Molecular Diagnosis, Antimicrobial Resistance Profiles and Disease Patterns of Gram-Positive Pathogens Recovered from Clinical Infections in Major Ha’il Hospitals
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ha’il Province and All Its Socioeconomic Strata
2.2. Study Designs, Data Sources, and Statistical Analysis
2.3. GeneXpert Molecular Detection Systems
2.4. Classifications as Multi-, Extremely- and Pan-Drug Resistant Bacteria (MDR, XDR, and PDR)
- (i)
- An MRSA is always considered MDR by virtue of being an MRSA.
- (ii)
- Non-susceptible to ≥1 agent in ≥3 antimicrobial categories.
- (iii)
- XDR: non-susceptible to ≥1 agent in all but ≥2 categories.
- (iv)
- PDR: non-susceptible to all antimicrobial agents listed. Oxacillin or cefoxitin represents all other b-lactams (and cephamycins) and resistance to either of these predicts non-susceptibility to all categories of b-lactam antimicrobials listed in this document, with the exception of the anti MRSA cephalosporins (i.e., all categories of penicillin, cephalosporins, b-lactamase inhibitors and carbapenems currently approved).
2.5. Molecular Detection of Spa Gene, mecA, and mec (SCCmec)
3. Results
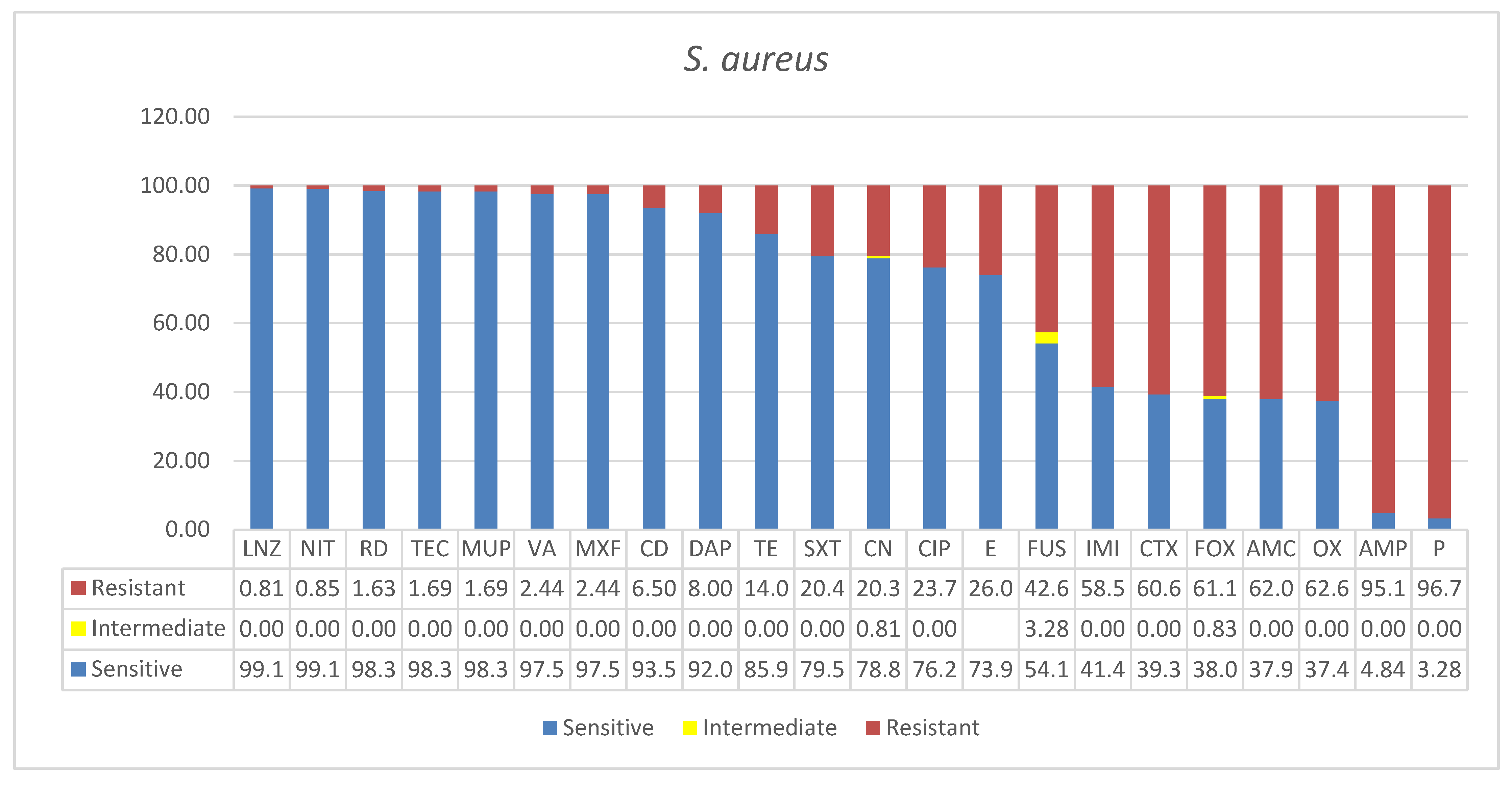
3.1. Staphylococcus aureus
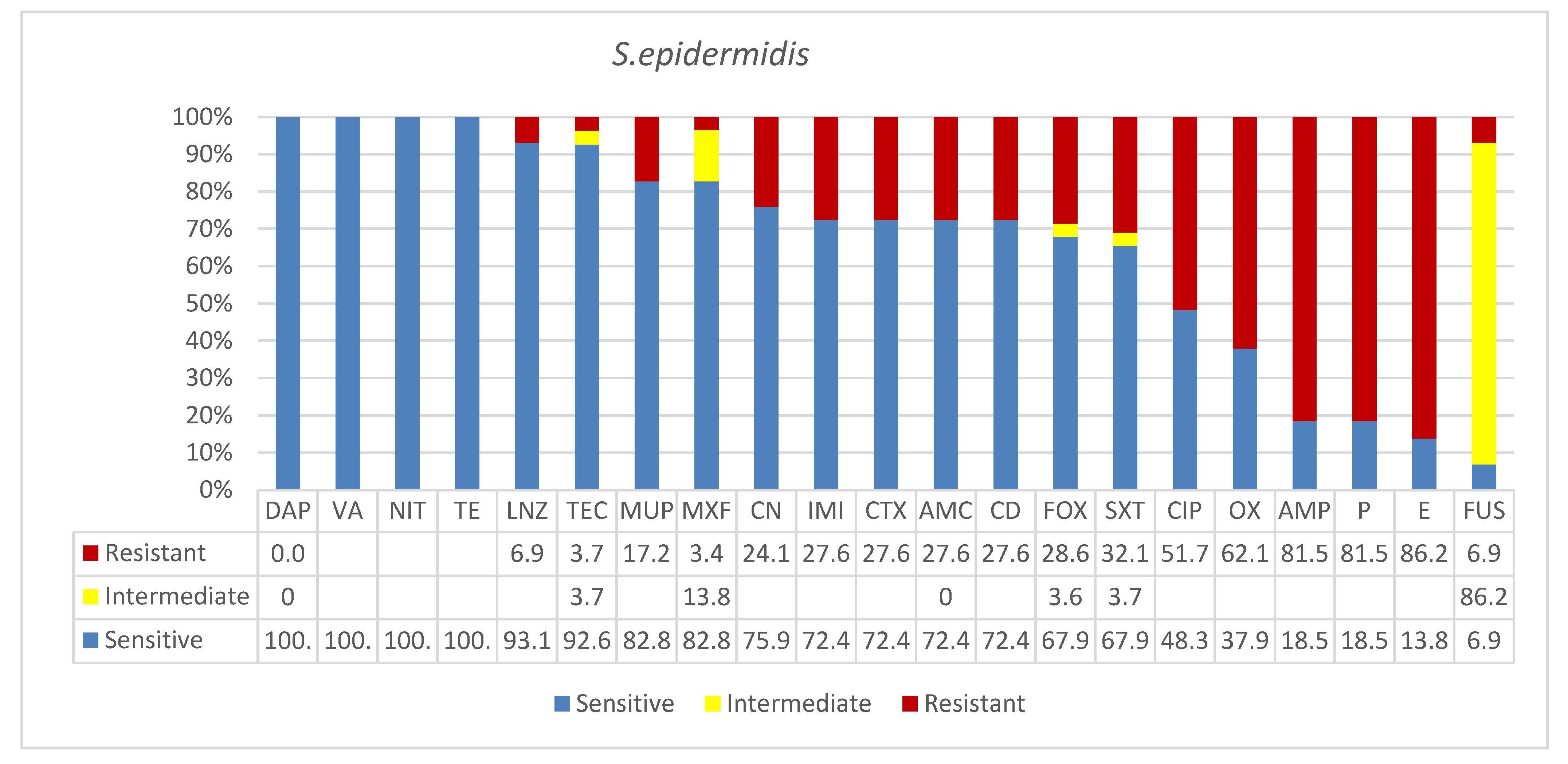
3.2. Staphylococcus epidermidis
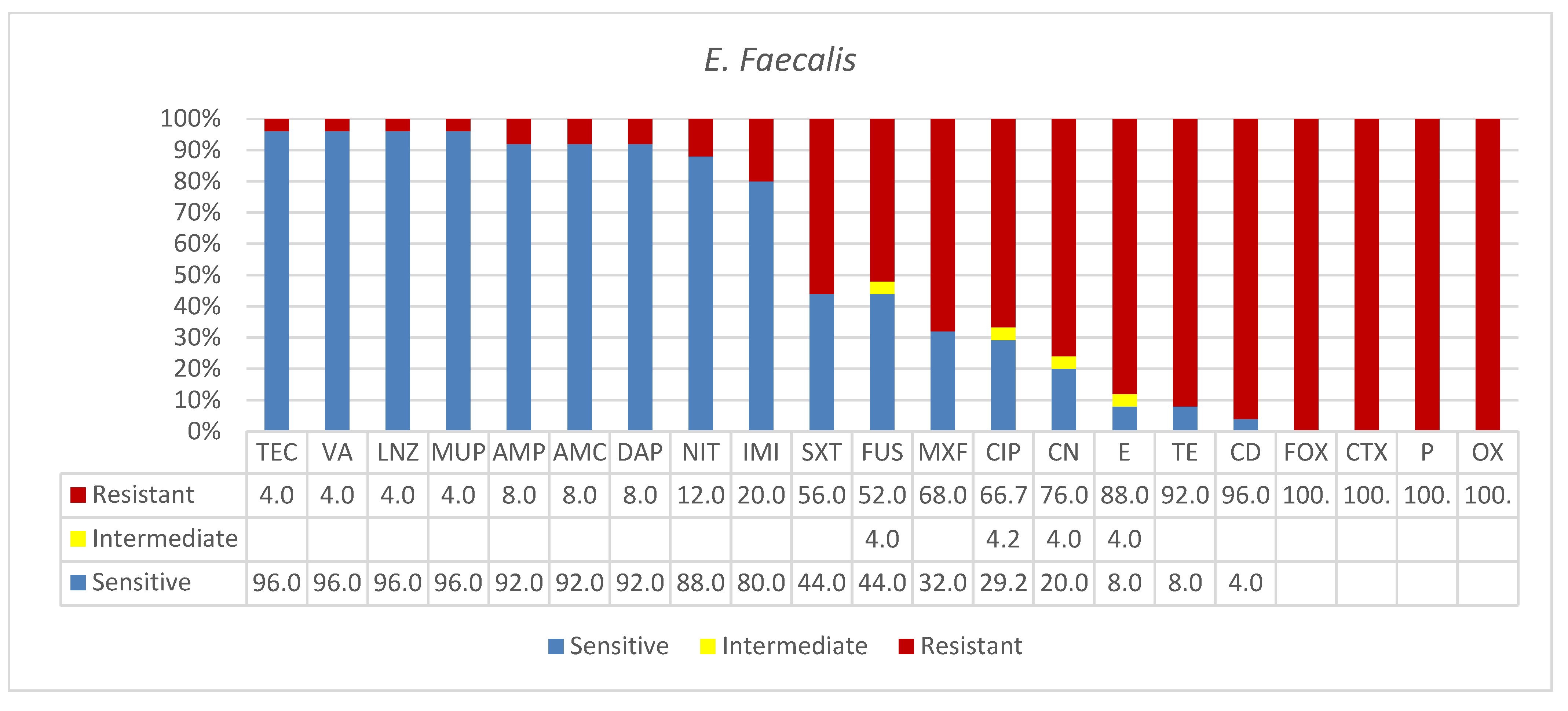
3.3. Enterococcus faecalis
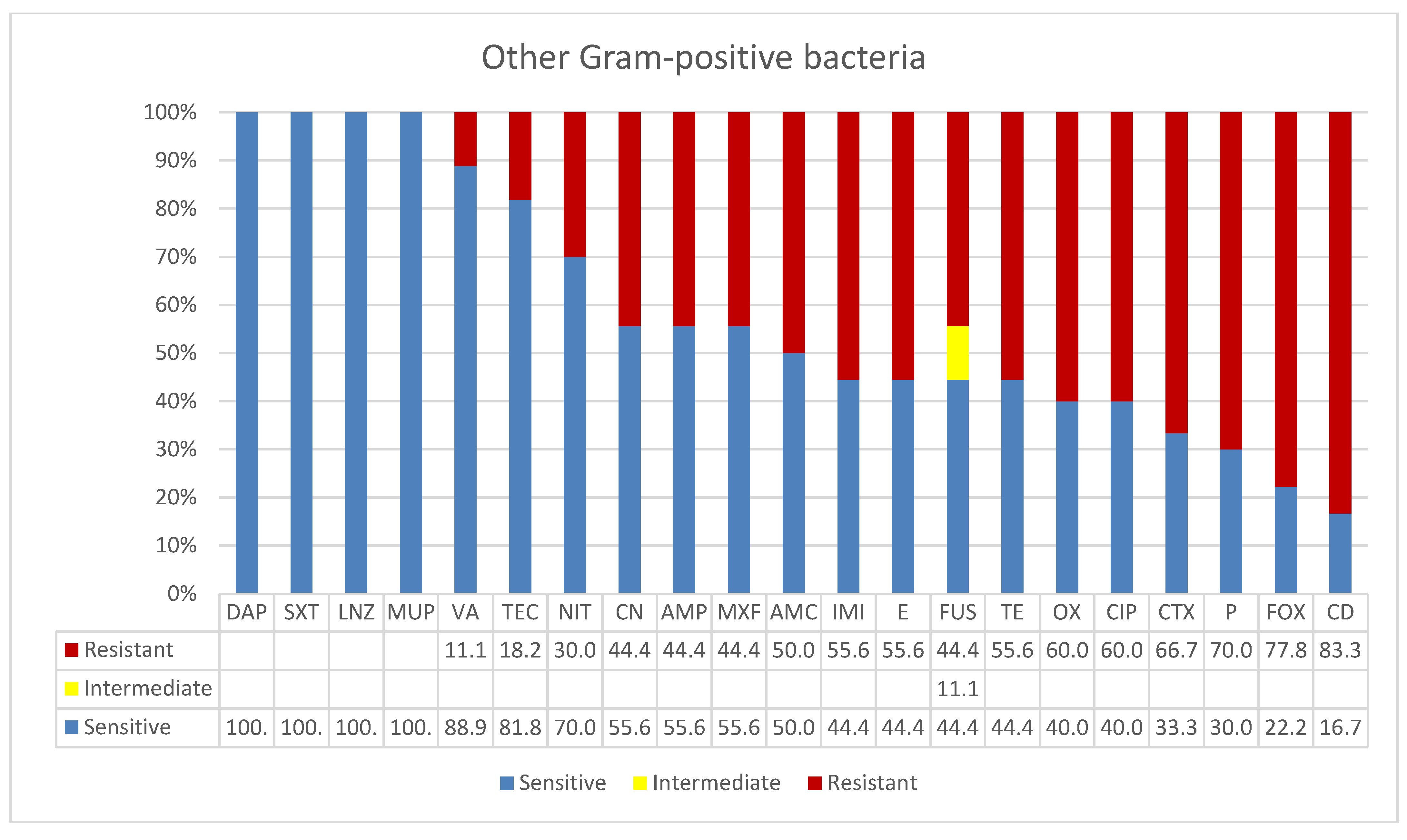
3.4. Other Gram-Positive Bacteria
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abussaud, M.J. Incidence of wound infection in three different departments and the antibiotic sensitivity pattern of the isolates in a Saudi Arabian hospital. Acta Microbiol. Et Immunol. Hung. 1996, 43, 301–305. [Google Scholar]
- El-Mahdy, T.S.; Al-Agamy, M.H.; Emara, M.; Barakat, A.; Goering, R.V. Complex Clonal Diversity of Staphylococcus aureus Nasal Colonization among Community Personnel, Healthcare Workers, and Clinical Students in the Eastern Province, Saudi Arabia. BioMed Res. Int. 2018, 2018, 4208762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenthal, V.D.; Belkebir, S.; Zand, F.; Afeef, M.; Tanzi, V.L.; Al-Abdely, H.M.; El-Kholy, A.; AlKhawaja, S.A.A.; Demiroz, A.P.; Sayed, A.F.; et al. Six-year multicenter study on short-term peripheral venous catheters-related bloodstream infection rates in 246 intensive units of 83 hospitals in 52 cities of 14 countries of Middle East: Bahrain, Egypt, Iran, Jordan, Kingdom of Saudi Arabia, Kuwait, Lebanon, Morocco, Pakistan, Palestine, Sudan, Tunisia, Turkey, and United Arab Emirates-International Nosocomial Infection Control Consortium (INICC) findings. J. Infect. Public Health 2020, 13, 1134–1141. [Google Scholar] [CrossRef] [PubMed]
- Ghahremani, M.; Jazani, N.H.; Sharifi, Y. Emergence of vancomycin-intermediate and -resistant Staphylococcus aureus among methicillin-resistant S. aureus isolated from clinical specimens in the northwest of Iran. J. Global Antimicrob. Resist. 2018, 14, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Boswihi, S.S.; Udo, E.E.; Monecke, S.; Mathew, B.; Noronha, B.; Verghese, T.; Tappa, S.B. Emerging variants of methicillin-resistant Staphylococcus aureus genotypes in Kuwait hospitals. PLoS ONE 2018, 13, e0195933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senok, A.; Ehricht, R.; Monecke, S.; Al-Saedan, R.; Somily, A. Molecular characterization of methicillin-resistant Staphylococcus aureus in nosocomial infections in a tertiary-care facility: Emergence of new clonal complexes in Saudi Arabia. New Microbes New Infect. 2016, 14, 13–18. [Google Scholar] [CrossRef] [Green Version]
- Albarrag, A.; Shami, A.; Almutairi, A.; Alsudairi, S.; Aldakeel, S.; Al-Amodi, A. Prevalence and Molecular Genetics of Methicillin-Resistant Staphylococcus aureus Colonization in Nursing Homes in Saudi Arabia. Can. J. Infect. Dis. Med. Microbiol. 2020, 2020. [Google Scholar] [CrossRef]
- El-Deeb, W.; Fayez, M.; Elmoslemany, A.; Kandeel, M.; Zidan, K. Methicillin resistant Staphylococcus aureus among goat farms in Eastern province, Saudi Arabia: Prevalence and risk factors. Prev. Vet. Med. 2018, 156, 84–90. [Google Scholar] [CrossRef]
- Kourtis, A.P.; Hatfield, K.; Baggs, J.; Mu, Y.; See, I.; Epson, E.; Nadle, J.; Kainer, M.A.; Dumyati, G.; Petit, S.; et al. Vital Signs: Epidemiology and Recent Trends in Methicillin-Resistant and in Methicillin-Susceptible Staphylococcus aureus Bloodstream Infections—United States. MMWR Morb. Mortal. Wkly. Rep. 2019, 68, 214–219. [Google Scholar] [CrossRef] [Green Version]
- Yilmaz, M.; Elaldi, N.; Balkan, I.I.; Arslan, F.; Batırel, A.A.; Bakıcı, M.Z.; Gozel, M.G.; Alkan, S.; Çelik, A.D.; Yetkin, M.A. Mortality predictors of Staphylococcus aureus bacteremia: A prospective multicenter study. Ann. Clin. Microbiol. Antimicrob. 2016, 15, 7. [Google Scholar] [CrossRef] [Green Version]
- Van Hal, S.J.; Jensen, S.O.; Vaska, V.L.; Espedido, B.A.; Paterson, D.L.; Gosbell, I.B. Predictors of mortality in Staphylococcus aureus Bacteremia. Clin. Microbiol. Rev. 2012, 25, 362–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rokney, A.; Baum, M.; Ben-Shimol, S.; Sagi, O.; Anuka, E.; Agmon, V.; Greenberg, D.; Valinsky, L.; Danino, D. Dissemination of the Methicillin-resistant Staphylococcus aureus Pediatric Clone (ST5-T002-IV-PVL+) as a Major Cause of Community-associated Staphylococcal Infections in Bedouin Children, Southern Israel. Pediatric Infect. Dis. J. 2019, 38, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Earls, M.R.; Steinig, E.J.; Monecke, S.; Castruita, J.A.S.; Simbeck, A.; Schneider-Brachert, W.; Vremerǎ, T.; Dorneanu, O.S.; Loncaric, I.; Bes, M.; et al. Exploring the evolution and epidemiology of European CC1-MRSA-IV: Tracking a multidrug-resistant community-associated meticillin-resistant Staphylococcus aureus clone. Microb. Genom. 2021, 7, 601. [Google Scholar] [CrossRef] [PubMed]
- Gustave, C.A.; Tristan, A.; Martins-Simões, P.; Stegger, M.; Benito, Y.; Andersen, P.S.; Bes, M.; Le Hir, T.; Diep, B.A.; Uhlemann, A.C.; et al. Demographic fluctuation of community-acquired antibiotic-resistant Staphylococcus aureus lineages: Potential role of flimsy antibiotic exposure. ISME J. 2018, 12, 1879–1894. [Google Scholar] [CrossRef] [PubMed]
- Cardot Martin, E.; Michel, A.; Raynal, B.; Badiou, C.; Laurent, F.; Vandenesch, F.; Etienne, J.; Lina, G.; Dumitrescu, O. Community-acquired meticillin-resistant Staphylococcus aureus strain USA300 resists staphylococcal protein A modulation by antibiotics and antimicrobial peptides. Int. J. Antimicrob. Agents 2015, 45, 19–24. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus epidermidis--the “accidental” pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.H.; Park, M.D.; Otto, M. Host Response to Staphylococcus epidermidis Colonization and Infections. Front. Cell. Infect. Microbiol. 2017, 7, 90. [Google Scholar] [CrossRef] [Green Version]
- De Leon, S.P.; Wenzel, R.P. Hospital-acquired bloodstream infections with Staphylococcus epidermidis. Review of 100 cases. Am. J. Med. 1984, 77, 639–644. [Google Scholar] [CrossRef]
- Dong, Y.; Speer, C.P.; Glaser, K. Beyond sepsis: Staphylococcus epidermidis is an underestimated but significant contributor to neonatal morbidity. Virulence 2018, 9, 621–633. [Google Scholar] [CrossRef] [Green Version]
- O’Toole, R.F. The interface between COVID-19 and bacterial healthcare-associated infections. Clinical microbiology and infection: The official publication of the European Society of Clinical Microbiology and Infectious Diseases. Clin. Microbiol. Infect. 2021, 27, 90. [Google Scholar] [CrossRef]
- Dong, Y.; Speer, C.P. The role of Staphylococcus epidermidis in neonatal sepsis: Guarding angel or pathogenic devil? Int. J. Med. Microbiol. IJMM 2014, 304, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Cason, C.; D’accolti, M.; Campisciano, G.; Soffritti, I.; Ponis, G.; Mazzacane, S.; Maggiore, A.; Risso, F.M.; Comar, M.; Caselli, E. Microbial Contamination in Hospital Environment Has the Potential to Colonize Preterm Newborns’ Nasal Cavities. Pathogens 2021, 10, 615. [Google Scholar] [CrossRef]
- Lee, J.Y.H.; Monk, I.R.; Gonçalves da Silva, A.; Seemann, T.; Chua, K.Y.; Kearns, A.; Hill, R.; Woodford, N.; Bartels, M.D.; Strommenger, B.; et al. Global spread of three multidrug-resistant lineages of Staphylococcus epidermidis. Nat. Microbiol. 2018, 3, 1175–1185. [Google Scholar] [CrossRef] [PubMed]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Diseases 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef] [PubMed]
- Abat, C.; Raoult, D.; Rolain, J.M. Low Level of Resistance in Enterococci Isolated in Four Hospitals, Marseille, France. Microb. Drug Resist. 2016, 22, 218–222. [Google Scholar] [CrossRef]
- Jernigan, J.A.; Hatfield, K.M.; Wolford, H.; Nelson, R.E.; Olubajo, B.; Reddy, S.C.; McCarthy, N.; Paul, P.; McDonald, L.C.; Kallen, A.; et al. Multidrug-Resistant Bacterial Infections in U.S. Hospitalized Patients, 2012–2017. N. Engl. J. Med. 2020, 382, 1309–1319. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, T. Trends in bacterial resistance among perioperative infections in patients with primary ovarian cancer: A retrospective 20-year study at an affiliated hospital in South China. J. Int. Med. Res. 2020, 48, 300060520928780. [Google Scholar] [CrossRef]
- Khodabandeh, M.; Mohammadi, M.; Abdolsalehi, M.R.; Hasannejad-Bibalan, M.; Gholami, M.; Alvandimanesh, A.; Pournajaf, A.; Rajabnia, R. High-Level Aminoglycoside Resistance in Enterococcus Faecalis and Enterococcus Faecium; as a Serious Threat in Hospitals. Infect. Disord. Drug Targets 2020, 20, 223–228. [Google Scholar] [CrossRef]
- Flokas, M.E.; Karageorgos, S.A.; Detsis, M.; Alevizakos, M.; Mylonakis, E. Vancomycin-resistant enterococci colonisation, risk factors and risk for infection among hospitalised paediatric patients: A systematic review and meta-analysis. Int. J. Antimicrob. Agents 2017, 49, 565–572. [Google Scholar] [CrossRef]
- Herrera-Hidalgo, L.; de Alarcón, A.; López-Cortes, L.E.; Luque-Márquez, R.; López-Cortes, L.F.; Gutiérrez-Valencia, A.; Gil-Navarro, M.V. Enterococcus faecalis Endocarditis and Outpatient Treatment: A Systematic Review of Current Alternatives. Antibiotics 2020, 9, 657. [Google Scholar] [CrossRef] [PubMed]
- Al-Ahdal, M.N.; Abozaid, S.M.; Al-Shammary, H.F.; Bohol, M.F.; Al-Thawadi, S.I.; Al-Jaberi, A.A.; Senok, A.C.; Shibl, A.M.; Al-Qahtani, A.A. Characterization of Enterococcus faecium isolates and first report of vanB phenotype-vanA genotype incongruence in the Middle East. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 3223–3229. [Google Scholar] [CrossRef] [PubMed]
- Somily, A.M.; Al-Mohizea, M.M.; Absar, M.M.; Fatani, A.J.; Ridha, A.M.; Al-Ahdal, M.N.; Senok, A.C.; Al-Qahtani, A. Molecular epidemiology of vancomycin resistant enterococci in a tertiary care hospital in Saudi Arabia. Microb. Pathog. 2016, 97, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, M.; Al-Saafin, M. Overview of Prevalence, Characteristics, Risk Factors, Resistance, and Virulence of Vancomycin-Resistant Enterococci in Saudi Arabia. Microb. Drug Resist. 2019, 25, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.B.; Cockerill, F.R.; Alder, J.; Bradford, P.A.; Dudley, M.N.; Eliopoulos, G.M.; Hardy, D.J.; Hecht, D.W.; Hindler, J.A.; Powell, M.; et al. Performance Standards for Antimicrobial Susceptibility Testing an Informational Supplement for Global Application Developed through the Clinical and Laboratory Standards Institute Consensus Process, 26th ed. Available online: www.clsi.org (accessed on 8 December 2021).
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zipperer, A.; Konnerth, M.C.; Laux, C.; Berscheid, A.; Janek, D.; Weidenmaier, C.; Burian, M.; Schilling, N.A.; Slavetinsky, C.; Marschal, M.; et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 2016, 535, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Uslan, D.Z.; Crane, S.J.; Steckelberg, J.M.; Cockerill, F.R.; Sauver, J.L.S.; Wilson, W.R.; Baddour, L.M. Age- and sex-associated trends in bloodstream infection: A population-based study in Olmsted County, Minnesota. Arch. Intern. Med. 2007, 167, 834–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yahav, D.; Eliakim-Raz, N.; Leibovici, L.; Paul, M. Bloodstream infections in older patients. Virulence 2016, 7, 341–352. [Google Scholar] [CrossRef] [Green Version]
- Bandy, A.; Almaeen, A.H. Pathogenic spectrum of blood stream infections and resistance pattern in Gram-negative bacteria from Aljouf region of Saudi Arabia. PLoS ONE 2020, 15, e0233704. [Google Scholar] [CrossRef]
- Bouiller, K.; Ilic, D.; Wicky, P.H.; Cholley, P.; Chirouze, C.; Bertrand, X. Spread of clonal linezolid-resistant Staphylococcus epidermidis in an intensive care unit associated with linezolid exposure. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2020, 39, 1271–1277. [Google Scholar] [CrossRef]
- Weßels, C.; Strommenger, B.; Klare, I.; Bender, J.; Messler, S.; Mattner, F.; Krakau, M.; Werner, G.; Layer, F. Emergence and control of linezolid-resistant Staphylococcus epidermidis in an ICU of a German hospital. J. Antimicrob. Chemother. 2018, 73, 1185–1193. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Cave, R.; Chen, L.; Yangkyi, T.; Liu, Y.; Li, K.; Meng, G.; Niu, K.; Zhang, W.; Tang, N.; et al. Antibiotic resistance and molecular characteristics of methicillin-resistant Staphylococcus epidermidis recovered from hospital personnel in China. J. Global Antimicrob. Resist. 2020, 22, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Shah, H.N.; Misra, R.; Chen, J.; Zhang, W.; Liu, Y.; Cutler, R.R.; Mkrtchyan, H.V. The prevalence, antibiotic resistance and mecA characterization of coagulase negative staphylococci recovered from non-healthcare settings in London, UK. Antimicrob. Resist. Infect. Control 2018, 7, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alghoribi, M.F.; Balkhy, H.H.; Woodford, N.; Ellington, M.J. The role of whole genome sequencing in monitoring antimicrobial resistance: A biosafety and public health priority in the Arabian Peninsula. J. Infect. Public Health 2018, 11, 784–787. [Google Scholar] [CrossRef]
- Balkhi, B.; Mansy, W.; Alghadeer, S.; Alnuaim, A.; Alshehri, A.; Somily, A. Antimicrobial susceptibility of microorganisms causing Urinary Tract Infections in Saudi Arabia. J. Infect. Dev. Ctries. 2018, 12, 220–227. [Google Scholar] [CrossRef] [Green Version]
- Kamińska, W.; Grochowska, M.; Chmielarczyk, A.; Olszewska, A.; Skolimowska, G.; Dzierżanowska-Fangrat, K. Genetic typing of Enterococcus faecium VRE strains isolated in three hospitals in Warsaw and Siedlce in 2015-2016. Przeglad Epidemiologiczny 2019, 73, 49–60. [Google Scholar] [CrossRef]
- Conceição, N.; Rodrigues, W.F.; de Oliveira, K.L.P.; da Silva, L.E.P.; de Souza, L.R.C.; Barata, C.D.D.C.H.; de Oliveira, A.G. Beta-lactams susceptibility testing of penicillin-resistant, ampicillin-susceptible Enterococcus faecalis isolates: A comparative assessment of Etest and disk diffusion methods against broth dilution. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 43. [Google Scholar] [CrossRef] [PubMed]
| Ward a | Total Clinical Isolates of Gram-Positive Bacteria (n = 188) | |||||||
|---|---|---|---|---|---|---|---|---|
| S. aureus (65.4%, n = 123) | S. epidermidis (15.4%, n = 29) | E. faecalis (13%, n = 25) | Other-Gram-Positive Bacteria (6%, n = 11) | |||||
| n | % | n | % | n | % | n | % | |
| Intensive Care Unit ICU |
28 Blood, respiratory soft tissue infections (12) Wound/pus AE (16) | 25 |
11 Blood (11) | 38 |
10 Blood (8) Urine (2) | 40 |
1 Urine | 9 |
| COVID-19 Isolation Zones (ISO) OR COVID Ward (COW) | 1 | 1 | 0 | 0 | 1 | 4 | 0 | 0 |
| Surgical infections FMW, FSW, MMW, MSW, and AKU |
67 Pus (7), Wounds, skins, swabs, etc. (36) Throat and nasal collections (14) | 60 |
8 Blood (8) | 27.6 |
11 Blood (4) Urine (6) Wound (1) | 44 |
9 Urine (2) Blood (4 Wound (3) | 82 |
| Other | 27 | 14 | 10 (swabs (2), pus (1), vitreous (1), peritoneal fluid (1) Av necrosis (1), lab origin (2) | 34.5 | 3 | 12 | 1 | 9 |
| Total | 123 | 100% | 29 | 100% | 25 | 100% | 11 | 100% |
| Age | ||||||||
| Young 0–20 | 21 | 19 | 4 | 14 | 1 | 4 | 0 | 0 |
| Adults (21–49 yrs.) | 41 | 38 | 4 | 14 | 2 | 8 | 0 | 0 |
| Seniors (>50 yrs.) | 46 | 43 | 20 | 69 | 19 | 76 | 9 | 82 |
| Age unidentified | 15 | 12 | 1 | 3.4 | 3 | 12 | 2 | 18 |
| Gender | ||||||||
| Male | 70 | 63 | 16 | 55 | 16 | 64 | 3 | 27 |
| Female | 41 | 37 | 13 | 45 | 9 | 36 | 8 | 73 |
| Gender unidentified | 12 | 10 | 0 | 0 | 0 | 0 | 0 | 0 |
| MDR, XDR, PDR b |
MDR Satisfies both options for MDR: (i) by virtue of MRSA is (ii) non-susceptible to ≥1 agent in ≥3 antimicrobial categories | MDR | MDR | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Said, K.B.; Alsolami, A.; Khalifa, A.M.; Khalil, N.A.; Moursi, S.; Rakha, E.; Osman, A.; Rashidi, M.; Taha, T.E.; Bashir, A.I.; et al. Molecular Diagnosis, Antimicrobial Resistance Profiles and Disease Patterns of Gram-Positive Pathogens Recovered from Clinical Infections in Major Ha’il Hospitals. Microbiol. Res. 2022, 13, 49-63. https://doi.org/10.3390/microbiolres13010004
Said KB, Alsolami A, Khalifa AM, Khalil NA, Moursi S, Rakha E, Osman A, Rashidi M, Taha TE, Bashir AI, et al. Molecular Diagnosis, Antimicrobial Resistance Profiles and Disease Patterns of Gram-Positive Pathogens Recovered from Clinical Infections in Major Ha’il Hospitals. Microbiology Research. 2022; 13(1):49-63. https://doi.org/10.3390/microbiolres13010004
Chicago/Turabian StyleSaid, Kamaleldin B., Ahmed Alsolami, Amany M. Khalifa, Nuha A. Khalil, Soha Moursi, Ehab Rakha, Abuzar Osman, Musleh Rashidi, Taha E. Taha, Abdelhafiz I. Bashir, and et al. 2022. "Molecular Diagnosis, Antimicrobial Resistance Profiles and Disease Patterns of Gram-Positive Pathogens Recovered from Clinical Infections in Major Ha’il Hospitals" Microbiology Research 13, no. 1: 49-63. https://doi.org/10.3390/microbiolres13010004
APA StyleSaid, K. B., Alsolami, A., Khalifa, A. M., Khalil, N. A., Moursi, S., Rakha, E., Osman, A., Rashidi, M., Taha, T. E., Bashir, A. I., Moussa, S., Al Jadani, A., Nagi, H., Kuddus, M., Alrashedi, O. M., Alharbi, A. S., Alfaraj, A., Mustafa, R., & on behalf of the Ha’il COM Research Unit Group. (2022). Molecular Diagnosis, Antimicrobial Resistance Profiles and Disease Patterns of Gram-Positive Pathogens Recovered from Clinical Infections in Major Ha’il Hospitals. Microbiology Research, 13(1), 49-63. https://doi.org/10.3390/microbiolres13010004







