Keratinocytes and Activation of TREM-1 Pathway in Cutaneous Leishmaniasis Lesions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Transcriptome and Pathway Analysis
2.2. Analysis of the TREM-1 Signaling Pathways
2.3. Patients and Ethics Statement
2.4. Cell Culture and Infection
2.5. Quantitative Real Time Polymerase Chain Reaction (RT-qPCR)
2.6. Immunohistochemistry
2.7. Statistical Analysis
3. Results
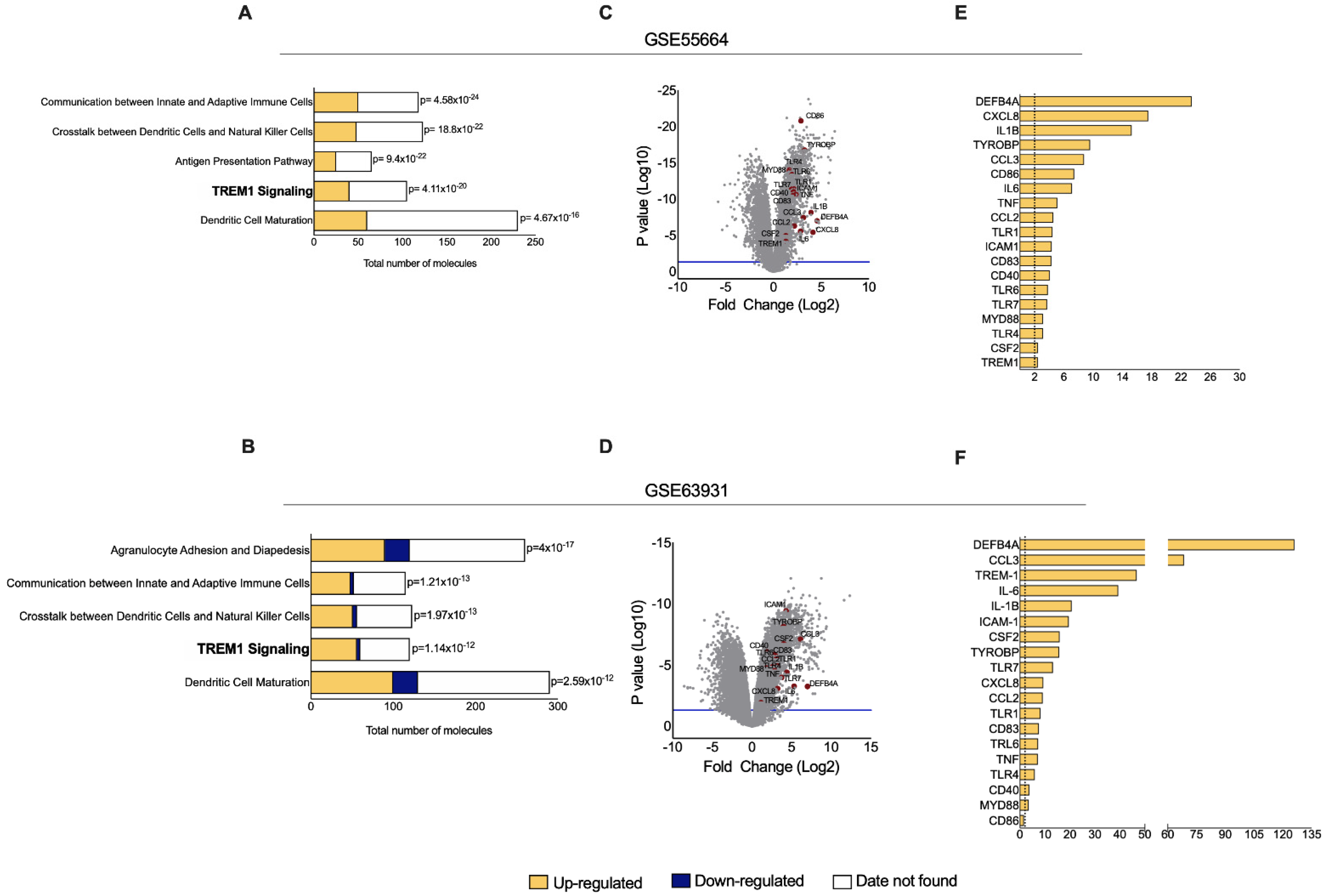
3.1. TREM-1 Signaling Pathway Is Significantly Up-Regulated in CL
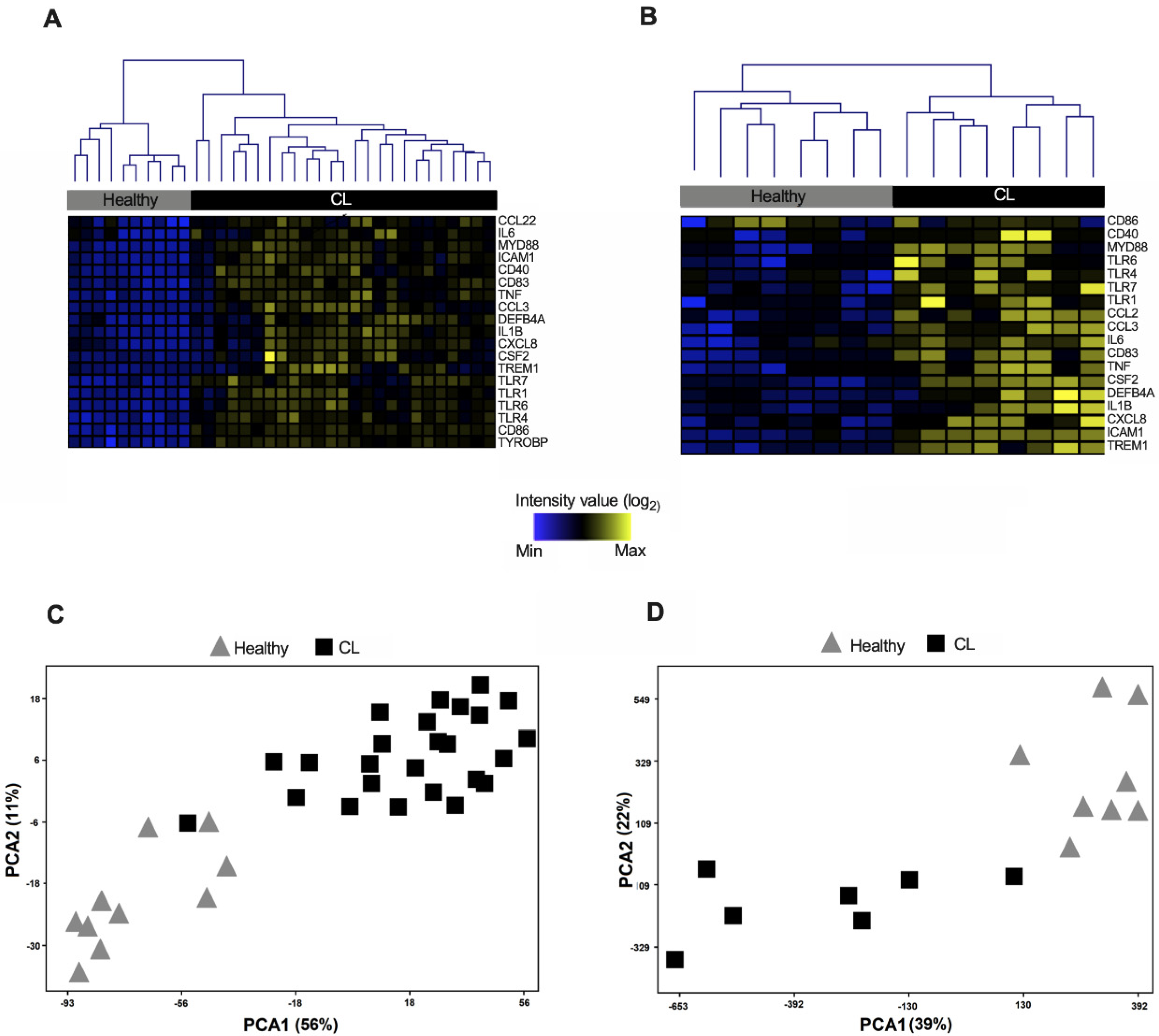
3.2. Expression Profile of Genes from the TREM-1 Pathway Distinguishes CL Samples from Healthy Controls
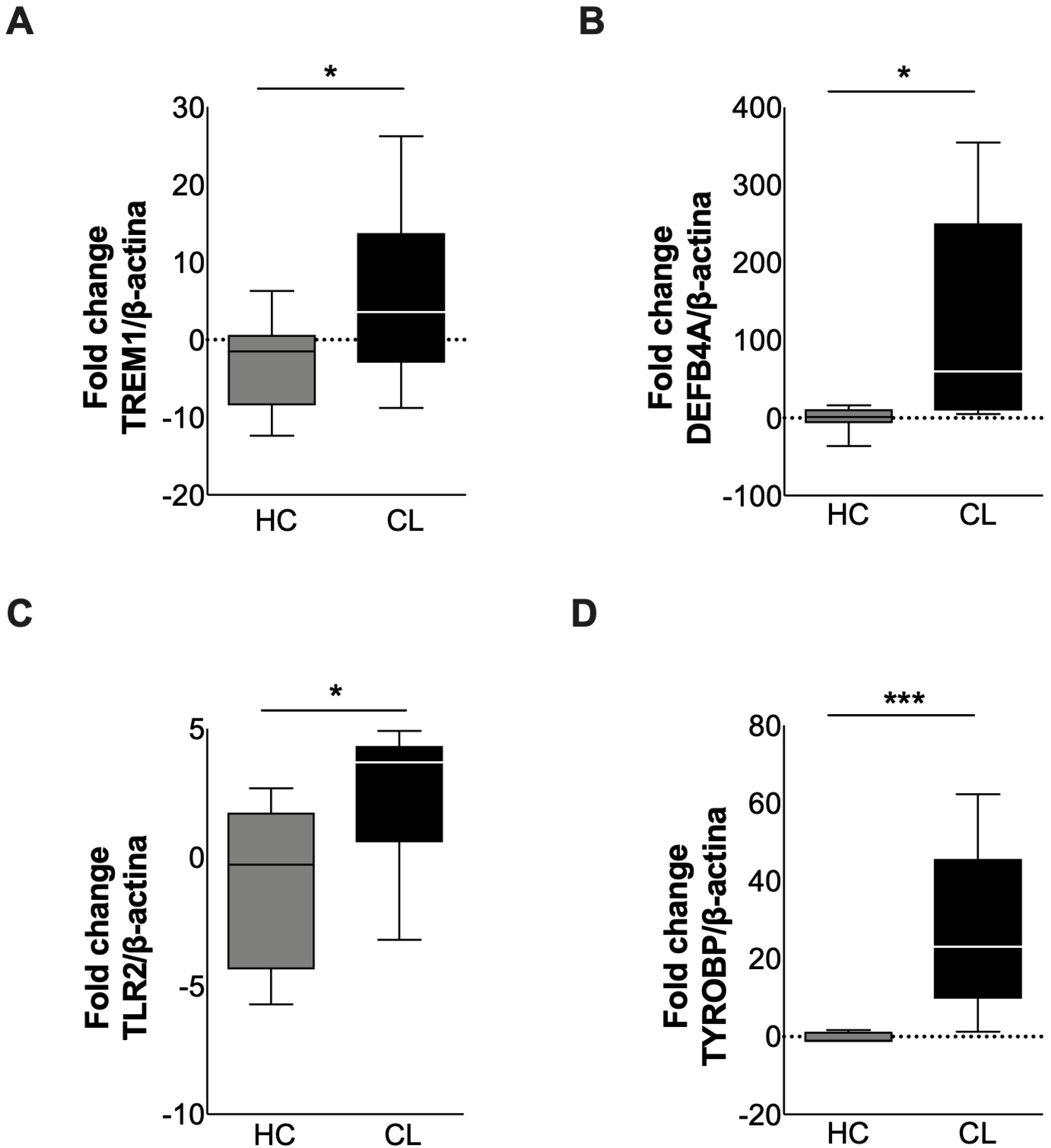
3.3. Validation of Differentially Expressed Genes from TREM-1 Pathway in Active Lesions of CL
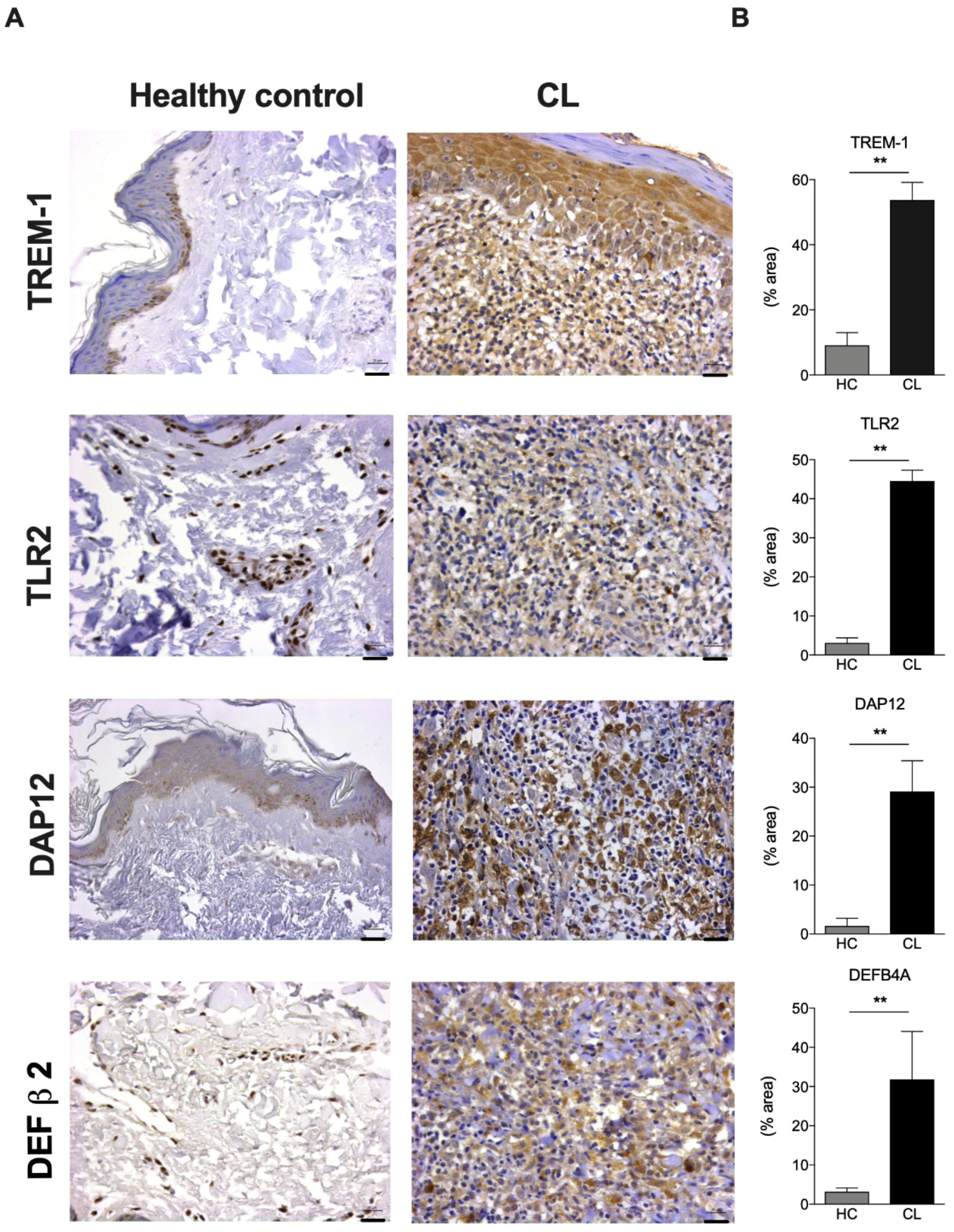
3.4. TREM-1 Protein Is Highly Expressed in the Epidermis of CL Lesions from Patients Infected with L. braziliensis
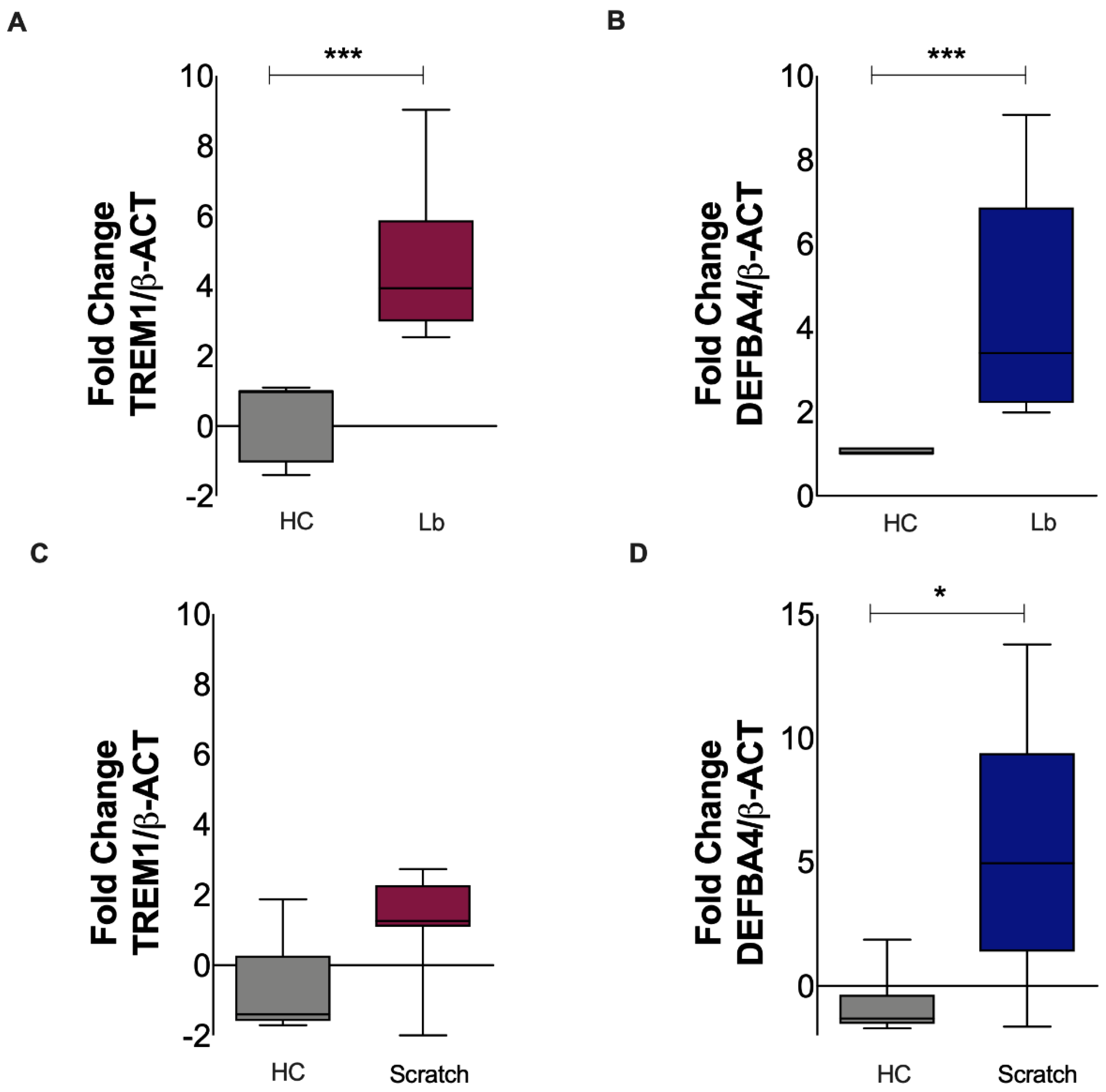
3.5. Gene Expression of TREM-1 Is Increased in Keratinocytes Exposed to L. braziliensis but Not to Mechanical Damage
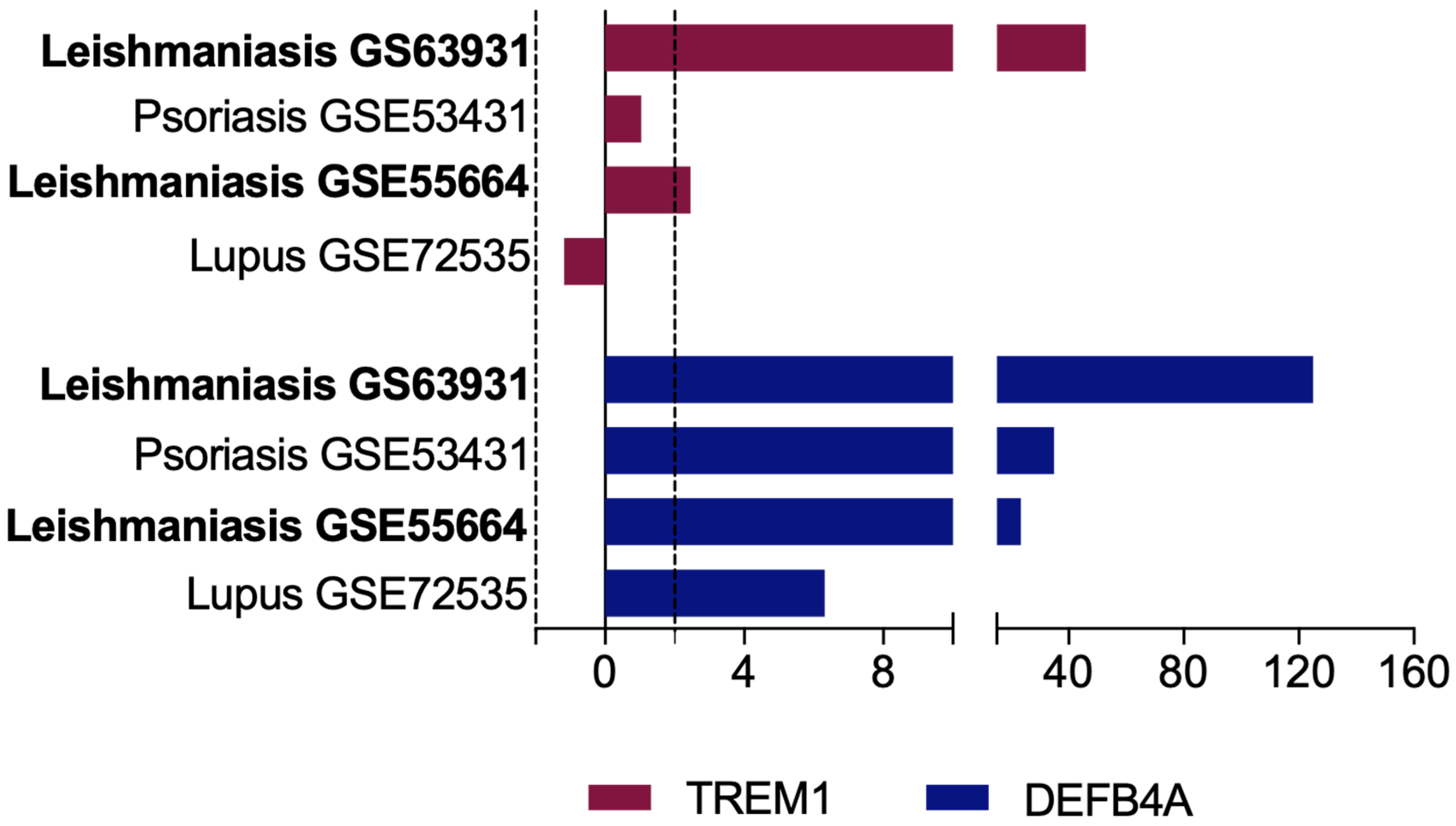
3.6. Overexpression of TREM-1 Is Specific of CL and Is Not Observed in Other Non-Infectious Inflammatory Skin Diseases
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaye, P.; Scott, P. Leishmaniasis: Complexity at the host-pathogen interface. Nat. Rev. Microbiol. 2011, 9, 604–615. [Google Scholar] [CrossRef]
- WHO Leishmaniasis. Available online: https://www.who.int/news-room/fact-sheets/detail/leishmaniasis (accessed on 5 January 2021).
- Scott, P.; Novais, F.O. Cutaneous leishmaniasis: Immune responses in protection and pathogenesis. Nat. Rev. Immunol. 2016, 16, 581–592. [Google Scholar] [CrossRef]
- Carvalho, L.P.; Passos, S.; Schriefer, A.; Carvalho, E.M. Protective and pathologic immune responses in human tegumentary leishmaniasis. Front. Immunol. 2012, 3, 301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scorza, B.M.; Carvalho, E.M.; Wilson, M.E. Cutaneous Manifestations of Human and Murine Leishmaniasis. Int. J. Mol. Sci. 2017, 18, 1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, B.; Schuster, S.; Zysset, D.; Rihs, S.; Dickgreber, N.; Schürch, C.; Riether, C.; Siegrist, M.; Schneider, C.; Pawelski, H.; et al. TREM-1 deficiency can attenuate disease severity without affecting pathogen clearance. PLoS Pathog. 2014, 10, e1003900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouchon, A.; Dietrich, J.; Colonna, M. Cutting Edge: Inflammatory Responses Can Be Triggered by TREM-1, a Novel Receptor Expressed on Neutrophils and Monocytes. J. Immunol. 2000, 164, 4991–4995. [Google Scholar] [CrossRef]
- Colonna, M. TREMs in the immune system and beyond. Nat. Rev. Immunol. 2003, 3, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Bouchon, A.; Facchetti, F.; Weigand, M.A.; Colonna, M. TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature 2001, 410, 1103–1107. [Google Scholar] [CrossRef]
- Klesney-tait, J.; Turnbull, I.R.; Colonna, M. The TREM receptor family and signal integration. Nat. Immunol. 2006, 7, 1266–1273. [Google Scholar] [CrossRef]
- Carneiro, M.W.; Fukutani, K.F.; Andrade, B.B.; Curvelo, R.P.; Cristal, J.R.; Carvalho, A.M.; Barral, A.; Van Weyenbergh, J.; Barral-Netto, M.; de Oliveira, C.I. Gene Expression Profile of High IFN-γ Producers Stimulated with Leishmania braziliensis Identifies Genes Associated with Cutaneous Leishmaniasis. PLoS Negl. Trop. Dis. 2016, 10, e0005116. [Google Scholar] [CrossRef] [Green Version]
- Nunes, S.; Silva, I.B.; Ampuero, M.R.; de Noronha, A.L.L.; de Souza, L.C.L.; Correia, T.C.; Khouri, R.; Boaventura, V.S.; Barral, A.; Ramos, P.I.P.; et al. Integrated analysis reveals that miR-193b, miR-671, and TREM-1 correlate with a good response to treatment of human Localized cutaneous leishmaniasis caused by Leishmania braziliensis. Front. Immunol. 2018, 9, 640. [Google Scholar] [CrossRef] [PubMed]
- Bomfim, L.G.S.; Magalhães, L.S.; Santos-Filho, M.A.A.; Peres, N.T.A.; Corrêa, C.B.; Tanajura, D.M.; Silva, A.M.; Lipscomb, M.W.; Borges, V.M.; Jesus, A.R.; et al. Leishmania infantum Induces the Release of sTREM-1 in Visceral Leishmaniasis. Front. Microbiol. 2017, 8, 2265. [Google Scholar] [CrossRef] [PubMed]
- Scorza, B.M.; Wacker, M.A.; Messingham, K.; Kim, P.; Klingelhutz, A.; Fairley, J.; Wilson, M.E. Differential Activation of Human Keratinocytes by Leishmania Species Causing Localized or Disseminated Disease. J. Invest. Dermatol. 2017, 137, 2149–2156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogers, M.; Kropf, P.; Choi, B.S.; Dillon, R.; Podinovskaia, M.; Bates, P.; Müller, I. Proteophosophoglycans regurgitated by Leishmania-infected sand flies target the L-arginine metabolism of host macrophages to promote parasite survival. PLoS Pathog. 2009, 5, e1000555. [Google Scholar] [CrossRef] [Green Version]
- Pivarcsi, A.; Kemény, L.; Dobozy, A. Innate immune functions of the keratinocytes: A review. Acta Microbiol. Immunol. Hung. 2004, 51, 303–310. [Google Scholar] [CrossRef]
- Ehrchen, J.M.; Roebrock, K.; Foell, D.; Nippe, N.; Von Stebut, E.; Johannes, M.; Viemann, D.; Varga, G.; Mu, C.; Schuberth, J.; et al. Keratinocytes Determine Th1 Immunity during Early Experimental Leishmaniasis. PLoS Pathog. 2010, 6, e1000871. [Google Scholar] [CrossRef]
- Novais, F.O.; Carvalho, L.P.; Passos, S.; Roos, D.S.; Carvalho, E.M.; Scott, P.; Beiting, D.P. Genomic Profiling of Human Leishmania braziliensis Lesions Identifies Transcriptional Modules Associated with Cutaneous Immunopathology. J. Investig. Dermatol. 2015, 135, 94–101. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, P.R.S.; Dessein, H.; Romano, A.; Cabantous, S.; De Brito, M.E.F.; Santoro, F.; Pitta, M.G.R.; Pereira, V.; Lain, C.; Rodrigues, V.; et al. IL2RA Genetic Variants Reduce IL-2—Dependent Responses and Aggravate Human Cutaneous Leishmaniasis. J. Immunol. 2015, 194, 2664–2672. [Google Scholar] [CrossRef] [Green Version]
- Ulusan, Ö.; Mert, U.; Sadıqova, A.; Öztürk, S.; Caner, A. Identification of gene expression profiles in Leishmania major infection by integrated bioinformatics analyses. Acta Trop. 2020, 208, 105517. [Google Scholar] [CrossRef]
- Maiese, A.; Bolino, G.; Mastracchio, A.; Frati, P.; Fineschi, V. An immunohistochemical study of the diagnostic value of TREM-1 as marker for fatal sepsis cases. Biotech. Histochem. 2019, 94, 159–166. [Google Scholar] [CrossRef]
- Fontana, R.; Raccosta, L.; Rovati, L.; Steffensen, K.R.; Jakobsson, T.; Melloni, G.; Bandiera, A.; Bergamini, A.; Maggioni, D.; Doglioni, C.; et al. Nuclear receptor ligands induce TREM-1 expression on dendritic cells: Analysis of their role in tumors. Oncoimmunology 2019, 8, 1554967. [Google Scholar] [CrossRef] [Green Version]
- Amrun, S.N.; Tan, J.J.L.; Rickett, N.Y.; Cox, J.A.; Lee, B.; Michael, J.G.; Solomon, T.; Perera, D.; Ooi, M.H.; Hiscox, J.A.; et al. TREM-1 activation is a potential key regulator in driving severe pathogenesis of enterovirus A71 infection. Sci. Rep. 2020, 10, 3810. [Google Scholar] [CrossRef]
- Pedro, V.; Neto, S.; De Carvalho, J.C.S.; Pimentel, V.E. Prognostic value of sTREM-1 in COVID-19 patients: A biomarker for disease severity and mortality. medRxiv 2020. [Google Scholar] [CrossRef]
- Tammaro, A.; Derive, M.; Gibot, S.; Leemans, J.C.; Florquin, S.; Dessing, M.C. TREM-1 and its potential ligands in non-infectious diseases: From biology to clinical perspectives. Pharmacol. Ther. 2017, 177, 81–95. [Google Scholar] [CrossRef]
- Adukpo, S.; Gyan, B.A.; Ofori, M.F.; Dodoo, D.; Velavan, T.P.; Meyer, C.G. Triggering receptor expressed on myeloid cells 1 (TREM-1) and cytokine gene variants in complicated and uncomplicated malaria. Trop. Med. Int. Health 2016, 21, 1592–1601. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; He, J.; Zheng, H.; Huang, S.; Lu, F. Association of TREM-1, IL-1β, IL-33/ST2, and TLR Expressions with the Pathogenesis of Ocular Toxoplasmosis in Mouse Models on Different Genetic Backgrounds. Front. Microbiol. 2019, 10, 2264. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.C.; Lin, C.N.; Chen, Y.J.; Chang, F.S.; Tsaihong, J.C.; Lee, K.M. Triggering receptor expressed on myeloid cells (TREM)-1 participates in Schistosoma mansoni inflammatory responses. Parasite Immunol. 2011, 33, 276–286. [Google Scholar] [CrossRef]
- Erdman, L.K.; Dhabangi, A.; Musoke, C.; Conroy, A.L.; Hawkes, M.; Higgins, S.; Rajwans, N.; Wolofsky, K.T.; Streiner, D.L.; Liles, W.C.; et al. Combinations of host biomarkers predict mortality among Ugandan children with severe malaria: A retrospective case-control study. PLoS ONE 2011, 6, e17440. [Google Scholar] [CrossRef]
- Matejuk, A. Skin Immunity. Arch. Immunol. Ther. Exp. 2017, 66, 45–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsepkolenko, A.; Tsepkolenko, V.; Dash, S.; Mishra, A.; Bader, A.; Melerzanov, A.; Giri, S. The regenerative potential of skin and the immune system. Clin. Cosmet. Investig. Dermatol. 2019, 12, 519–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santiago-Téllez, A.; Castrillón-Rivera, L.E.; Palma-Ramos, A.; Bello-López, J.M.; Sainz-Espuñes, T.; Contreras-Paredes, A.; Luna-Herrera, J.; Castañeda-Sánchez, J.I. Keratinocyte infection by Actinomadura madurae triggers an inflammatory response. Trans. R. Soc. Trop. Med. Hyg. 2019, 113, 392–398. [Google Scholar] [CrossRef]
- Koike, S.; Yamasaki, K. Melanogenesis connection with innate immunity and toll-like receptors. Int. J. Mol. Sci. 2020, 21, 9769. [Google Scholar] [CrossRef]
- Shimada-Omori, R.; Yamasaki, K.; Koike, S.; Yamauchi, T.; Aiba, S. TLR3 augments glucocorticoid-synthetic enzymes expression in epidermal keratinocytes; Implications of glucocorticoid metabolism in rosacea epidermis. J. Dermatol. Sci. 2020, 100, 58–66. [Google Scholar] [CrossRef]
- Altonsy, M.O.; Kurwa, H.A.; Lauzon, G.J.; Amrein, M.; Gerber, A.N.; Almishri, W.; Mydlarski, P.R. Corynebacterium tuberculostearicum, a human skin colonizer, induces the canonical nuclear factor-κB inflammatory signaling pathway in human skin cells. Immunity, Inflamm. Dis. 2020, 8, 62–79. [Google Scholar] [CrossRef] [Green Version]
- Sánchez Luquez, K.Y.; Zerpa, O.; Paz-Villarraga, C.A.; Fernández-Mestre, M. Genetic variability of molecules involved in the disease pathogenesis in Leishmania infection. Exp. Parasitol. 2020, 218, 108007. [Google Scholar] [CrossRef]
- Regli, I.B.; Passelli, K.; Martínez-Salazar, B.; Amore, J.; Hurrell, B.P.; Müller, A.J.; Tacchini-Cottier, F. TLR7 Sensing by Neutrophils Is Critical for the Control of Cutaneous Leishmaniasis. Cell Rep. 2020, 31, 107746. [Google Scholar] [CrossRef] [PubMed]
- Tavares, N.M.; Araújo-Santos, T.; Afonso, L.; Nogueira, P.M.; Lopes, U.G.; Soares, R.P.; Bozza, P.T.; Bandeira-Melo, C.; Borges, V.M.; Brodskyn, C. Understanding the Mechanisms Controlling Leishmania amazonensis Infection In Vitro: The Role of LTB4 Derived From Human Neutrophils. J. Infect. Dis. 2014, 210, 656–666. [Google Scholar] [CrossRef]
- Tessarz, A.S.; Cerwenka, A. The TREM-1/DAP12 pathway. Immunol. Lett. 2008, 116, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Ornatowska, M.; Joo, M.S.; Sadikot, R.T. TREM-1 expression in macrophages is regulated at transcriptional level by NF-κB and PU.1. Eur. J. Immunol. 2007, 37, 2300–2308. [Google Scholar] [CrossRef]
- Ganz, T. Defensins: Antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003, 3, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Arts, R.J.W.; Joosten, L.A.B.; Van Der Meer, J.W.M.; Netea, M.G. TREM-1: Intracellular signaling pathways and interaction with pattern recognition receptors. J. Leukoc. Biol. 2013, 93, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Prado-Montes de Oca, E. Human β-defensin 1: A restless warrior against allergies, infections and cancer. Int. J. Biochem. Cell Biol. 2010, 42, 800–804. [Google Scholar] [CrossRef] [PubMed]
- Wilmes, M.; Sahl, H.G. Defensin-based anti-infective strategies. Int. J. Med. Microbiol. 2014, 304, 93–99. [Google Scholar] [CrossRef] [PubMed]
| Gene | Primer Sequence |
|---|---|
| TREM1 | GAACTCCGAGCTGCAACTAAA (F) TCTAGCGT GTAGTCACATTTCAC (R) |
| TYROBP | ACTGAGACCGAGTCGCCTTAT (F) ATACGGCCTCTGTGTGTTGAG (R) |
| TLR2 | CCTACTGGGTGGAGAACCTTAT (F) CAGGAATGAAGTCCCGCTTATG (R) |
| DEFB4A | CGC CTA TAC CAC CAA AAA CAC (F) TCC TGG TGA AGC TCC CA (R) |
| ACTB | CCT TGC ACA TGC CGG AG (F) ACA GAG CCT CGC CTT TG (R) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nunes, S.; Ampuero, M.R.; Bonyek-Silva, Í.; Lima, R.; Lima, F.R.; Arruda, S.M.; Khouri, R.; Oliveira, P.R.S.; Barral, A.; Boaventura, V.S.; et al. Keratinocytes and Activation of TREM-1 Pathway in Cutaneous Leishmaniasis Lesions. Microbiol. Res. 2021, 12, 765-778. https://doi.org/10.3390/microbiolres12040056
Nunes S, Ampuero MR, Bonyek-Silva Í, Lima R, Lima FR, Arruda SM, Khouri R, Oliveira PRS, Barral A, Boaventura VS, et al. Keratinocytes and Activation of TREM-1 Pathway in Cutaneous Leishmaniasis Lesions. Microbiology Research. 2021; 12(4):765-778. https://doi.org/10.3390/microbiolres12040056
Chicago/Turabian StyleNunes, Sara, Mariana Rosa Ampuero, Ícaro Bonyek-Silva, Reinan Lima, Filipe Rocha Lima, Sérgio Marcos Arruda, Ricardo Khouri, Pablo Rafael Silveira Oliveira, Aldina Barral, Viviane Sampaio Boaventura, and et al. 2021. "Keratinocytes and Activation of TREM-1 Pathway in Cutaneous Leishmaniasis Lesions" Microbiology Research 12, no. 4: 765-778. https://doi.org/10.3390/microbiolres12040056
APA StyleNunes, S., Ampuero, M. R., Bonyek-Silva, Í., Lima, R., Lima, F. R., Arruda, S. M., Khouri, R., Oliveira, P. R. S., Barral, A., Boaventura, V. S., Brodskyn, C. I., & Tavares, N. M. (2021). Keratinocytes and Activation of TREM-1 Pathway in Cutaneous Leishmaniasis Lesions. Microbiology Research, 12(4), 765-778. https://doi.org/10.3390/microbiolres12040056







