Abstract
Background: Plant associated endophytic microbes play an important role in plant’s growth and development. After seed germination, the seed associated endophytes rapidly colonize the seedlings and help in their growth and protection against pathogens. This study was aimed to understand the diversity in the endophytic microbial population associated with the seeds of papaya (Carica papaya) and its wild relatives from Vasconcellea genus (family: Caricaceae). The species of Vasconcellea genus are widely used to introgress virus resistance in cultivated varieties of papaya. Hence, the diversity of seed associated endophytic microbes and their gene functional analysis was carried out through next generation sequencing of the microbial 16S rRNA and ITS sequences. Results: The 16S rRNA amplicon analysis revealed that the number of operational taxonomic units (OTUs) was higher for the endophytic bacteria, ranging between 144–204 when compared to 41–69 OTUs for the endophytic fungi. The bacterial phylum Proteobacteria was the most abundant seed associated phylum, with 64.7–72.8% abundance, across all four species of Caricaceae family, followed by Firmicutes (13.6–26.1%), Patescibacteria (1.1–2%) and Actinobacteria (0.7–2.7%). With respect to the diversity of bacteria by abundance indices, Vasconcellea goudotiana had the highest OTUs of 204, followed by 177 in V. cauliflora, 156 in V. cundinamarcensis, and 144 in C. papaya. The alpha diversity indices and functional analysis revealed the differences in the OTUs and the functional annotations among the above four plant species. The fungal OTUs were in the range of 41–69; however, only a small fraction of them could be taxonomically classified. Conclusion: Our microbiome studies reveal the differences in the seed associated endophytic microbial community across the four plant species of Caricaceae family. This study also unravels the composition of endophytic microbial population associated with the seeds of different plant species of Caricaceae family and their gene functions. It also provides an insight into both culturable and nonculturable endophytic microbes. Further this study reveals that domestication of Carica papaya might have resulted into reduced microbial diversity when compared to their wild relatives from Vasconcellea genus.
1. Introduction
Papaya (Carica papaya L.) is a popular fruit crop of tropical and subtropical regions of the world [1] with an annual production of more than 10 million tons [2,3]. Papaya belongs to the family Caricaceae consisting of 6 genera and 35 species. Among the six genera, Carica and Vasconcellea (wild highland relatives of C. papaya) are the two most important genera [4,5]. Papaya fruit is highly nutritive and can fulfill the standard recommended daily requirements of vitamins (A, C, B9, B3, B1, B2), iron, potassium, calcium, and fiber [3,6,7]. They are also cultivated for proteolytic enzymes (papain) derived from the milky latex, used for food, textile, leather and pharmaceutical industries [8,9]. The mountain papaya from the Andes (Vasconcellea spp.) is an underexploited wild relative of papaya, despite having several desirable traits that include cold tolerance, disease resistance, and stronger proteolytic enzyme activity [5,10]. Several efforts are made to introgress these traits into C. papaya varieties. However, the incompatibility and fertility issues have restricted the papaya breeding efforts [11,12,13,14]. It may also be possible that the microbial endophytes associated with Vasconcellea spp., the wild relatives of papaya, may be helping in tolerance to various biotic and abiotic stresses. In recent years, the use of biocontrol agents, such as the endophytic microbes for biological control has significantly helped in protecting plants against pathogens, insect pests, abiotic stresses and overall increase in crop yield [15].
The association of the endophytic microbes with the plants is estimated to have happened >400 million years ago, coinciding with the colonization of plants on the land [16,17]. The association of endophytic microbes with the plants, is known to give resistance against both plant pests and pathogens [18,19,20,21]. This association also keeps the plant pathogens at check by production of a wide range of alkaloids such as ergot, pyrrolizidine, etc. [22,23]. The beneficial bacterial and fungal endophytes are more common and highly diverse, that belong to the genera Enterobacter, Colletotrichum, Phomopsis, Phyllosticta and Cladosporium [24]. Whereas the specialized endophytic genera, Pseudomonas [25,26,27], Bacillus [28,29], Paenibacillus [30,31] and Streptomyces [28,32] can suppress the wide range of plant pathogens. As the endophytes thrive in the plant tissues, their vertical transmission results in movement of the microbiome to the seeds [33]. Studies have revealed that in most of the plant species, the specific microbiota is transferred from mother plants to their seeds, that aids in their germination, and subsequent colonization of the emerging plants, thus resulting in vertical transmission of the endophytes. The conventional methods of microbe isolation are not enough to understand the actual structural diversity and abundance of the entire microbiome, since many obligate microbes and those in low titers are missed out. The obligate bacteria that cannot be cultured represent 26 phyla out of the 52 major lineages within the domain bacteria, and in some plant systems, these unculturable microbes may be in high abundance [34,35]. To address this problem, the use of deep sequencing of the host-associated microbial niches using the metagenomic approach is more reliable and promising for studies in the field of microbial ecology, evolution and diversity [31,36,37,38,39]. Metagenomic studies by NGS can reveal the signature pattern of functional genes from the matching sequences from databases, namely the Kyoto Encyclopedia of Genes and Genomes (KEGG) from Kyoto University [40] and the Clusters of Orthologous Groups (COGs) from the National Center for Biotechnology Information (NCBI) [41].
Hitherto, the studies on the diversity of seed associated endophytes have not been explored by next generation sequencing, in the two genera of the Caricaceae family, namely Carica and Vasconcellea. Our study is focused on the structure of the seed associated microbiome of the cultivated papaya (Carica papaya) and its wild relatives (Vasconcellea spp.) by next generation sequencing. The data throws light on to the composition and variations of the endophytic microbiome among the four plant species of the Caricaceae family.
2. Materials and Methods
2.1. Source of the Seeds
In this study, the seeds from the four plant species, namely C. papaya (var. Arka Prabhat), Vasconcellea cundinamarcensis, V. cauliflora and V. goudotiana, were used (Figure 1). All the seed samples were collected from ICAR-Indian Institute of Horticultural Research (IIHR), Hessaraghatta lake post, Bengaluru, Karnataka, India (13.1348° N, 77.4960° E), except for the seeds of V. cundinamarcensis that were collected from Government Botanical Garden, Ooty, Tamil Nadu, India (11.4189° N, 76.7114° E). The sites of collection are shown on the map India (Springer copyrights, [42]) using raster package in R studio [43] for geography of the collection sites (Figure S1).
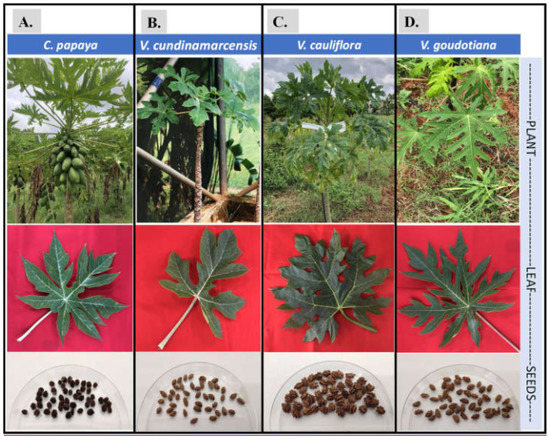
Figure 1.
Pictures of the plants, their leaves and the seeds of each of the four sample plant species, namely (A) C. papaya; (B) V. cundinamarcensis; (C) V. cauliflora; and (D) V. goudotiana, used in this study. The pictures of the established plants are shown in the top row, and their respective leaf and seed samples are in the middle and bottom row.
2.2. Isolation of DNA from the Seeds
The seeds were washed with sterile distilled water and then subjected to mercuric chloride (0.05%) treatment for 5 min for surface sterilization. Later, the seed coat was removed using a sterile surgical scalpel under aseptic condition [44]. The resultant seed pulp was transferred into a sterile 1.5 mL micro-centrifuge tubes and homogenized using sterile micro pestles. Then, the DNA was isolated using DNASure® Plant Mini Kit (Genetix Biotech Asia Pvt. Ltd., New Delhi, India) by following the manufacturer’s protocol. The DNA extracts were checked for their quality and quantity using Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA) in the Qubit 4 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). For each of the four samples, net 20 ng of DNA was used for MiSeq library preparation.
2.3. Preparation of MiSeq Library in Illumina, Quality Check and Sequencing
The amplicon libraries were prepared using Nextera XT Index Kit (Illumina Inc., San Diego, CA, USA) as per the 16S rRNA metagenomic sequencing library preparation protocol (Part # 15044223 Rev. B). The size of the library was chosen to obtain 2 × 300 bp paired ends. The primers were designed for the polymerase chain reaction (PCR) based amplification of the bacterial 16S V3–V4 and fungal ITS regions (Table S1) and the PCR amplicons were produced.
The QC passed amplicons with the Illumina adaptors were amplified as per the standard Illumina protocol. The amplicon libraries were purified by AMPure XP beads (Beckman Coulter, Brea, CA, USA) and quantified using Qubit Fluorometer. The amplified libraries were analyzed on the 4200 Tape Station system (Agilent Technologies, Santa Clara, CA, USA) using D1000 Screen tape as per manufacturer’s instructions for library size. The molarity was calculated for the library using the maximum peak size. After obtaining the mean peak size from the Tape Station profile, libraries were loaded onto MiSeq (20 pM) for cluster generation and sequencing. Paired-end sequencing was performed for template fragmentation for sequencing in both forward and reverse directions. The kit reagents were used in the binding of samples to complementary adapter oligos on a paired-end flow cell. The adapters were designed to allow selective cleavage of the forward strands after re-synthesis of the reverse strand during sequencing. The copied reverse strands were then used to sequence from the opposite end of the fragment [45].
2.4. Pipelines and Metagenomic Analysis
The amplicon sequencing analysis of Illumina paired-end reads were managed through roadmap (ampliseq v1.1.2) (https://github.com/nf-core/ampliseq; accessed on 30 July 2021) by means of DADA2 [46] and QIIME2 [47] integrated in the nf-core framework [48], which support data analysis that follows FAIR principles [49]. Several bio tools were used in sequences to analyze the raw data. Firstly, we used FASTQC [50] to check the quality of the raw data, and next MultiQC was applied to summarize the same [51]. Cut adapt plugin removes the primer fragments of the sequencing reads [52]. Further, we used DADA2 plugin for quality filter of the sequences, to remove the noise, combine them and finally the chimeric sequences were removed. We downloaded the SILVA database (Release 138) [53] and the sequences that were flanked with the forward primer [5′-GCCTACGGGNGGCWGCAG-3′] and reverse primer [5′-ACTACHVGGGTATCTAATCC-3′] were pulled out, followed by QIIME2 Naïve Bayes feature accuracy classifier [54]. The fitted classifier of the taxon was used to classify the representative sequences. The nf-core ampliseq pipeline was executed with standard parameters, with DADA2 quality settings with --trunclenf and --trunclenr parameters of 277 and 215, respectively. The nf-core/ampliseq pipelines helped in removing the amplicon sequence variants that were annotated as mitochondria or chloroplasts. Relative abundance was calculated based on the taxonomic classification. An iterative abundance of the taxa was plotted using bar plot with respect to their samples. For each sample, using Chao1 and Abundance-based Coverage Estimator indices, we estimated the alpha diversity that correlated with the species richness, whereas taxa richness and distribution were measured using Shannon and Simpson indices. The Beta diversity indices, distance matrices for the Jaccard, BrayCurtis, unweighted UniFrac, and weighted UniFrac were measured for visualization of the principal co-ordinates analysis. The cleaned reads were analyzed using SILVAngs (https://ngs.arb-silva.de/silvangs/; with default parameters; accessed on 29 June 2021). Further downstream analysis was performed by using the filtered biom and phylogenetic tree files using Shiny-Phyloseq [55] (https://github.com/joey711/shiny-phyloseq; accessed on 18 July 2019). The heatmap was plotted using the ampvis2 R package [56].
Predictive functional analysis of bacterial communities was done using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States 2 (PICRUSt2) [57]. To predict functions, the representative sequences were placed into a reference tree using the place_seqs.py utility. The BIOM (Biological Observation Matrix) table was then used as an input to PICRUSt to derive relative Kyoto Encyclopedia of Genes and Genomes (KEGG).
2.5. Fungal ITS Metagenomic Analysis
Analysis of fungal internal transcribed spacer (ITS) sequences from the Illumina sequencing platform was done using PIPITS v2.7 [58], which is reported to perform better than QIIME2 and Galaxy [59]. The illumina fastq sequences were assembled with VSEARCH [60] and quality-filtering was done with fastx through the PIPITS command pispino_createreadpairslist. The ITSx [61] was executed through the PIPITS command pipits_funits. Chimera filtering and clustering was done using VSEARCH through the PIPITS command pipits_process. The classic tabular OTU table was converted into a BIOM format, and taxonomy was assigned with UNITE [RDP Classifier].
2.6. Data Availability and Graphs
All the codes, scripts, expression and taxonomy data are provided as supplementary files and are uploaded in github: https://github.com/karthiksnarayan/Seed-metagenomics_papaya.git; accessed on 30 July 2021. The fastq reads from the samples were named as C-papaya_R1 and C-papaya_R2 for 16S rRNA. While the fastq reads for ITS were named as C-papaya-ITS_R1 and C-papaya-ITS_R2. A similar naming pattern was followed for other samples, namely, V. cundinamarcensis, V. cauliflora and V. goudotiana. These samples were registered in NCBI-GenBank, with BioSample accession numbers BioProject: PRJNA707250 with accession numbers from SRR13926680 to SRR13926687. The project details and the associated files are deposited in the Open Science Framework repository (https://osf.io/x92ng/; accessed on 29 July 2021). The graphs were built using package “ggplot2” in R studio (V1.3.1093). The statistical analysis was also performed in R studio using phyloseq package for Constrained Analysis of Principal Coordinates (CAP). The flow of the analysis is diagrammatically represented for the amplicon assays in supplementary Figure S2.
3. Results
The amplicons of V3–4 regions of the bacterial 16S rRNA and the fungal ITS regions of the seed associated endophytic microbes from C. papaya and the three Vasconcellea spp. were successfully sequenced on the MiSeq platform. All the samples resulted in data size that ranged between 53 and 91 Mb (Table 1). The sequence data (Number of reads and Mb) were recorded low for both the ITS and the 16S rRNA amplicons in C. papaya when compared to the three Vasconcellea spp. The highest number of ITS reads were obtained from V. cundinamarcensis with 90,803,894 bases accounting for 186,838 reads, resulting in a size of 91 Mb, followed by V. goudotiana and V. cauliflora. Whereas, for V3–V4 regions of 16S rRNA, the reads were highest for Vasconcellea goudotiana followed by V. cauliflora and V. cundinamarcensis (Table 1).

Table 1.
Summary of the raw reads of amplicon sequences derived from the V3–V4 region of bacterial 16S rRNA and the fungal ITS region from the seed associated endophytic microbes.
3.1. Structural Composition and Alpha Diversity of the Seed Associated Endophytic Microbes
The highest bacterial OTUs of 204 were obtained from the seeds of V. goudotiana and the lowest of 144 from seeds of C. papaya. Overall, 422 distinct OTUs were recorded from the seed samples of four different species from the SILVA database. The α-diversity indices based by abundance (Chao1) and by non-parametric method (ACE) placed V. goudotiana at the highest level, whereas the indices plotted C. papaya at the lowest level. The richness and the diversity of the samples were estimated using Shannon and Simpson indices. The Shannon index ranged between 3.90 and 3. The highest and the lowest Shannon index for the endophytic bacterial microbiome were observed in the seeds of V. cundinamarcensis and V. cauliflora, respectively. The Simpson index ranged between 0.955 and 0.861. The highest and lowest Simpson index in the endophytic bacterial microbiome were observed in the seeds of C. papaya and V. cauliflora, respectively. From the four seed samples, the relative abundance of bacterial and fungal communities with α-diversity indices are compared and presented in Figure 1. As revealed by the number of reads in 16S rRNA amplicon sequences from the different species, the phylum Proteobacteria was the most abundant (64.7–72.8%) and was at highest level in V. cundinamarcensis (72.8%). The other bacterial phyla that are represented in all the samples were Firmicutes, Bacteroidetes, Planctomycetes and Actinobacteria, in the order of highest to lowest abundance. The phylum Campilobacterota was present at low levels only in the seeds of V. goudotiana and V. cauliflora. The levels of beneficial phylum such as the Firmicutes varied among the seeds of different species. The abundance of Firmicutes was higher in the seed samples of V. cauliflora and V. goudotiana, accounting for 26.1% and 25% abundance, respectively. Whereas the abundance of Firmicutes was low in the seeds of C. papaya and V. cundinamarcensis, accounting for 14.8% and 13.6%, respectively. For the bacterial taxonomic unit “Family”, the Enterobacteriaceae was most abundant in the seeds of V. cauliflora (57.0%) and V. goudotiana (57.3%), while the most abundant bacterial family in the seeds of V. cundinamarcensis (17.2%) and C. papaya (16.7%) was Chromabacteriaceae. The other bacterial classes that were found in the seeds of all the plant species are Betaproteobacteria, Bacilli, Alphaproteobacteria, Flavobacteriia, Actinobacteria, Sphingobacteriia and Verrucomicrobiae, in the order of their highest to lowest abundance. For the taxonomical level “Order”, the order Enterobacteriales was most abundant in the seeds of V. cauliflora (60.93%) and V. goudotiana (60.98%), while the order Neisseriales was most abundant in the seeds of two other species, namely C. papaya (20.81%) and V. cundinamarcensis (20.83%).
The kingdom fungi is the highest level of taxonomic unit for the ITS amplicons. The highest number of fungal OTUs of 69 were obtained from the seeds of V. goudotiana and the lowest number of fungal OTUs of 43 were from the seeds of C. papaya. Overall, only 91 distinct fungal OTUs were recorded from the seed samples of the four different species, by using the SILVA database (Release 138). Since the number of fungal OTUs recorded were low, the α-diversity was recorded lower for fungi than the bacterial indices. The relative abundance of the fungal communities with α-diversity indices from the four seed samples are compared and presented in Figure 1F. Based on the abundance (Chao1) by non-parametric method (ACE), the α-diversity indices were highest for V. goudotiana, while it was lowest for V. cundinamarcensis. The richness and the diversity of the endophytic fungi in all the four seed samples were estimated using Shannon and Simpson indices. The Shannon index ranged between 1.80 and 0.87. The highest and the lowest α-diversity for the endophytic fungi, as revealed by the Shannon index were recorded in the seeds of V. goudotiana and V. cundinamarcensis, respectively. The Simpson index ranged between 0.76 and 0.54. The highest and lowest α-diversity in the endophytic fungal microbiome as revealed by the Simpson index were observed in the seeds of V. goudotiana and V. cundinamarcensis, respectively. In both the Shannon and Simpson indices, C. papaya and V. cundinamarcensis were plotted lower than the other two seed samples (Figure 2F). Only 10 fungal OTUs matched to the taxonomic unit phylum from a total of 91 OTUs retrieved from the ITS amplicons from the seeds of four distinct plant species. The sequence analysis could identify only two phyla namely Ascomycota and Basidiomycota, and the phylum Ascomycota was abundant in the seeds of all the four species. Whereas the phylum Basidiomycota was recorded at low levels in the seeds of C. papaya and V. goudotiana with an abundance of 13% and 8%, respectively. The highest identified fungal OTU reads in the ITS amplicons represented the genus Pyxine with 314 reads, accounting for 83% abundance that could only be traced to the seed samples of V. goudotiana (Figure 2D). For further information about the distribution across different taxonomical levels for the 4 seed samples, we have provided the supplementary files for both 16S rRNA (Supplementary File S1: Taxonomy_16S) and ITS sequences (Supplementary File S2: Taxonomy_ITS).
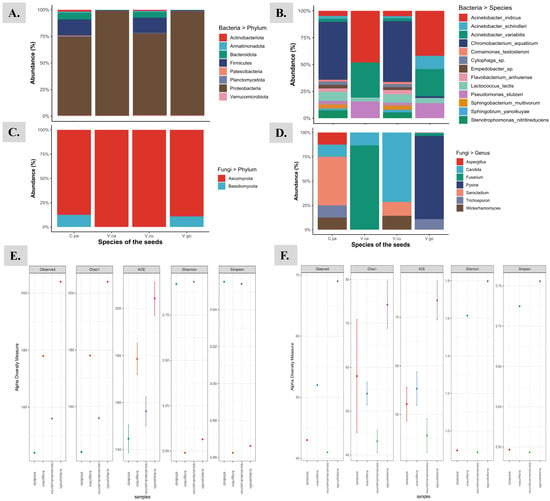
Figure 2.
The graphical representation of the taxonomical distribution of endophytic bacteria (A,B) and fungi (C,D) associated with the seeds of following plant species: 1. Carica papaya (C.pa); 2. Vasconcellea cauliflora (V.ca); 3. Vasconcellea cundinamarcensis (V.cu); and 4. Vasconcellea goudotiana (V.go). (A) represents the bacterial taxonomic unit “phylum”; (B) represents the bacterial taxonomic unit “species”; (C) represents the fungal taxonomic unit “phylum”; and (D) represents the fungal taxonomic unit “genus”. The bacterial (E) and fungal (F) alpha diversity measures were plotted with observed OTUs.
3.2. Functional Annotations
In the present study, functional analysis of bacterial 16S rRNA amplicons from seeds of four different species of the Caricaceae family, i.e., V. cauliflora, V. goudotiana, V. cundinamarcensis and C. papaya, are characterized for their functional genes by KEGG orthologous (KO) analysis. In total, 6385 KO were recorded, and the individual KO reads and their description are presented in the Supplementary File 3: Functional_KO_genes. The sum of the reads ranged between 1,013,340 and 1,015,395. The highest read of 9072 was observed for the KO TC.FEV:OM (iron complex outer membrane receptor proteins), in Vasconcellea cundinamarcensis (V. cu) and for the same, the lowest (3134) KO was observed in V. goudotiana (V. go) (Figure 3). Under metabolism, there were many reads for biosynthesis of various amino acids, vitamins, steroid, lipids, enzymes, nucleotides, secondary metabolites and others (Supplementary file 4: KEGG_Annotations). The sample V. goudotiana displayed 341,471 reads and the maximum reads were involved in the production of various alkaloids, flavone, flavanol, penicillin and streptomycin. The species V. goudotiana displayed an abundance of reads in biosynthesis of novobiocin, an aminocoumarin, penicillin and cephalosporin. Clavulanic acid is a beta-lactamase inhibitor of bacteria, and it was more abundant in V. cundinamarcensis than in other species (Figure 3). The variation in the reads of the biosynthetic pathways of the seed associated endophytic microbiome was significant for all the four plant species.
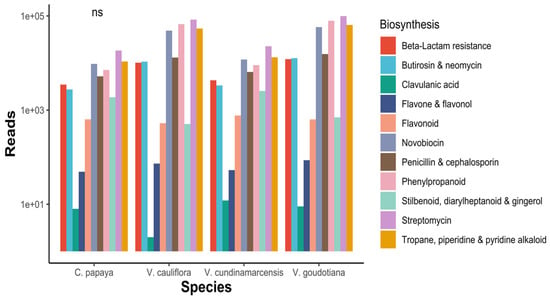
Figure 3.
Datamining the distribution of secondary metabolite biosynthesis related to KEGG functional annotations. ns: not significant.
3.3. Statistical Analysis
The metagenomic analysis revealed 4222 OTU IDs in bacteria. Using phyloseq, a Constrained Analysis of Principal coordinates (CAP) was performed for the OTUs table from the biome and plotted using the Bray Curtis measures. The CAP analysis placed the seed samples at 97% and 2% distance in the CAP1 and CAP2 axes, respectively (Figure 4A). The cultivated C. papaya sample was placed distinctly away from the three Vasconcellea spp. samples. Further, the microbiomes associated with the seeds of two wild species namely, V. goudotiana and V. cauliflora, were plotted close to each other. The beta diversity plotted the seed samples for Axis 1 to 3, with a value of 98.57%, 1.153% and 0.272%, respectively. The beta diversity also revealed that the microbiomes associated with the seeds of the two wild species, V. goudotiana and V. cauliflora, were plotted close to each other, reflecting their similarity (Figure 4B).
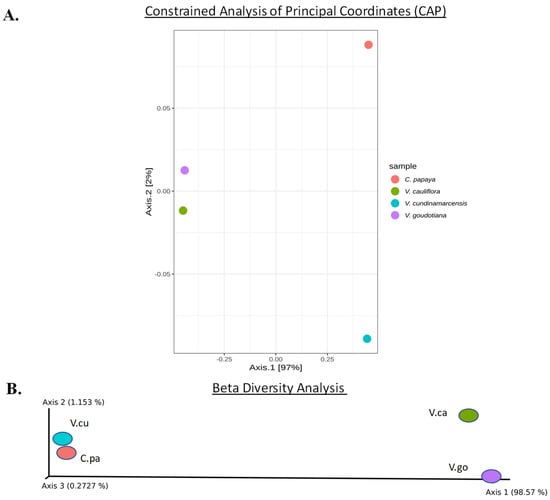
Figure 4.
(A) The Constrained Analysis of Principal Coordinates (CAP) was carried for the seed associated endophytic bacteria and their systematic patterns of variations among the OTUs are plotted. (B) The Beta diversity of the seed associated microbial samples using weighted UniFrac measures, and the visualization of the principal co-ordinates analysis. The color code of the dot represents sample species as 1. Carica papaya (C.pa); 2. Vasconcellea cauliflora (V.ca); 3. Vasconcellea cundinamarcensis (V.cu); and 4. Vasconcellea goudotiana (V.go).
4. Discussion
The beneficial role of the plant associated endophytic microbes in the growth and development of plants is well established. However, there are not enough studies on the structural and functional diversity of seed associated endophytes that are vertically transmitted in the plants. Although, some of the seed-borne endophytes can be isolated and characterized in vitro, but there are many that cannot be isolated and cultured for further characterization. In the present study, the seed associated endophytes from a cultivated papaya variety and three of its wild relatives from the Vasconcellea genus, all of them belonging to the Caricaceae family, were explored by next generation sequencing. The structural and functional annotations indicated that the cultivated species C. papaya showed the lowest bacterial OTUs when compared to the three of its wild relatives (Vasconcellea spp.) from the Caricaceae family. Further, the number of reads and the data obtained was low for C. papaya when compared to the other three species. The presence of a rich diversity of the endophytic microbes in Vasconcellea spp. may partly be responsible for the desirable traits, such as the tolerance to biotic and abiotic stresses [5,10]. The most desirable trait present in some of the Vasconcellea spp. is resistance to Papaya ring spot virus (PRSV), whereas the cultivated papaya varieties are highly susceptible to PRSV, worldwide [3]. Breeding efforts for introgression of virus resistance trait did not yield fruitful results in papaya. Jayavalli et al. [62] crossed V. cauliflora with six Indian varieties of papaya, which resulted in F1-progeny with only 23.6% of the plants free of viral symptoms. However, the remaining plants exhibited mosaic and other characteristic ring-spot viral symptoms such as the deformation of fruits. Magdalita [62] crossed C. papaya × V. cauliflora, and the resulting 114 hybrid plant lines were screened with two isolates of PRSV-P from Australia in the greenhouse studies. Post-inoculation of PRSV, only 22 hybrid plants survived, in which there was no development of viral symptoms and were tested negative by ELISA. When these 22 hybrid plants were tested in the field against PRSV infection, only 12 hybrids survived and were free from the symptoms. The progeny papaya plants derived from the inter-species crosses are difficult to raise because of slow proliferation and growth rate, and high rate of mortality upon hardening and acclimatization.
The current study unravels the differences in the endophytic microbes associated with the seeds of C. papaya and three of its wild relatives from genus Vasconcellea. Further, the sequences obtained from the amplicons derived from the seed associated microbiome defined the functional classifications that includes genes involved in metabolism, defense, etc. This is the first such study that employed the next generation sequencing technology to elucidate the seed associated microbiome structure and their functional characterization. With these preliminary findings, we anticipate that the nature of endophytic microbial population residing within the plants can be an indicator of adaptive measure for incorporating desirable traits into the cultivated varieties of papaya. After the micro-propagation of the plant tissues, microbial incorporation can be done in the papaya seedlings during the hardening stage to boost the growth and development and to incorporate stress tolerance. These practices are in practice for bio hardening of micropropagated banana plantlets, through the application of beneficial microbes at both the primary and secondary stages of hardening for the promotion of plant growth [56]. This treatment also resulted in the reduced incidence in soft rot of banana [63]. Here, we believe that such microbial introduction will improve their chances of survival in the field and can withstand the pressure from abiotic and biotic pressures. Thus, exploration and comparison of microbial consortium in the seeds of cultivated C. papaya (variety: Arka Prabhath) with its wild relatives Vasconcellea spp. will help in boosting the plant yields through application of beneficial microbial consortium. There are not many metagenomic studies carried out for papaya; however, one of the studies was done for the in vitro cultured papaya field shoots representing three varieties of papaya (Arka Surya, Arka Prabhath and Red Lady) [44]. However, the seed associated endophytic microbial structure and their roles remained unanswered for both the cultivated and wild relatives of papaya.
In the seed associated microbiome of the cultivated papaya (Arka Prabhath) and three of its wild relative species from Vasconcellea genus, the most abundant bacterial phyla were Proteobacteria and Firmicutes. Previous microbial metagenomic studies by Thomas et al. [44] in different cultivated papaya varieties had also revealed that the phyla Proteobacteria was the most abundant. About 33% of Bacillus distribution remained unclassified, which corresponds to 0.5% of Firmicutes in C. papaya, whereas in case of V. cauliflora, V. cundinamarcensis and V. goudotiana, the genus Bacillus showed an abundance of 0.3%, 1% and 0.4%, respectively. In the previous reports of metagenomic studies of endophytes associated with the cultivated papaya varieties, the genus Bacillus also remained unclassified beyond the genus level [44]. The Bacillus species namely Bacillus subtilis was isolated from the in vitro samples of the three papaya varieties namely Arka Surya, Arka Prabhath, and Red Lady [44]. The Bacillus species are reported to be cultivable bacteria that reduced the disease incidence by 74.94% over the control, for rhizome rot in banana [63]. Another species, Bacillus firmus from the phylum Firmicutes, is reported to be beneficial bacteria in rapeseed and mustard for their role in promotion of both root and shoot growth [64]. The previous reports had also revealed that the seeds of papaya have Bacillus spp., which readily colonize the roots upon germination [44]. It was also hypothesized that the colonization by Bacillus sp. will protect the papaya plants from crown rot disease. However, this report did not reveal anything about the microbiome structure and their heritability through the seeds [65]. In addition, the role of secondary metabolites from Bacillus subtilis, has been shown to have an antagonistic effect on the plant pathogens [66]. This bacterium produces a wide range of antimicrobial lipopeptide compounds such as the urfactins, iturins, fengycins, polyketides and non-ribosomal peptides, all of which have antimicrobial activity [66,67,68]. These antimicrobials released by Bacillus subtilis are toxic to nematodes and plant pathogens, but are considered to be safe to mammals as per the standards of US Food and Drug Administration (US FDA) [69]. The use of B. subtilis as a biocontrol agent can substantially replace [70] the use of toxic nematicides [71]. The weighted β-diversity measured by the abundance of OTUs revealed that none of the plant species are close to each other with reference to their seed associated endophytic population. The comparison of the branch lengths of the OTUs by CAP analysis of the samples revealed that the two species, V. cauliflora and V. goudotiana are closer to each other. Further, these seed associated microbial samples showed similar functional signatures, but not the same levels of abundance. The factor Biosynthesis of secondary metabolite genes in functional analysis showed significant variations among the endophytic population associated with the seeds of the four plant species. The samples that show maximum abundance of the recorded class of secondary metabolite gene annotations were from the species V. cauliflora and V. goudotiana. However, the diversity of fungi was significantly low for further detailed analysis. Pyxine sp. was observed only in V. goudotiana and is reported to be a lichenized fungus. However, for the fungal family Saccharomycetales Incertae sedis that produces indole acetic acid (IAA), the recorded reads were very low. The next highest reads after Pyxine sp. belonged to the OTU of Trichosporon genus with 40 reads traced in V. goudotiana. The Trichosporon genus is reported to be a rhizosphere-based plant growth-promoting yeast that can degrade various nitrogen sources and produce auxin [72].
5. Conclusions
This comparative study of microbiomes associated with the seeds of the cultivated Carica papaya and its four wild relatives from Caricaseae family (Vasconcellea spp.) precisely determines the population structure of the endophytic microbes. The characterization of the functional genes by KEGG showed that there was a high abundance of genes involved in secondary metabolite pathways in the seed associated endophytes from the genus Vasconcellea than the genus Carica. This is the first report on the seed associated endophytic microbial diversity and their functional gene structures between the cultivated C. papaya and its wild relative species from Vasconcellea genus (mountain papaya). The functional distribution of genes from the microbes partly, if not completely, may help the Vasconcellea species for their desirable traits. Furthermore, this study indicates that the domestication of plant species (e.g., Carica papaya) may result in reduced endophytic microbial diversity than their corresponding wild relative species (Vasconcellea spp.), which show higher microbial diversity.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microbiolres12040057/s1. Figure S1. Collection sites of papaya and their wild relatives namely, Carica papaya Va. Arka prabhath; Vasconcellea cauliflora; Vasconcellea cundinamarcensis; and Vasconcellea goudotiana from Southern India. Figure S2. Pipelines of next generation sequencing. Figure S3. Top 10 functional orthologues genes across the samples. Table S1. Primers used in this study for the PCR amplifications of bacterial 16S rRNA and the fungal ITS sequences from the DNA extracts of the seed pulps.
Author Contributions
B.L.P. Conceptualized the work, designed experiments, supervised the entire study, compiled and edited the manuscript; K.S.N. assisted in coordination of the data analysis and both K.S.N. and A.M.G. in drafting of the manuscript. All the authors have read and agreed to publish these findings. All authors have read and agreed to the published version of the manuscript.
Funding
The authors acknowledge the financial support from ICAR (Govt. of India) network project on “Application of microorganisms in agriculture and allied sectors (AMAAS)”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All codes, scripts, expression and taxonomy data are provided as supplementary files and are uploaded in github: https://github.com/karthiksnarayan/Seed-metagenomics_papaya.git accessed on 30 July 2021. These samples were registered in NCBI-GenBank, with BioSample accession numbers BioProject: PRJNA707250 with accession numbers from SRR13926680 to SRR13926687. The project details and the associated files are deposited in the Open Science Framework repository (https://osf.io/x92ng/; accessed on 29 July 2021).
Acknowledgments
We are grateful to the ICAR (India) funded network program AMAAS for the financial support. We thank C. Vasugi (IIHR) for providing the seeds.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Saran, P.L. Papaya; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Altendorf, S. Major Tropical Fruits Review 2017; FAO: Rome, Italy, 2019; Available online: http://www.fao.org/fileadmin/templates/est/COMM_MARKETS_MONITORING/Tropical_Fruits/Documents/CA2895EN.pdf (accessed on 29 July 2021).
- Mishra, R.; Gaur, R.K.; Patil, B.L. Current knowledge of viruses infecting papaya and their transgenic management. In Plant Viruses: Evolution and Management; Springer: Berlin/Heidelberg, Germany, 2016; pp. 189–203. [Google Scholar]
- Yamamoto, H.Y. Comparison of the Carotenoids in Yellow-and Red-Fleshed Carica papaya. Nature 1964, 201, 1049–1050. [Google Scholar] [CrossRef]
- Antunes Carvalho, F.; Renner, S.S. A Dated Phylogeny of the Papaya Family (Caricaceae) Reveals the Crop’s Closest Relatives and the Family’s Biogeographic History. Mol. Phylogenet. Evol. 2012, 65, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Liebman, B. Nutritional Aspects of Fruit. Nut Action Newslett. 1992, 1, 10–11. [Google Scholar]
- Chandrika, U.G.; Jansz, E.R.; Wickramasinghe, S.M.D.N.; Warnasuriya, N.D. Carotenoids in Yellow- and Red-fleshed Papaya (Carica papaya L.). J. Sci. Food Agric. 2003, 83, 1279–1282. [Google Scholar] [CrossRef]
- Jiménez-Coello, M.; Guzman-Marín, E.; Ortega-Pacheco, A.; Perez-Gutiérrez, S.; Acosta-Viana, K.Y. Assessment of the Anti-Protozoal Activity of Crude Carica papaya Seed Extract against Trypanosoma Cruzi. Molecules 2013, 18, 12621–12632. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Lucas, J.; Castañeda, D.; Hormigo, D. New Trends for a Classical Enzyme: Papain, a Biotechnological Success Story in the Food Industry. Trends Food Sci. Technol. 2017, 68, 91–101. [Google Scholar] [CrossRef]
- Scheldeman, X.; Willemen, L.; Coppens d’Eeckenbrugge, G.; Romeijn-Peeters, E.; Restrepo, M.T.; Romero Motoche, J.; Jiménez, D.; Lobo, M.; Medina, C.I.; Reyes, C.; et al. Distribution, diversity and environmental adaptation of highland papayas (Vasconcellea spp.) in tropical and subtropical America. In Plant Conservation and Biodiversity; Hawksworth, D.L., Bull, A.T., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 293–310. ISBN 978-1-4020-6444-9. [Google Scholar]
- Sawant, A.C. Crossing Relationships in the Genus Carica. Evolution 1958, 12, 263–266. [Google Scholar] [CrossRef]
- Dinesh, M.R.; Rekha, A.; Ravishankar, K.V.; Praveen, K.S.; Santosh, L.C. Breaking the Intergeneric Crossing Barrier in Papaya Using Sucrose Treatment. Sci. Hortic. 2007, 114, 33–36. [Google Scholar] [CrossRef]
- Siar, S.V.; Beligan, G.A.; Sajise, A.J.C.; Villegas, V.N.; Drew, R.A. Papaya Ringspot Virus Resistance in Carica Papaya via Introgression from Vasconcellea quercifolia. Euphytica 2011, 181, 159–168. [Google Scholar] [CrossRef]
- O’Brien, C.M.; Drew, R.A. Marker-Assisted Hybridisation and Backcrossing between Vasconcellea Species and Carica Papaya for PRSV-P Resistance. In Proceedings of the International Symposium on Molecular Markers in Horticulture 859, Corvallis, OR, USA, 25–27 September 2010. [Google Scholar]
- Gouda, S.; Das, G.; Sen, S.K.; Shin, H.S.; Patra, J.K. Endophytes: A Treasure House of Bioactive Compounds of Medicinal Importance. Front. Microbiol. 2016, 7, 1538. [Google Scholar] [CrossRef] [Green Version]
- Krings, M.; Taylor, T.N.; Hass, H.; Kerp, H.; Dotzler, N.; Hermsen, E.J. Fungal Endophytes in a 400-Million-Yr-Old Land Plant: Infection Pathways, Spatial Distribution, and Host Responses. New Phytol. 2007, 174, 648–657. [Google Scholar] [CrossRef] [PubMed]
- Redecker, D.; Kodner, R.; Graham, L.E. Glomalean Fungi from the Ordovician. Science 2000, 289, 1920–1921. [Google Scholar] [CrossRef] [Green Version]
- Clay, K. Fungal Endophytes of Grasses: A Defensive Mutualism between Plants and Fungi. Ecology 1988, 69, 10–16. [Google Scholar] [CrossRef]
- Clay, K. Fungal Endophytes of Plants: Biological and Chemical Diversity. Nat. Toxins 1993, 1, 147–149. [Google Scholar] [CrossRef]
- Breen, J.P. Acremonium Endophyte Interactions with Enhanced Plant Resistance to Insects. Annu. Rev. Entomol. 1994, 39, 401–423. [Google Scholar] [CrossRef]
- Jaber, L.R.; Vidal, S. Fungal Endophyte Negative Effects on Herbivory Are Enhanced on Intact Plants and Maintained in a Subsequent Generation. Ecol. Entomol. 2010, 35, 25–36. [Google Scholar] [CrossRef]
- Leuchtmann, A.; Schmidt, D.; Bush, L.P. Different Levels of Protective Alkaloids in Grasses with Stroma-Forming and Seed-Transmitted Epichloe/Neotyphodium Endophytes. J. Chem. Ecol. 2000, 26, 1025–1036. [Google Scholar] [CrossRef]
- Faeth, S.H.; Saari, S. Fungal Grass Endophytes and Arthropod Communities: Lessons from Plant Defence Theory and Multitrophic Interactions. Fungal Ecol. 2012, 5, 364–371. [Google Scholar] [CrossRef] [Green Version]
- Nair, D.N.; Padmavathy, S. Impact of Endophytic Microorganisms on Plants, Environment and Humans. Sci. World J. 2014, 2014, 250693. [Google Scholar] [CrossRef] [Green Version]
- Mendes, R.; Kruijt, M.; De Bruijn, I.; Dekkers, E.; Van Der Voort, M.; Schneider, J.H.M.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A.H.M.; et al. Deciphering the Rhizosphere Microbiome for Disease-Suppressive Bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; Weller, D.M. Natural Plant Protection by 2,4-Diacetylphloroglucinol-Producing Pseudomonas Spp. in Take-All Decline Soils. Mol. Plant-Microbe Interact. 1998, 11, 144–152. [Google Scholar] [CrossRef] [Green Version]
- Kloepper, J.W.; Leong, J.; Teintze, M.; Schroth, M.N. Enhanced Plant Growth by Siderophores Produced by Plant Growth-Promoting Rhizobacteria. Nature 1980, 286, 885–886. [Google Scholar] [CrossRef]
- Cha, J.Y.; Han, S.; Hong, H.J.; Cho, H.; Kim, D.; Kwon, Y.; Kwon, S.K.; Crusemann, M.; Bok Lee, Y.; Kim, J.F.; et al. Microbial and Biochemical Basis of a Fusarium Wilt-Suppressive Soil. ISME J. 2016, 10, 119–129. [Google Scholar] [CrossRef] [Green Version]
- Cazorla, F.M.; Romero, D.; Pérez-García, A.; Lugtenberg, B.J.J.; De Vicente, A.; Bloemberg, G. Isolation and Characterization of Antagonistic Bacillus Subtilis Strains from the Avocado Rhizoplane Displaying Biocontrol Activity. J. Appl. Microbiol. 2007, 103, 1950–1959. [Google Scholar] [CrossRef] [PubMed]
- Haggag, W.M. Colonization of Exopolysaccharide-Producing Paenibacillus Polymyxa on Peanut Roots for Enhancing Resistance against Crown Rot Disease. Afr. J. Biotechnol. 2007, 6, 13. [Google Scholar] [CrossRef]
- Prasannakumar, M.K.; Mahesh, H.B.; Desai, R.U.; Kunduru, B.; Narayan, K.S.; Teli, K.; Puneeth, M.E.; Rajadurai, R.C.; Parivallal, B.; Babu, G.V. Metagenome Sequencing of Fingermillet-Associated Microbial Consortia Provides Insights into Structural and Functional Diversity of Endophytes. 3 Biotech 2020, 10, 15. [Google Scholar] [CrossRef]
- Liu, D.; Anderson, N.A.; Kinkel, L.L. Selection and Characterization of Strains of Streptomyces Suppressive to the Potato Scab Pathogen. Can. J. Microbiol. 1996, 42, 487–502. [Google Scholar] [CrossRef]
- Vujanovic, V.; Germida, J.J. Seed Endosymbiosis: A Vital Relationship in Providing Prenatal Care to Plants. Can. J. Plant Sci. 2017, 97, 972–998. [Google Scholar] [CrossRef] [Green Version]
- Handelsman, J. Metagenomics: Application of Genomics to Uncultured Microorganisms. Microbiol. Mol. Biol. Rev. 2005, 69, 195. [Google Scholar] [CrossRef] [Green Version]
- Hongoh, Y. Diversity and Genomes of Uncultured Microbial Symbionts in the Termite Gut. Biosci. Biotechnol. Biochem. 2010, 74, 1145–1151. [Google Scholar] [CrossRef]
- Adu-Oppong, B.; Gasparrini, A.J.; Dantas, G. Genomic and Functional Techniques to Mine the Microbiome for Novel Antimicrobials and Antimicrobial Resistance Genes. Ann. N. Y. Acad. Sci. 2017, 1388, 42–58. [Google Scholar] [CrossRef] [Green Version]
- Anantharaman, K.; Brown, C.T.; Hug, L.A.; Sharon, I.; Castelle, C.J.; Probst, A.J.; Thomas, B.C.; Singh, A.; Wilkins, M.J.; Karaoz, U.; et al. Thousands of Microbial Genomes Shed Light on Interconnected Biogeochemical Processes in an Aquifer System. Nat. Commun. 2016, 7, 13219. [Google Scholar] [CrossRef]
- Zaheer, R.; Noyes, N.; Ortega Polo, R.; Cook, S.R.; Marinier, E.; Van Domselaar, G.; Belk, K.E.; Morley, P.S.; McAllister, T.A. Impact of Sequencing Depth on the Characterization of the Microbiome and Resistome. Sci. Rep. 2018, 8, 5890. [Google Scholar] [CrossRef] [PubMed]
- Pereira-Marques, J.; Hout, A.; Ferreira, R.M.; Weber, M.; Pinto-Ribeiro, I.; Van Doorn, L.J.; Knetsch, C.W.; Figueiredo, C. Impact of Host DNA and Sequencing Depth on the Taxonomic Resolution of Whole Metagenome Sequencing for Microbiome Analysis. Front. Microbiol. 2019, 10, 1277. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galperin, M.Y.; Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Expanded Microbial Genome Coverage and Improved Protein Family Annotation in the COG Database. Nucleic Acids Res. 2015, 43, D261–D269. [Google Scholar] [CrossRef] [PubMed]
- Sankar Narayan, K.; Esack, E.R.; Radhapriya, P.; Gopal, V.B.; Muthu, S.; Perumal, P. Impact of Geography on Adaptation of Phyllanthus Amarus Seeds. 3 Biotech 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Hijmans, R.J.; van Etten, J.; Cheng, J.; Mattiuzzi, M.; Sumner, M.; Greenberg, J.A.; Lamigueiro, O.P.; Bevan, A.; Racine, E.B.; Shortridge, A. Package ‘Raster’. R Package 2015, 269, 734. [Google Scholar]
- Thomas, P.; Agrawal, M.; Bharathkumar, C.B. Diverse Cellular Colonizing Endophytic Bacteria in Field Shoots and in Vitro Cultured Papaya with Physiological and Functional Implications. Physiol. Plant. 2019, 166, 729–747. [Google Scholar] [CrossRef] [PubMed]
- Padmanaban, A. End to End Sample Quality Control for Next Generation Sequencing Library Preparation and SureSelect Target Enrichment on the Agilent 2200 TapeStation System Application Note; Agilent Technologies: Bangalore, India, 2015. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Ewels, P.A.; Peltzer, A.; Fillinger, S.; Patel, H.; Alneberg, J.; Wilm, A.; Garcia, M.U.; di Tommaso, P.; Nahnsen, S. The Nf-Core Framework for Community-Curated Bioinformatics Pipelines. Nat. Biotechnol. 2020, 38, 276–278. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.-W.; da Silva Santos, L.B.; Bourne, P.E. The FAIR Guiding Principles for Scientific Data Management and Stewardship. Sci. Data 2016, 3, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data 2010; Babraham Bioinformatics: Cambridge, UK, 2017. [Google Scholar]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2’s Q2-Feature-Classifier Plugin. Microbiome 2018, 6, 1–17. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, K.S.; Kirkegaard, R.H.; Karst, S.M.; Albertsen, M. Ampvis2: An R Package to Analyse and Visualise 16S RRNA Amplicon Data. BioRxiv 2018. [Google Scholar] [CrossRef] [Green Version]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2: An Improved and Customizable Approach for Metagenome Inference. BioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Gweon, H.S.; Oliver, A.; Taylor, J.; Booth, T.; Gibbs, M.; Read, D.S.; Griffiths, R.I.; Schonrogge, K. PIPITS: An Automated Pipeline for Analyses of Fungal Internal Transcribed Spacer Sequences from the I Llumina Sequencing Platform. Methods Ecol. Evol. 2015, 6, 973–980. [Google Scholar] [CrossRef]
- Anslan, S.; Nilsson, R.H.; Wurzbacher, C.; Baldrian, P.; Tedersoo, L.; Bahram, M. Great Differences in Performance and Outcome of High-Throughput Sequencing Data Analysis Platforms for Fungal Metabarcoding. MycoKeys 2018, 39, 29–40. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Ryberg, M.; Hartmann, M.; Branco, S.; Wang, Z.; Godhe, A.; de Wit, P.; Sánchez-García, M.; Ebersberger, I.; de Sousa, F. Improved Software Detection and Extraction of ITS1 and ITS 2 from Ribosomal ITS Sequences of Fungi and Other Eukaryotes for Analysis of Environmental Sequencing Data. Methods Ecol. Evol. 2013, 4, 914–919. [Google Scholar] [CrossRef]
- Magdalita, P.M.; Persley, D.M.; Godwin, I.D.; Drew, R.A.; Adkins, S.W. Screening Carica Papaya× C. Cauliflora Hybrids for Resistance to Papaya Ringspot Virus-type P. Plant Pathol. 1997, 46, 837–841. [Google Scholar] [CrossRef] [Green Version]
- Rajamanickam, S.; Karthikeyan, G.; Kavino, M.; Manoranjitham, S.K. Biohardening of Micropropagated Banana Using Endophytic Bacteria to Induce Plant Growth Promotion and Restrain Rhizome Rot Disease Caused by Pectobacterium Carotovorum Subsp. Carotovorum. Sci. Hortic. 2018, 231, 179–187. [Google Scholar] [CrossRef]
- Ghosh, S.; Penterman, J.N.; Little, R.D.; Chavez, R.; Glick, B.R. Three Newly Isolated Plant Growth-Promoting Bacilli Facilitate the Seedling Growth of Canola, Brassica Campestris. Plant Physiol. Biochem. 2003, 41, 277–281. [Google Scholar] [CrossRef]
- Rivarez, M.P.S.; Parac, E.P.; Dimasingkil, S.F.M.; Magdalita, P.M. Defense Biopriming and Antimicrobial Activity of Endophytic Bacteria and Associated Bacillus Species Contribute to Bacterial Crown Rot Tolerance in Papaya. BioxRiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Kaspar, F.; Neubauer, P.; Gimpel, M. Bioactive Secondary Metabolites from Bacillus Subtilis: A Comprehensive Review. J. Nat. Prod. 2019, 82, 2038–2053. [Google Scholar] [CrossRef] [PubMed]
- Kakinuma, A.; Sugino, H.; Isono, M.; Tamura, G.; Arima, K. Determination of Fatty Acid in Surfactin and Elucidation of the Total Structure of Surfactin. Agric. Biol. Chem. 1969, 33, 973–976. [Google Scholar] [CrossRef]
- Caulier, S.; Nannan, C.; Gillis, A.; Licciardi, F.; Bragard, C.; Mahillon, J. Overview of the Antimicrobial Compounds Produced by Members of the Bacillus Subtilis Group. Front. Microbiol. 2019, 10, 302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denner, H.W.B. The Legislative Aspects of the Use of Industrial Enzymes in the Manufacture of Food and Food Ingredients; Nature Press: New York, NY, USA, 1983; Volume 111. [Google Scholar]
- Amruta, N.; Prasanna Kumar, M.K.; Puneeth, M.E.; Sarika, G.; Kandikattu, H.K.; Vishwanath, K.; Narayanaswamy, S. Exploring the Potentiality of Novel Rhizospheric Bacterial Strains against the Rice Blast Fungus Magnaporthe Oryzae. Plant Pathol. J. 2018, 34, 126–138. [Google Scholar] [CrossRef] [PubMed]
- Cawoy, H.; Bettiol, W.; Fickers, P.; Onge, M. Bacillus-Based Biological Control of Plant Diseases. In Pesticides in the Modern World—Pesticides Use and Management; Stoytcheva, M., Ed.; Books on Demand: Norderstedt, Germany, 2011. [Google Scholar]
- El-Maraghy, S.S.; Tohamy, T.A.; Hussein, K.A. Expression of SidD Gene and Physiological Characterization of the Rhizosphere Plant Growth-Promoting Yeasts. Heliyon 2020, 6, e04384. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).