Molecular Characterisation and Phylogenetic Analysis of Dermatophytic Fungi Isolated from Tinea Capitis in Northwest Nigeria Using Sequence of the 28S rRNA
Abstract
:Lay Abstract
1. Introduction
2. Materials and Methods
2.1. Study Centre
2.2. Dermatophyte Clinical Strains
2.3. Dermatophyte DNA Extraction and PCR Amplification
2.4. Sequencing
2.5. Nucleotide Blast
2.6. Sequence Analysis
3. Results
4. Discussion
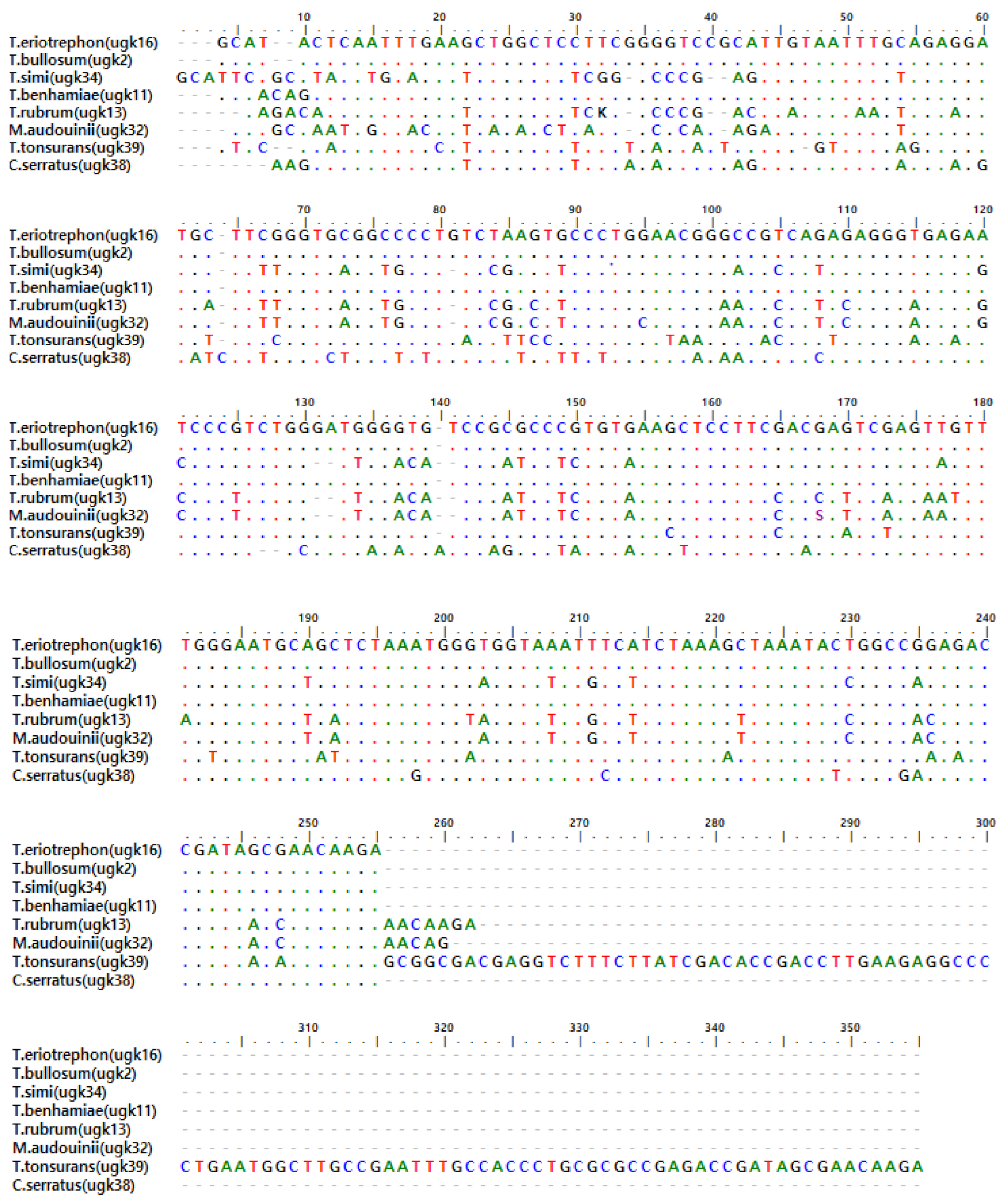
5. Sequences Analysis
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Adeiza, S.S.; Onaolapo, J.A.; Olayinka, B.O. Prospective nucleotide sequence analysis of methicillin resistant Staphylococcus aureus isolates from Sokoto state. Microbiol. Med. 2020, 35. [Google Scholar] [CrossRef]
- Ahmadi, B.; Mirhendi, H.; Makimura, K.; de Hoog, G.S.; Shidfar, M.R.; Nouripour-Sisakht, S.; Jalalizand, N. Phylogenetic analysis of dermatophyte species using DNA sequence polymorphism in calmodulin gene. Sabouraudia 2016, 54, 500–514. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.; Ahmed, Z.; Nasreen, S. Prevalence of tinea capitis and asymptomatic carriage amongst school going children. J. Pak. Assoc. Dermatol. 2017, 16, 215–219. [Google Scholar]
- Ansari, S.; Hedayati, M.T.; Zomorodian, K.; Pakshir, K.; Badali, H.; Rafiei, A.; Ravandeh, M.; Seyedmousavi, S. Molecular characterization and in vitro antifungal susceptibility of 316 clinical isolates of dermatophytes in Iran. Mycopathologia 2016, 181, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Arenas, R.; del Rocío Reyes-Montes, M.; Duarte-Escalante, E.; Frías-De-León, M.G.; Martínez-Herrera, E. Dermatophytes and Dermatophytosis. In Current Progress in Medical Mycology; Springer: New York, NY, USA, 2017; pp. 381–425. [Google Scholar]
- Cai, W.; Lu, C.; Li, X.; Zhang, J.; Zhan, P.; Xi, L.; Sun, J.; Yu, X. Epidemiology of superficial fungal infections in Guangdong, southern China: A retrospective study from 2004 to 2014. Mycopathologia 2016, 181, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Chisanga, M.; Muhamadali, H.; Ellis, D.I.; Goodacre, R. Enhancing disease diagnosis: Biomedical applications of surface-enhanced Raman scattering. Appl. Sci. 2019, 9, 1163. [Google Scholar] [CrossRef] [Green Version]
- Dogo, J.; Afegbua, S.L.; Dung, E.C. Prevalence of Tinea capitis among school children in Nok community of Kaduna state, Nigeria. J. Pathog. 2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, A.K.; Mays, R.R.; Versteeg, S.G.; Shear, N.H.; Piguet, V. Update on current approaches to diagnosis and treatment of onychomycosis. Expert Rev. Anti Infect. Ther. 2018, 16, 929–938. [Google Scholar] [CrossRef] [PubMed]
- Gyimah, N. Social Housing Systems: Perspective of Ghana, Nigeria, United Kingdom, and Netherland. Niger. UK Netherland 2020, 1, 11–12. [Google Scholar] [CrossRef]
- Hay, R.J.; Ashbee, H.R. Fungal infections. In Rook’s Textbook of Dermatology, 9th ed.; John Wiley & Sons: Chichester, UK, 2016; pp. 1–110. [Google Scholar]
- Kim, J.Y.; Choe, Y.B.; Ahn, K.J.; Lee, Y.W. Identification of dermatophytes using multiplex polymerase chain reaction. Ann. Dermatol. 2011, 23, 304–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mochizuki, T.; Takeda, K.; Anzawa, K. Molecular markers useful for intraspecies subtyping and strain differentiation of dermatophytes. Mycopathologia 2017, 182, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Moreno, L.F.; Ahmed, A.A.; Brankovics, B.; Cuomo, C.A.; Menken, S.B.; Taj-Aldeen, S.J.; Faidah, H.; Stielow, J.B.; Teixeira, M.; Prenafeta-Boldú, F.X.; et al. Genomic understanding of an infectious brain disease from the desert. G3 Genes Genomes Genet. 2018, 8, 909–922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moser, S.A.; Wicker, J. Commercial Methods for Identification and Susceptibility Testing of Fungi. In Commercial Methods in Clinical Microbiology, International ed.; John Wiley and Sons: Hoboken, NJ, USA, 2016; pp. 214–271. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1002/9781119021872.ch13 (accessed on 4 March 2021).
- Sacheli, R.; Menatong, X.; Labarbe, C.; Crützen, C.; Harag, S.; André, J.; Evrard, S.; Lagrou, K.; Laffineur, K.; Rousseaux, D. Belgian national survey on tinea capitis: Epidemiology and molecular investigations. Mycoses Suppl. 2019, 476. Available online: https://orbi.uliege.be/bitstream/2268/239981/1/Timm%202019%20Sacheli%20survey%20031019.pdf (accessed on 4 March 2021).
- Sánchez, M.J.I.; Pico, A.M.P.; Tejedor, F.M.; Sánchez, M.J.I.; Acevedo, R.M. Using a polymerase chain reaction as a complementary test to improve the detection of dermatophyte fungus in nails. J. Am. Podiatr. Med Assoc. 2014, 104, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Sen, S.S.; Rasul, E.S. Correspondence-Dermatophytosis in Assam, India. 2006. Available online: https://www.ijmm.org/article.asp?issn=0255-0857;year=2006;volume=24;issue=1;spage=77;epage=78;aulast=Sen (accessed on 4 March 2021).
- Shimoyama, H.; Satoh, K.; Makimura, K.; Sei, Y. Epidemiological survey of onychomycosis pathogens in Japan by real-time PCR. Med Mycol. 2019, 57, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Willinger, B. What Is the Target? Clinical Mycology and Diagnostics. In Clinically Relevant Mycoses; Springer: New York, NY, USA, 2019; pp. 3–24. [Google Scholar]
- Wingfield, D.S.S.; Hald, M.; Arendrup, M.C.; Hjorth, S.V.; Kofoed, K. Darier Disease Complicated by Terbinafine-resistant Trichophyton rubrum: A Case Report. Acta Derm. Venereol. 2017, 97, 139–140. [Google Scholar]

| Accession Number | Organism | Isolate ID (Ugk) |
|---|---|---|
| MT893932–MT893941 | Trichophyton eriotrephon | 1, 7, 10, 14, 16, 19, 20, 22, 31, 40 |
| MT8939342–MT893943 | Trichophyton bullosum | 2, 23 |
| MT893944–MT893955 | Trichophyton simii | 3, 4, 5, 6, 8, 9, 15,2 4, 26, 29, 34, 37 |
| MT893956 | Trichophyton benhamiae | 11 |
| MT893957 | Trichophyton rubrum | 13 |
| MT893958–MT893960 | Trichophyton tonsurans | 33, 35, 39 |
| MT893961–MT893962 | Microsporum audouinii | 30, 32 |
| MT893963 | Ctenomyces serratus | 38 |
| Species (Tested Strain Number) | LS (bp) | Range of Intra-Species Variations | Intra-Species Conserved Region |
|---|---|---|---|
| T. eriotrephon (10) | 247–253 | 0–81 | 221–234 |
| T. bullosum (2) | 248–249 | 41 | 114–137 |
| T. simii (12) | 239–251 | 2–42 | 61–81, 121–155, 194–212 |
| T. benhamiae (1) | 250 | - | - |
| T. rubrum (1) | 248 | - | - |
| T. tonsurans (3) | 247–347 | 13–192 | 17–26,152–161 |
| M. audouinii (2) | 248–251 | 30 | 60–80, 90–157,172–190,193–249 |
| C. serratus (1) | 246 | - | - |
| Seq-> | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ugk1 | ID | |||||||||||||||||||||||||||||||
| 2 | ugk2 | 20 | ID | ||||||||||||||||||||||||||||||
| 3 | ugk3 | 66 | 56 | ID | |||||||||||||||||||||||||||||
| 4 | ugk4 | 66 | 56 | 2 | ID | ||||||||||||||||||||||||||||
| 5 | ugk5 | 71 | 59 | 10 | 10 | ID | |||||||||||||||||||||||||||
| 6 | ugk6 | 66 | 57 | 2 | 2 | 9 | ID | ||||||||||||||||||||||||||
| 7 | ugk7 | 72 | 60 | 89 | 90 | 93 | 89 | ID | |||||||||||||||||||||||||
| 8 | ugk8 | 66 | 57 | 2 | 4 | 9 | 2 | 88 | ID | ||||||||||||||||||||||||
| 9 | ugk9 | 66 | 58 | 6 | 6 | 12 | 8 | 94 | 8 | ID | |||||||||||||||||||||||
| 10 | ugk10 | 20 | 1 | 57 | 57 | 58 | 56 | 59 | 56 | 59 | ID | ||||||||||||||||||||||
| 11 | ugk11 | 22 | 7 | 58 | 58 | 60 | 57 | 62 | 58 | 57 | 7 | ID | |||||||||||||||||||||
| 12 | ugk13 | 94 | 85 | 35 | 37 | 43 | 35 | 91 | 35 | 41 | 84 | 88 | ID | ||||||||||||||||||||
| 13 | ugk14 | 81 | 68 | 91 | 91 | 97 | 90 | 38 | 91 | 96 | 67 | 73 | 100 | ID | |||||||||||||||||||
| 14 | ugk15 | 68 | 59 | 6 | 5 | 6 | 4 | 91 | 5 | 10 | 58 | 59 | 39 | 93 | ID | ||||||||||||||||||
| 15 | ugk16 | 20 | 0 | 56 | 56 | 59 | 57 | 60 | 57 | 58 | 1 | 7 | 85 | 68 | 59 | ID | |||||||||||||||||
| 16 | ugk19 | 23 | 4 | 58 | 58 | 59 | 57 | 62 | 57 | 60 | 3 | 10 | 85 | 69 | 59 | 4 | ID | ||||||||||||||||
| 17 | ugk20 | 21 | 6 | 58 | 58 | 60 | 58 | 60 | 58 | 60 | 6 | 7 | 88 | 71 | 60 | 6 | 9 | ID | |||||||||||||||
| 18 | ugk22 | 20 | 1 | 57 | 57 | 58 | 56 | 59 | 56 | 59 | 0 | 7 | 84 | 67 | 58 | 1 | 3 | 6 | ID | ||||||||||||||
| 19 | ugk23 | 56 | 41 | 75 | 74 | 76 | 75 | 30 | 76 | 78 | 42 | 42 | 91 | 44 | 76 | 41 | 45 | 42 | 42 | ID | |||||||||||||
| 20 | ugk24 | 83 | 74 | 21 | 23 | 29 | 21 | 88 | 21 | 27 | 73 | 75 | 17 | 98 | 25 | 74 | 74 | 75 | 73 | 84 | ID | ||||||||||||
| 21 | ugk26 | 81 | 71 | 17 | 17 | 26 | 19 | 92 | 19 | 21 | 72 | 74 | 25 | 98 | 22 | 71 | 73 | 73 | 72 | 82 | 15 | ID | |||||||||||
| 22 | ugk29 | 90 | 81 | 34 | 33 | 38 | 32 | 91 | 34 | 39 | 80 | 84 | 12 | 95 | 33 | 81 | 81 | 83 | 80 | 87 | 19 | 24 | ID | ||||||||||
| 23 | ugk30 | 94 | 86 | 38 | 40 | 44 | 39 | 99 | 39 | 36 | 86 | 83 | 15 | 109 | 42 | 86 | 87 | 87 | 86 | 93 | 20 | 28 | 21 | ID | |||||||||
| 24 | ugk31 | 24 | 10 | 57 | 56 | 63 | 58 | 56 | 59 | 55 | 11 | 14 | 81 | 64 | 61 | 10 | 14 | 13 | 11 | 38 | 71 | 67 | 77 | 80 | ID | ||||||||
| 25 | ugk32 | 88 | 80 | 42 | 42 | 44 | 41 | 102 | 42 | 44 | 79 | 84 | 34 | 107 | 43 | 80 | 80 | 83 | 79 | 91 | 33 | 37 | 29 | 35 | 79 | ID | |||||||
| 26 | ugk33 | 92 | 83 | 33 | 33 | 39 | 31 | 93 | 33 | 38 | 82 | 82 | 13 | 99 | 34 | 83 | 83 | 84 | 82 | 86 | 16 | 23 | 9 | 19 | 79 | 33 | ID | ||||||
| 27 | ugk34 | 66 | 62 | 12 | 11 | 14 | 12 | 99 | 13 | 7 | 62 | 61 | 46 | 99 | 13 | 62 | 63 | 64 | 62 | 83 | 32 | 27 | 42 | 39 | 61 | 44 | 42 | ID | |||||
| 28 | ugk35 | 92 | 82 | 34 | 34 | 43 | 36 | 94 | 36 | 38 | 83 | 87 | 12 | 101 | 39 | 82 | 84 | 86 | 83 | 88 | 19 | 22 | 13 | 16 | 77 | 32 | 13 | 44 | ID | ||||
| 29 | ugk37 | 95 | 86 | 40 | 42 | 46 | 41 | 99 | 41 | 38 | 86 | 84 | 19 | 108 | 44 | 86 | 87 | 88 | 86 | 92 | 24 | 28 | 23 | 10 | 80 | 37 | 21 | 42 | 16 | ID | |||
| 30 | ugk38 | 62 | 52 | 70 | 69 | 71 | 68 | 88 | 69 | 74 | 51 | 54 | 96 | 89 | 68 | 52 | 53 | 54 | 51 | 73 | 87 | 85 | 93 | 100 | 54 | 98 | 94 | 78 | 95 | 100 | ID | ||
| 31 | ugk39 | 164 | 152 | 187 | 186 | 188 | 185 | 145 | 186 | 191 | 151 | 153 | 190 | 158 | 186 | 152 | 154 | 153 | 151 | 136 | 183 | 186 | 189 | 196 | 149 | 197 | 187 | 195 | 192 | 196 | 176 | ID | |
| 32 | ugk40 | 22 | 5 | 56 | 56 | 60 | 56 | 60 | 57 | 57 | 5 | 9 | 85 | 68 | 58 | 5 | 8 | 9 | 5 | 41 | 73 | 71 | 81 | 84 | 11 | 80 | 80 | 60 | 83 | 84 | 52 | 150 | ID |
| Age (Years) | ||||
|---|---|---|---|---|
| Sex | <5 | >5 (6–10) | Total | Percentage (%) |
| Males | 8 | 22 | 30 | 75 |
| Females | 4 | 6 | 10 | 25 |
| Total | 12 | 28 | 40 | 100 |
| Percentage (%) | 30 | 70 | 100 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ungo-kore, H.Y.; Ehinmidu, J.O.; Onaolapo, J.A.; Olonitola, O.S. Molecular Characterisation and Phylogenetic Analysis of Dermatophytic Fungi Isolated from Tinea Capitis in Northwest Nigeria Using Sequence of the 28S rRNA. Microbiol. Res. 2021, 12, 646-655. https://doi.org/10.3390/microbiolres12030046
Ungo-kore HY, Ehinmidu JO, Onaolapo JA, Olonitola OS. Molecular Characterisation and Phylogenetic Analysis of Dermatophytic Fungi Isolated from Tinea Capitis in Northwest Nigeria Using Sequence of the 28S rRNA. Microbiology Research. 2021; 12(3):646-655. https://doi.org/10.3390/microbiolres12030046
Chicago/Turabian StyleUngo-kore, Hussain Yahaya, Joseph Olorunmola Ehinmidu, Josiah Ademola Onaolapo, and Olayeni Stephen Olonitola. 2021. "Molecular Characterisation and Phylogenetic Analysis of Dermatophytic Fungi Isolated from Tinea Capitis in Northwest Nigeria Using Sequence of the 28S rRNA" Microbiology Research 12, no. 3: 646-655. https://doi.org/10.3390/microbiolres12030046
APA StyleUngo-kore, H. Y., Ehinmidu, J. O., Onaolapo, J. A., & Olonitola, O. S. (2021). Molecular Characterisation and Phylogenetic Analysis of Dermatophytic Fungi Isolated from Tinea Capitis in Northwest Nigeria Using Sequence of the 28S rRNA. Microbiology Research, 12(3), 646-655. https://doi.org/10.3390/microbiolres12030046






