibility Testing
All of the unique
P. mirabilis pulse-type/participant strains and one isolate per participant from untyped households were tested for antimicrobial resistance by disk diffusion. Susceptibility testing was conducted according to the CLSI guidelines for the following antimicrobials: ampicillin 10 μg, amoxicillin/clavulanate 30 μg, cefoxitin 30 μg, cefotaxime 30 μg, meropenem 10 μg, ciprofloxacin 5 μg, gentamicin 10 μg, amikacin 30 μg, tetracycline 30 μg, chloramphenicol 30 μg and trimethoprim/sulfamethoxazole 25 μg (Oxoid, Hampshire, UK) [
17,
18]. These antimicrobials were selected as they are antimicrobial classes with clinical relevance for human and veterinary medicine, especially for the treatment of UTI.
Human CLSI breakpoints were used since Veterinary CLSI breakpoints are either not determined for the tested antimicrobials or are the same as those that are recommended in the Human CLSI guidelines [
17,
18]. The only exception is the gentamicin breakpoint that varies by 1mm in the intermedium/susceptible categorization [
17,
18].
2.7. Virulence Genes
All of the
P. mirabilis that were tested for antimicrobial resistance were also screened for the presence of urothelial cell adhesion fimbriae (UCA/NAF [
ucaA]), mannose-resistant
Proteus-like fimbriae (MR/P [
mrpA]),
Proteus mirabilis fimbriae (PMF [
pmfA]) and HpmA/HpmB hemolysin (
HpmA and
HpmB) codifying genes by PCR using the previously described primers [
19,
20].
2.8. Data Analysis
Fisher’s exact test was used to compare the groups using the SAS statistical software package for Windows, version 9.3 (SAS Institute Inc., Cary, NC, USA).
The BioNumerics software, version 6.6 (bioMérieux, Marcy-l’Étoile, France), was used to compare the P. mirabilis restriction PFGE-patterns using the Dice/UPGMA clustering method with a tolerance of 1.5% and a clustering cut-off of 80%.
3. Results
All of the human participants were above 18 years of age. Women were over-represented (70.8%, n = 17/24) and 33.3% (n = 8/24) of the human participants had undergone an antimicrobial treatment in the prior year. The companion animals had ages ranging from 2 months to 17 years, 19.2% of the animals (n = 5/26) underwent an antimicrobial treatment in the prior year and 57.7% (n = 15/26) were females. Only one cat had access to the outdoors, 83.3% (n = 15/18) of the dogs lived indoors while the remaining dogs lived in private yards.
A total of 11 participants from 8 households had fecal colonization by
P. mirabilis (
Figure 1). The use of the tetrathionate broth was necessary for the detection of
P. mirabilis in 36% (
n = 4/11) of the fecal samples of human and animal origin. Nevertheless, all of the fecal samples (
n = 50) were negative for
P. mirabilis growth in the cefotaxime- and meropenem-supplemented agar plates. When considering each participant species by itself, the
P. mirabilis fecal colonization was significantly higher (
p = 0.0329) in dogs (44.4%,
n = 8/18) than in humans (12.5%,
n = 3/24). None of the cats had detectable
P. mirabilis in their fecal samples, despite the use of the tetrathionate broth prior to plating.
Most of the positive households included at least one colonized dog (87.5%,
n = 7/8) (
Figure 1). Interestingly, the only household containing solely a colonized human (H2) also included a non-colonized cat. Five out of the eight positive households had a single participant colonized and, therefore, the within-household sharing of
P. mirabilis between these human–animal pairs was absent (
Figure 1). Two of the three households with multiple colonized participants included colonized human–dog pairs (H17, H18), while the third household (H14) had two positive dogs (
Figure 1).
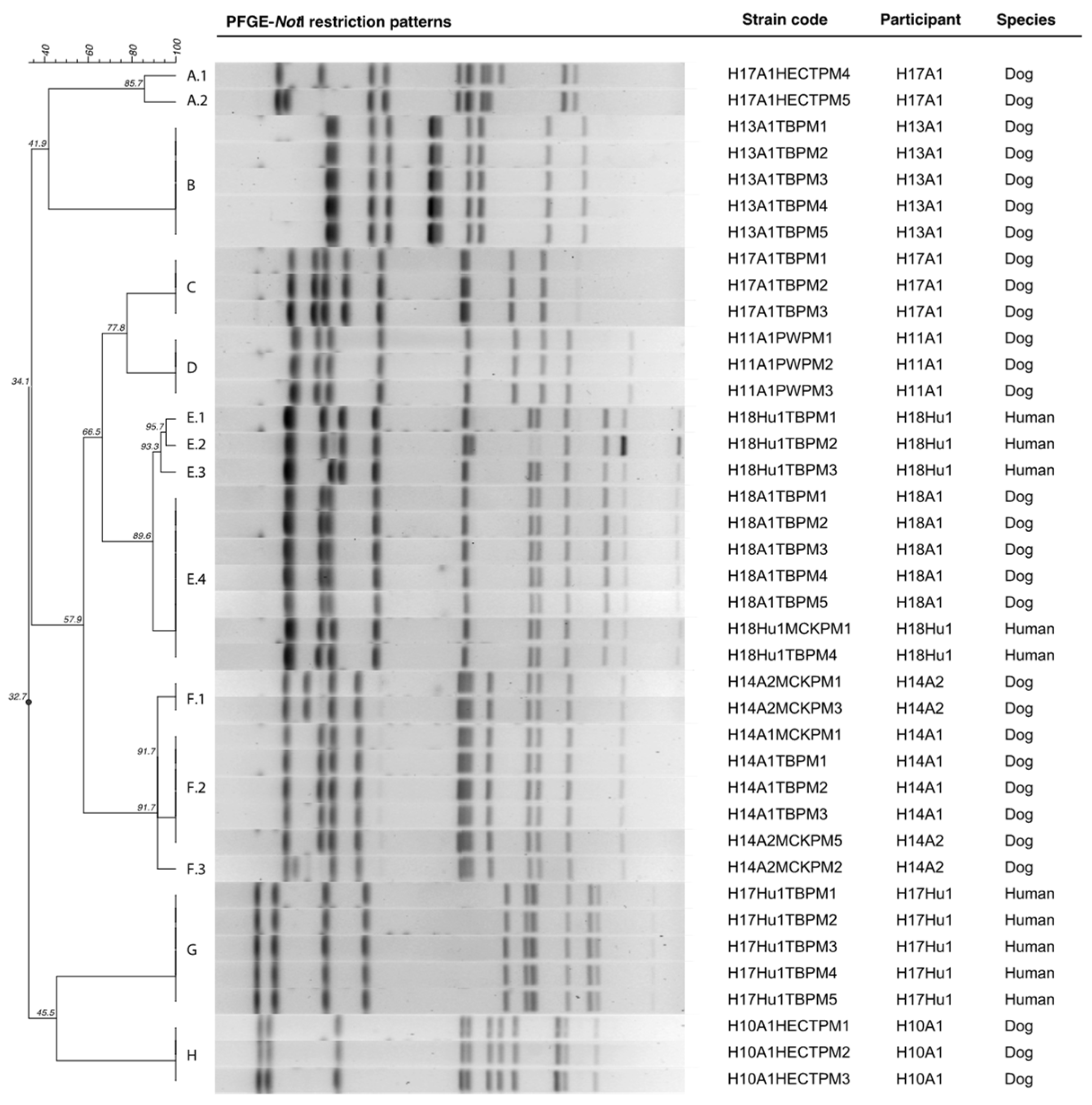
PFGE analysis was conducted in 39
P. mirabilis isolated from 3 households with multiple colonized participants (
n = 28) and from 3 households with single colonized participants (
n = 11). A total of 8
P. mirabilis PFGE clusters (from A to H) were identified. None of the
P. mirabilis from the different households clustered together, therefore
P. mirabilis was not shared across households (
Figure 2).
The PFGE analysis of the multiple
P. mirabilis isolates that were obtained from each participant revealed that two dogs (H14A2 and H17A1) and one human (H18Hu1) had several variants of the same colonizing strain (
Figure 2,
Table 1). Following the criteria outlined by Tenover et al., these variants were found to be consistent with single genetic events and therefore these strains are likely to be closely related [
21].
Another interesting finding was that only one dog (H17A1) was colonized by several unrelated
P. mirabilis strains (
Figure 2,
Table 1).
The H17 household human–dog pair (H17Hu1 and H17A1) was colonized by unrelated
P. mirabilis stains (
Figure 2,
Table 1). Conversely, the H18 household human–dog pair (H18Hu1 and H18A1) was colonized by indistinguishable and closely related
P. mirabilis strains (89–100% Dice/UPGMA index), thus showing that
P. mirabilis are likely shared between humans and animals that are living in close contact (
Figure 2,
Table 1). It should be noted that although the
P. mirabilis strains from the dog H18A1 (H18A1TBPM1) and the human H18Hu1 (H18Hu1TBPM4) were considered to be indistinguishable by DICE/UPGMA, the resistance phenotype varied in the gentamicin-susceptibility (
Figure 2,
Table 1). Furthermore, at visual inspection, the second and third restriction bands from the top seem to vary slightly. Nevertheless, according to the criteria put forward by Tenover et al., these variants are likely to be related [
21]. The detection of
P. mirabilis colonization in dog H18A1 required the use of tetrathionate broth while in the human H18Hu1 fecal sample this was not necessary. Both of the colonized humans from household H17 and H18 reported to frequently allow the colonized dogs to lick their faces.
Indistinguishable and closely related
P. mirabilis strains (91.7–100% Dice/UPGMA index) were also shared by the two dogs living in household H14 (
Figure 2,
Table 1). Here, the dog H14A2 presented a higher fecal burden than the dog H14A1.
Overall,
P. mirabilis human–dog sharing occurred in 5.5% (
n = 1/18) of all of the included households and in 12.8% (
n = 1/8) of the positive households. If only the households that included dogs were to be considered, these percentages increase to 7.7% (
n = 1/13) and 14.3% (
n = 1/7), respectively (
Figure 1). Furthermore, if the potential within-household human–dog sharing pairs are considered, 11.1% (
n = 1/9) or 4.2% (
n = 1/24) of these shared
P. mirabilis strains, depending on whether households without colonized participants are included.
A total of 18
P. mirabilis strains were screened for antimicrobial resistance and for the presence of virulence genes (
Table 1).
Most of the
P. mirabilis strains (77.7%,
n = 14/18) were fully susceptible to all of the tested antimicrobials except for tetracycline, to which
P. mirabilis is intrinsically resistant. Only the dog H17A1 was colonized by a
P. mirabilis strain with acquired resistance to more than three antimicrobial categories (
Table 1).
The HmpA/HmpB hemolysin and the MR/P and PMF fimbriae codifying genes were present in all of the strains. Furthermore, only one strain lacked
ucaA (
Table 1). All of the PFGE-indistinguishable and closely related
P. mirabilis strains also presented an identical antimicrobial resistance phenotype and virulence genotype, with the exception of the H18Hu E.4 (H18Hu1TBPM4)
P. mirabilis variant.
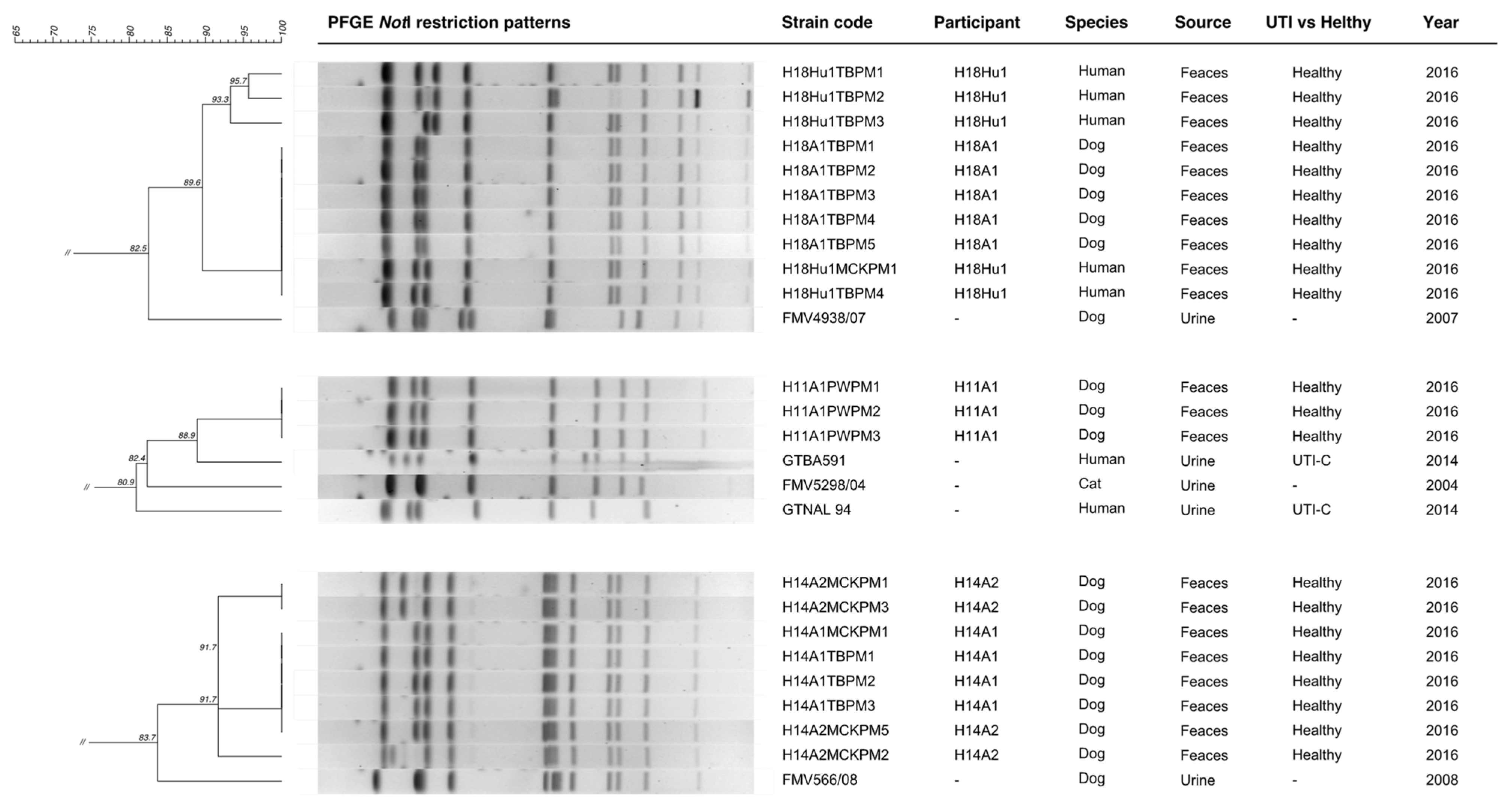
When comparing the fecal
P. mirabilis that were isolated in this study with the previously characterized collection of clinical
P. mirabilis from companion animals and humans with UTI [
6], the fecal strains from the H11 (pulse-type D), H14 (pulse-type F) and H18 (pulse-type E) households clustered with at least one uropathogenic clinical strain (
Figure 3).
The P. mirabilis that was colonizing the dog H11A1 clustered with two of the clinical strains that were causing UTI in human community patients in 2014 (GTBA591 and GTNAL94) and with one clinical strain that was isolated from a cat with a UTI in 2004 (FMV5298/04). Data concerning the prior history of UTI in the dog H11A1 or in his household members were not disclosed.
The P. mirabilis strains from the human (H18Hu1) and the dog (H18A1) living in household H18 were 82.5% similar (Dice/UPGMA) to the P. mirabilis clinical strain FMV4938/07 that was isolated from a dog with a UTI in 2007. Neither human H18Hu1 nor dog H18A1 had a previous history of UTI.
Finally, the P. mirabilis strain that was shared by both dogs of the household H14 showed 83.7% similarity with a clinical strain that was isolated from a dog with a UTI in 2008 (FMV566/08). Both of the dogs from the H14 household were never diagnosed with UTI.
4. Discussion
To date, only a few studies have reported data on the
P. mirabilis fecal colonization of healthy humans and companion animals [
1,
12,
13]. Moreover, to the best of our knowledge, this is the first study that focuses on humans and companion animals that are living in close contact.
P. mirabilis is an important cause of UTI and the gut has been identified as its major reservoir [
1,
2]. In fact, several studies associate the fecal colonization by
P. mirabilis with cross and auto-infections [
1]. Little is known regarding the role of healthy dogs and cats as reservoirs of
P. mirabilis. Only 6% of feral cats from Grenada, with unknown health status, were colonized by
P. mirabilis in the rectum [
22]. The absence of colonized healthy domestic cats in the present study may, therefore, be explained by the small sample size. The high
P. mirabilis colonization rate of the healthy dogs agrees with one early study conducted in enclosed beagles (42.8%) [
12]. However, another early study that focused on dogs with UTI failed to detect
P. mirabilis in the fecal samples of the control group that was composed of healthy dogs [
13]. The rate of
P. mirabilis colonization in healthy humans ranges from 4% to 45% according to the study and to the gastro-intestinal section that was considered [
1,
23]. It is important to note that
P. mirabilis colonization frequencies may vary between studies due to methodological differences. Furthermore, the normal
E. coli fecal burden may disguise and limit the detection of less frequent bacteria. Hence, it is possible that
P. mirabilis colonization frequencies are being underestimated.
The growing concerns regarding the transmission of pathogenic bacteria between companion animals and humans [
4,
5] are validated by studies that are reporting that
E. coli and
Klebsiella pneumoniae strains may be shared between companion animals and humans from the same household [
8,
10,
24].
P. mirabilis is a common uropathogen among cats, dogs and humans [
1,
2,
3]; however, there is a lack of studies evaluating the colonization of epidemiologically related companion animals and humans. One important finding from this study was the detection of within-household
P. mirabilis sharing between human–dog and dog–dog pairs. These results suggest that dogs may be reservoirs of
P. mirabilis to humans and vice versa, but also that dogs are likely reservoirs to other dogs. These outcomes are also relevant in infection settings since dogs with UTI that is caused by
P. mirabilis are known to have a significantly higher
P. mirabilis fecal burden [
13] which, hypothetically, may increase the transmission risk through direct or indirect contact. Care must be taken when interpreting these results since the fecal
P. mirabilis strains from humans and dogs that were isolated in this study were not compared using the most state-of-the-art methods that are now available, namely whole genome sequencing. Nevertheless, following the criteria that were proposed by Tenover et al. [
21] and considering that these strains were epidemiologically related, these findings support that humans and dogs may share
P. mirabilis strains.
The indistinguishable P. mirabilis strains from the dog H18A1 and human H18Hu1 showed distinct gentamicin-susceptibility which may be related to the acquisition or loss of the gentamicin-resistance mechanisms after gut colonization. The study design that was used in this work does not allow us to determine the P. mirabilis transmission dynamics and directionality; however, since P. mirabilis was only detected without the use of tetrathionate broth in the colonized human (H18Hu1), it could be speculated that he had a higher P. mirabilis fecal burden and was more likely to transmit it. The same rationale applies to the household H14 dog–dog sharing. A common source of acquisition, such as a fomite, could also have been involved in the co-colonization of both household members. Future studies using whole genome sequencing and, preferably, a longitudinal design are warranted in order to clarify the P. mirabilis transmission dynamics and colonization persistence in humans and dogs.
E. coli within-household sharing has been found to be significantly more common among dog–human pairs that report to allow the dog to lick the human’s face [
10]. It was not possible to establish such association in the current study due to the sample size. The presence of several variants of the same pulse-type colonizing a single host suggests that within-host
P. mirabilis evolution may be occurring, which is a known phenomenon already reported in other bacteria [
25].
Previous studies on
E. coli found that 17%, 9.8%, 8.8% or 3.5% of households had human–dog pairs sharing indistinguishable or closely related
E. coli clones by PFGE (94–100% PFGE profile similarity) [
7,
8,
9,
10]. The different sharing frequencies reported in these studies may be explained by the different number of participants per household included and by the different number of
E. coli isolates typed per participant. Overall, a higher number of participants per household and a higher number of isolates per participant usually lead to the detection of a higher percentage of sharing [
7,
8,
9,
10]. It was interesting to notice that the human–dog
P. mirabilis sharing frequency that was detected in this study is within a similar range as that which has been previously published for
E. coli. On the other hand, unlike in
E. coli studies [
9,
10], the
P. mirabilis strains were not shared across-households. Likely, this was related to the small sample size.
Johnson et al. reported that adult humans shared
E. coli strains more frequently with cats than with dogs. Regarding
P. mirabilis, the opposite was noted [
7].
The detection of multidrug-resistant CMY-2-producing
P. mirabilis is increasing in companion animals with UTI that are living in the same geographic region where the present study was conducted [
3]. CMY-2-like β-lactamases are also important in human infection in Europe [
26]. Furthermore, antimicrobial use in the prior year is a known risk factor for gut colonization by third-generation cephalosporin-resistant bacteria [
27,
28]. Despite the inclusion of participants that underwent antimicrobial use in the prior year, third-generation cephalosporin and meropenem-resistant
P. mirabilis were not detected. Although the sample size from the current study is a limitation that could explain this result, this is considered to be a positive finding.
The gut of catheterized humans with UTI is usually colonized by the index uropathogenic
P. mirabilis strain [
1,
29]. The clonal relatedness between
P. mirabilis clinical strains from individuals with UTI and
P. mirabilis strains colonizing healthy individuals is poorly studied. In the present study, some of the
P. mirabilis strains from the fecal samples (from household H11, H14 and H18 colonized participants) clustered with UTI clinical strains from unrelated patients with UTI. Regarding the households H14 and H18, none of the colonized participants have had a prior UTI in their life. These findings suggest the role of the gut as a reservoir of uropathogenic
P. mirabilis in healthy individuals. The detection of
P. mirabilis colonizing a dog that clustered with clinical
P. mirabilis isolated from a human community-associated UTI also points to the possible role of dogs as reservoirs of uropathogenic strains to humans. Regretfully, it was not possible to obtain epidemiological data on this dog concerning prior UTI or contact with infected individuals.
Notably, almost all of the
P. mirabilis that were found colonizing healthy companion animals and humans codified for HmpA/HmpB hemolysin and UCA/NAF, PMP and MR/P fimbriae, which are important virulence factors for UTI [
2].