Abstract
Bacteria of the genus Wolbachia are widely distributed in arthropods, particularly in ants; nevertheless, it is still little explored with the Multilocus Sequence Typing (MLST) methodology, especially in the genus Solenopsis, which includes species native to South America. Ants from this genus have species distributed in a cosmopolitan way with some of them being native to South America. In Brazil, they are widely spread and preferentially associated with areas of human activity. This study aimed to investigate the diversity of Wolbachia in ants of the genus Solenopsis through the MLST approach and their phylogenetic relationship, including the relationship between mtDNA from the host and the related Wolbachia strain. We also tested the geographic correlation between the strains to infer transmission and distributional patterns. Fifteen new strains and eleven previously unknown alleles were obtained by sequencing and analyzing the five genes that make up the MLST. The phylogenetic relationship between the strains showed a polyphyletic pattern, indicative of the complexity of the evolutionary history of these bacteria in the analyzed species. We detected the correlation of host’s mitochondrial DNA with Wolbachia diversity which imply that related strains exist in related hosts, strongly suggesting the occurrence of vertical transfer. We found no specificity of the Wolbachia strain for a given geographic region that could indicate either that there is no horizontal transfer of the strain from the environment for the host or that the human action could be shuffling the distribution of the Solenopsis ants and the endosymbiont Wolbachia, as well. Our study highlights the complexity and novelty of Wolbachia diversity with this specific group of ants and the need for further studies that focus on understanding of this intricate relationship.
1. Introduction
Wolbachia (Alphaproteobacteria: Rickettsiales) is an endosymbiont widely distributed in insects, and estimates by Zug and Hammerstein [1] indicate that around 40% of arthropods are infected. Particularly, in ants, this infection rate is around 34% [2], being indicative of the most prevalent heritable symbiont in ants [3]. Its ability to reproductively alter its hosts has made it the subject of many studies in several insect species. However, in ants, its performance in reproductive alterations is not yet widely known, especially because of obstacles in such experimental procedures [2]. Still, few studies have been successful in understanding the effects of Wolbachia on Formicidae, with acceleration of the colony life cycle observed in Monomorium pharaonis and nutritional supplementation of vitamin B detected in Tapinoma melanocephalum in the presence of Wolbachia [4,5].
Studies involving Wolbachia occurrence in ants are numerous (reviewed by References [2,6,7]), but most of them aimed the analysis of the wsp gene (Wolbachia Surface Protein), which, according to Baldo et al. [8], undergoes extensive recombination. Through a more robust and rigorous approach to study the evolution of Wolbachia, the MLST (Multilocus Sequence Typing) developed by Baldo et al. [9] had several studies with Formicidae [7,9,10,11,12,13,14]; however, studies in specific ant genera are still restricted.
The genus Solenopsis Westwood, 1840, despite being highly diverse, having about 194 valid species [15], is famous mainly for the species S. saevissima and S. invicta known as fire ants, highly distributed in South America, where they are considered native [16]. Several studies have already been carried out examining the distribution and prevalence of Wolbachia infections using the wsp methodology in native populations of the species Solenopsis invicta, Buren, and other species of the genus, as well as the effects of Wolbachia in the variation of mitochondrial DNA in these species [17,18,19,20,21,22,23]. Previous research focusing on wsp genes from Solenopsis populations from Brazil documented high frequency of Wolbachia infection with 51% of the nest infected and several multi-infected nests [20]. In Russell’s [2] compilation about publications of PCR screening from Wolbachia, he indicated that Solenopsis occupies the third position in prevalence of Wolbachia in Formicidae, with around 60% of frequency. Despite this high frequency, we still have a very limited view of the phylogeny and diversity of Wolbachia in Solenopsis, with all the studies in the genus carried out so far based on the wsp gene.
Baldo et al. [9] was the first study to characterize strains of Wolbachia in Solenopsis using the MLST methodology and identified the ST29 in S. invicta, although the focus of the study was not on the genus or species. In addition, to our knowledge there is no further investigations regarding Wolbachia diversity with a focus exclusively on native Solenopsis using MLST approach, which highlights the need to deepen studies with a more precise methodology to reconstruct the evolution and review the diversity of Wolbachia strains in Solenopsis species. Although genomic approches are more robust and suitable to undertand Wolbachia dynamics and the real strain diversity (see References [24,25,26]), the difficulty of accessing many genomes due to the high costs makes MLST a more accessible method for initial surveys of specific groups of host and larger samples. Furthermore, the number of Wolbachia ant genomes are still restricted [26,27], and the MLST has an extensive database for initial comparative analyses.
Given this scenario, the goals of this study were: (a) to investigate the diversity of Wolbachia strains associated with the Solenopsis ant genus from South America through Multilocus Sequence Typing (MLST) analysis; (b) to test the hypothesis of the relationship between the host’s DNAmt and the associated Wolbachia strain; and (c) to assess whether there is geographical correlation of the host and strains of Wolbachia in order to infer patterns of transmission and distribution. This is the first study that took into account samples of Solenopsis in its native area, and the data of this symbiotic interaction are interesting from an evolutionary point of view.
2. Materials and Methods
2.1. Ant Samples and Wolbachia Isolation
One hundred and fourteen nest samples from native populations of Solenopsis were used, from which 33 had previously shown presence of Wolbachia through the sequencing of the wsp gene [20]. Additionally, five samples of S. invicta nests previously analyzed by Souza et al. [23] were added to our analysis. It is important to clarify that these samples did not show multiple infections through the previous analysis of the wsp gene, since the premise of single infection is necessary for the application of the MLST methodology according to Baldo et al. [9]. The ant sample used were collected directly from the nests and kept in ethanol and frozen until DNA extraction that was done using the whole ant body (samples from Reference [20] were pooled with five individuals per nest). For the wsp gene amplification, we used the control primer EF1α-532F (5′-AGGCAAATGTCTTATTGAAG-3′) and EF1α-610R (5′-GCGGGTGCGAAGGTAACAAC-3′) [22] and wsp81F (5′-TGGTCCATTAAGTGATGAAGAAAC-3′) and wsp691R (5′-AAAAATTAAACGCTACTCCA-3′) [28]. The first two sets amplify a fragment of 400 bp of the nuclear gene EF1α (elongation factor), used as a control for the reaction, and the presence of its product and the absence of wsp fragment most likely reflects the absence of the bacteria [22]. In cases with absence of EF1α and wsp gene fragments, the genomic DNA was diluted, and the PCR protocol repeated.
Lastly, these 38 nest samples were analyzed for the strain of the bacterium present using the MLST methodology following the standard protocol of Baldo et al. [9], where five genes of MLST approach were amplified and sequenced (coxA, gatB, hcpA, ftsZ, and fbpA). Table 1 details the collection locations and species. The morphological identification of ants was based on Pitts [29] and Trager [30].

Table 1.
Wolbachia associated with Solenopsis analyzed with the respective nest collection codes, geographic coordinates, and definition of the wsp for each sample, as well as the COI, Sequence type (ST), and the corresponding alleles. * indicates new alleles or STs found in the present study. wsp: Wolbachia surface protein gene; HVR: hypervariable region of the wsp gene.id: identification provided for each submission. ST: sequence type (equivalent to a strain or a haplotype). COI: cytochrome oxidase I. gatB: Glutamyl-tRNA(Gln) amidotransferase, subunit B. coxA: Cytochrome coxidase, subunit I. hcpA: Conserved hypothetical protein. ftsZ: Cell division protein. fbpA: Fructose-bisphosphate aldolase. na = does not apply. p = partial definition of wsp. For more details on location, see Figure 2.
For MLST amplification, a final reaction volume of 25 μL was used, about 1.5 μL of genomic DNA, 1X buffer, 0.2 mM dNTPs, 1.5 mM MgCl2, 2.5 μL of each primer with dilution 1 μM, and 0.5U of Taq DNA polymerase (Invitrogen, Waltham, MA USA). The thermal cycler was programmed following the one suggested by the Wolbachia MLST Database (http://pubmlst.org/wolbachia/ (accessed on 28 June 2021) [31]: 94 °C for 2 min, 37 cycles of 94 °C for 30 s, specific annealing temperature for each primer combination for 45 s, 72 °C for 1.5 min, and a final cycle of 72 °C for 10 min. The confirmation of the amplifications was visualized on a 1% agarose gel. In some cases, it was necessary to use alternative primers, which are indicated for individuals with double infections (supergroups A and B) by Baldo et al. [9] and described in the Wolbachia MLST Database. In cases where the existing primer combinations did not produce positive PCR reactions, the nested PCR was used, with the template DNA being the dilution of the product of the first PCR in the proportion of 1:9.
In cases where the existing primer combinations for the ftsZ gene generated as a sequencing product a fragment with a size smaller than necessary for the application of the MLST approach, a new primer combination was designed using Gene Runner Version 3.05 (Hastings Software Inc. Hastings, NY, USA) and NCBI Primer-BLAST tool [32]. The new primer pair designed (ftsZnewF 5′- CATATGCTTTTCATTACAGCAGGAATGGGC—3′, and ftsZnewR 5′- CGCAGCTTCCGCAGCACTAA—3′) amplifies a region of the ftsZ gene of about 480 bp with annealing temperature of 60 °C.
2.2. DNA Sequencing
To purify the amplified products the GFXTM PCR DNA and Gel Band Purification Kit (GE Healthcare) was used following the manufacturer’s recommendations. The result of the purification was quantified in NanoDrop® 2000 (Thermo Scientific, Waltham, MA, USA). The sequencing reactions were performed with BigDye Terminator chemistry (Applied Biosystem Inc., Waltham, MA, USA) and sequenced in a 3130 Genetic Analyzer automatic sequencer (Applied Biosystem Inc., Waltham, MA, USA).
2.3. Sequence Editing, Allele Identification and Sequence Typing
The sequences obtained from the five MLST genes were initially analyzed separately using the BioEdit program [33], aligned using the ClustalW application [34], and manually edited. Allele identification and sequence typing (the definition of a sequence type or ST by the MLST approach [9] is equivalent to a strain or a haplotype) was performed on the Wolbachia MLST Database. In cases where the allele was not registered in the database, the new allele found was deposited according to the rules of the Wolbachia MLST Database. The same was done to register new STs. The records for each allele and ST in the Wolbachia MLST Database, as well as the access code for each COI (from Martins et al. [35]), are shown in Table 1.
2.4. Phylogenetic Reconstruction and Mantel Test
The MLST concatenated data (2079 bp) generated in the present study, together with data from the Wolbachia MLST Database related to Formicidae deposited until the moment of the analysis, were used for partitioned phylogenetic reconstruction. PartitionFinder2 (2.1.1) [36] was used to choose the best model of molecular evolution that returned GTR + G. Bayesian inference was carried out by MrBayes (3.2.6) on the Cipres Portal [37] for phylogenetic reconstruction with the Markov chain Monte Carlo analysis for 1,000,000 generations with sampling every 1000 generations and discarding the first 25% of trees as burnin. ST78 was used as an outgroup because Opistophthalmus chaperi (supergroup F) strain shows similarity with strains associated with ants but has not been found associated with ants [38].
Genetic distance analyzes in pairs were performed with the host’s COI data and the concatenated data of the five genes of the endosymbiont sequenced in this work using the R software [39] http://www.R-project.org (accessed on 16 October 2017) with APE package [40], using k80 distance method [41]. A geographic distance matrix paired between the samples was calculated with the data obtained by GPS transformed into UTM, and the distance calculated through euclidean distance, using R software with rgdal package [42]. To test the correlation, Mantel test was performed with R software with vegan package [43]. Geographic maps were reconstructed with the geographical coordinates of the collection points through the speciesMapper website [44], and the corresponding strains in each location were indicated on the maps.
3. Results
All samples of Solenopsis spp. infected with Wolbachia (n = 38) were successfully sequenced in at least one of the MLST genes. Table 1 summarizes the results obtained in addition to other data generated in previous studies (the wsp gene and the COI gene) that were added for analysis in the present study [20,23,35].
Of the 38 populations analyzed, 10 showed evidence of multiple infection after analyzing the sequences of MLST (samples E1713, E1725, E1726, E1727, E1739, E1746, E1789 in the ftsZ gene, E1645 in the fbpA gene, E1646 in the gatB gene, and Sol48 in the coxA and fbpA genes). It is important to note that the previous sequencing of the wsp gene [20] did not detect these multiple infections, since the electropherogram peaks were clearly unique to this gene, and these were only detected with the sequencing of the MLST genes, which indicates the ineffectiveness of using a single gene.
Several genes in the samples with multiple infections were successfully sequenced and had their alleles determined, but, in some cases, it was impossible to separate the correct combination of the genes of MLST for each sample. In a single sample, it was possible to detect the two strains because a single allele was variable, and the grouping of the five genes was possible (sample E1645, variation only in the fbpA gene, with 60 variable nucleotides, or 14%, corresponding to ST324 and ST325). For sample E1646, it was possible to sequence only four of the five alleles of MLST, and fbpA showed two different alleles that were separated using the alternative primers that are indicated for individuals with double infections (supergroups A and B) by Baldo et al. [9] and described in the Wolbachia MLST Database. gatB, however, was not determined because its sequencing resulted in several double peaks in eletropherogram, and it was not successfully separated with alternative primers. It should be noted that, in the case of sample E1646, even if success was achieved in sequencing the gatB genes, the combination of alleles would be undetermined, since there would be variation in more than one allele.
Among these alleles, a total of 11 new ones of MLST genes were found in the analyzed Solenopsis samples. An unprecedented allele was found for gatB (allele 196), two alleles for coxA (183 and 184), hcpA three alleles (207, 208 and 210), fbpA four alleles (251, 252, 253 and 254), and ftsZ one allele (160). Table 1 illustrates these alleles with an asterisk and in which samples they were found. These new alleles identified could be ant-specific because they were not found in groups other than ants.
Incomplete profiles were obtained for samples E1818, E1822 due to low yield in amplification reactions, and in samples E1725, E1726, E1727, Sol48, E1739, and E1646 due to the presence of multiple infection and impossibility of separation, despite the use of alternative primers for multiple infections.
The definitions of the sequence type (STs) through the Wolbachia MLST database were performed on 30 samples that had all five Wolbachia MLST genes sequenced, and the results are shown in Table 1. The characterizations of the allelic profiles revealed the presence of 16 STs in the samples of this study, of which 15 were new and registered and deposited in the Wolbachia MLST Database (ST314 to ST328 indicated with an asterisk in Table 1); ST 29, previously registered by Baldo et al. [9], was frequent in the samples analyzed in the present study. The “id” numbers of the samples studied here are 515 to 554, shown in Table 1.
The phylogenetic inference of the Wolbachia associated with Solenopsis show that the endosymbionts are distributed in a very diffuse way in the phylogenetic tree clustering with unrelated species and were not grouped in the same ant species and not even as to the geographic location (Figure 1 and Mantel Test results). It is important to highlight that, although most of the tips are grouped in a clade with polytomy, it is evident the Wolbachia grouping in New World and Old World Formicidae in the nested clades.
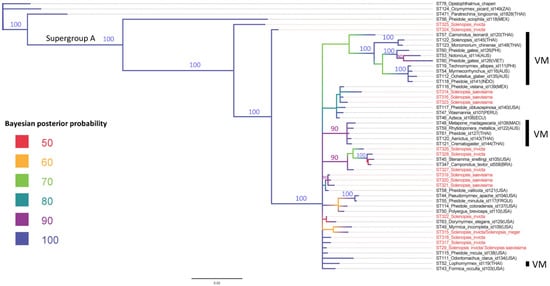
Figure 1.
Bayesian inference of strains of Wolbachia in Formicidae. Note that most of them belong to supergroup A, with the exceptions of ST124 and ST471, which are classified in supergroup F, ST56 is are classified in supergroup B, and ST78 as outgroup (ST78). The terminals marked in red correspond to the STs detected in the present study. VM = Old World. The subsequent Bayesian posterior probability values are indicated according to the colors of the branches. However, numbers equivalent to high Bayesian posterior probability support (100 and 90) were also added to the nodes. Lower numbers have not been added for ease of viewing.
In a deeper analysis regarding the correlation between the host’s mitochondrial DNA (COI gene) and Wolbachia diversity, our results indicated a significant correlation (Mantel test, r2 = 0.4476, p = 0.00005), which suggests that related strains exist in related hosts, a strong indication of the occurrence of vertical transfer. However, there was no significant correlation (Mantel test, r2 = 0.02175, p = 0.27281) between the geographic location of the host and the associated Wolbachia diversity. Figure 2 illustrates the location of each population and its respective strain.
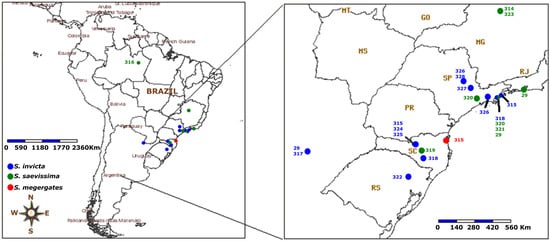
Figure 2.
Map illustrating the geographical location of each strain and the associated ant species. The numbers indicate the STs, the coloring is referring to the ant species. Source: speciesMapper [44].
4. Discussion
The present study sought to understand the complexity of the symbiotic interaction between Solenopsis species present in their native region and the associated Wolbachia diversity. Our data emphasizes the complexity and novelty of these interactions and being able to identify multiple infections in some samples (n = 10), and the detection of 16 strains, 15 of which are new, in addition to 11 new alleles, highlighting the ineffectiveness of the wsp to define the diversity of Wolbachia strains. The divergence between MLST and wsp anlysis was also found by Baldo et al. [45], supporting the conclusion that wsp alone is not indicated to predict the diversity of strains or divergence.
Some samples (n = 10) analyzed were determined to be multi-infected by detecting multiple peaks on the electropherogram, making it impossible in most of these samples to define the strains present by the multilocus approach. The exception was one sample in which it was possible to identify two close strains in the same nest. Cases of multiple infections have already been described in other studies with ants, such as Formica exsecta [46], Acromyrmex [47], Solenopsis invicta [20,48], Solenopsis saevissima [19], Solenopsis daguerrei [18], Formica rufa species group [49], Camponotus [12,50], Polyrhachis [51], and in Cephalotes atratus [11,13], therefore being relatively frequent.
Focusing on some specific strains, we must first highlight the presence of strains ST325 and ST324, both identified in Solenopsis invicta (Figure 1 and Figure 2). These strains of Wolbachia have ancestral characteristics of Supergroup A not only within the host Solenopsis but within the family Formicidae [38]. Another strain that deserves further investigation is the strain ST29 that has been observed several times in Solenopsis species in the native region (35% of our S. invicta samples and 36% of S. saevissima), and in several locations (n = 3) (Table 1 and Figure 2). In addition, considering that the ST29 is related to several COI haplotypes (Table 1), this may be an indication that the infection is old and already fixed in the analyzed populations, differing from what was observed by Zhang et al. [52], where two strains related to the same host COI haplotype are indicative of being a recent infection and in the process of spreading. Supporting our hypothesis, it is emphasized that most species of Solenopsis are native to the Neotropical region, with South America having a high diversity of species [29], and the southern portion of South America is likely the region of origin of the genus and coincidentally a region where the analyzed nests had, for the most part, infection by the ST29 strain. Therefore, if the origin of the genus is pointed to this region, the origin of the Supergroup A of Wolbachia present in Solenopsis [38] could indicate association with this region and may have diverged together with the genus. However, this hypothesis needs to be tested, especially with other genes or genome analyzes that would give more support to answer these questions and even more collecting locations and Solenopsis species.
Our data also recovered some strains shared by different species (Table 1, Figure 1 and Figure 2), that is, ST29 occurring in S. invicta and S. saevissima, and ST315 occurring in S. invicta and S. megergates. This scenario shows the possibility that these strains were acquired vertically in the common ancestor of these species. Results like these highlight the complexity of the symbiotic interactions that Solenopsis spp. engages with Wolbachia being highly diverse.
The phylogenetic inference of the Wolbachia distribution associated with Formicidae indicates the complex evolutionary history of this symbiotic interaction, with special emphasis on the Solenopsis STs identified in the present study. The Wolbachia associated with Formicidae did not group on the ant species, not even as to the geographic location of the nest (as evidenced in the Mantel Test), and is distributed in a very diffuse way in the phylogenetic tree, forming clusters with unrelated species. This lack of codivergence was already documented in several studies [11,12,14,38].
The lack of geographic correlation between the host and the strain found shown by the Mantel test is an indicative that there was no specificity of the Wolbachia strain for a given geographic region, or it may mean that: (1) there is no horizontal transfer of the strain of the environment for the host, or even that (2) the human action in modifying the environment and consequently the distribution of the Solenopsis populations shuffles the distribution of the endosymbiont Wolbachia, as well. Absences of correlation of this type have already been found in ants from Thailand [14], Linepthema micans from southern Brazil [53], in Hylyphantes graminicola spiders [54], and in spiders of the genus Agelenopsis and the species A. aperta [55]. The increased mobility of the host through human transport could be “mixing” Solenopsis colonies, and, consequently, Wolbachia strains in non-original geographical locations, that may be the cause of the lack of correlation between Wolbachia and the geographic location of populations analyzed. Supporting this hypothesis, there are data that suggest that Solenopsis populations are being moved by human activity in Brazil at a much higher frequency than previously suggested [35,56]. Another recent study that approached Wolbachia through the MLST methodology focused on the species of Cephalotes atratus and found a correlation between the diversity of strains with the geographic location of the host [11]. This could suggest that Cephalotes atratus, unlike Solenopsis spp., is not being impacted by human transport.
Despite the absence of correlation between geographic locations and strains of Wolbachia, in nests with sympatric occurrence, a tendency for similar or relatively close strains of Wolbachia to occur was observed (Table 1). Most nests in Corrientes (Argentina) have the same strain (ST29), with only one nest with a different strain (ST317), with variation only in the ftsZ gene compared to ST29. The same fact occurred with samples from Rio de Janeiro, RJ that also share the strains (ST29). Likewise, samples from Buritizeiro, MG have strains close to ST323 and ST314, with variation in the coxA and ftsZ genes. These data support the hypothesis of movement of propagules of the nests and, consequently, of the strains by human activity, as there are nearby nests that have related strains, being an indication that the nearby strains would occur in approximate geographical locations. In addition, the occurrence of related strains occurring in adjacent nests (which also have the same COI) constitutes evidence of vertical transfer of the strains, since related strains are occurring in related COI.
Therefore, it is assumed that the most frequent mode of transmission is the vertical, since Wolbachia is transmitted to offspring through the egg cytoplasm [57,58] and also because related hosts have related strains [11,14,55,59]. However, evidence obtained through the wsp gene and mitochondrial DNA patterns indicate that, in S. invicta, the evolutionary history of Wolbachia seems to be more complex, involving multiple invasions or horizontal transmission events [48]. Although horizontal transmission is not a common event in ants and appears to occasionally occur in related hosts [14,59], this complex history of evolutionary history, with both vertical and horizontal transmissions have recently been suggested in other species, using the multilocus approach, as, for example, in Baldo et al. [55]; Frost et al. [10]; Raychoudhury et al. [60]; Watanabe et al. [61]; and Zhang et al. [62], showing the complexity of the Wolbachia—host relationship and the need for further studies that lead to an understanding of this complicated relationship.
5. Conclusions
Our results highlight the complexity and novelty of Wolbachia diversity with one specific group of ants and unravel an unprecedent variety of 15 new strains detected and eleven previously unknown alleles using MLST approach. We found a polyphyletic pattern which we believe is an indicative of the generalized capacity of Wolbachia to infect this ant species. But we also found some related strains associated with related hosts which were indicative of vertical transfer and that the human activities could be shuffling the distribution pattern of the Solenopsis ants and the endosymbiont Wolbachia, as well. We believe that further studies should focus in specific groups of ants to help elucidate the complex history and evolutionary relationship of Wolbachia with ants. Moreover, given the recent studies on Wolbachia genome and the low resolution on MLST approach [24,25], new approaches, such as comparative genome studies, should change substantially our knowledge of this complex history and help understand the real strain diversity and the evolutionary dynamics of Wolbachia and its host.
Author Contributions
Conceptualization, C.M. and O.C.B.; formal analysis, C.M., M.d.O.R.; writing—original draft preparation, C.M., M.d.O.R., L.M.R.S., R.F.d.S.; writing—review and editing, C.M., M.d.O.R., L.M.R.S., R.F.d.S., O.C.B.; visualization, C.M., M.d.O.R.; supervision, O.C.B.; project administration, C.M.; funding acquisition, O.C.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Council for Scientific and Technological Development, FAPEPI (The Piauí Research Foundation), CAPES Foundation (process no. 007343/2014-00 for M.d.O.R.), and the Ministry of Education of Brazil.
Institutional Review Board Statement
This research was registered at the National System of Genetic Resource Management and Associated Traditional Knowledge (SisGen) under the number A3E46A2.
Data Availability Statement
The data is available at Wolbachia MLST Database (https://pubmlst.org/organisms/wolbachia-spp) under ids 515-554.
Acknowledgments
C.M. would like to give thanks to CNPq (National Council for Scientific and Technological Development) and FAPEPI (The Piauí Research Foundation) for their financial support. M.d.O.R. thanks CAPES Foundation and the Ministry of Education of Brazil for their financial support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zug, R.; Hammerstein, P. Still a host of hosts for Wolbachia: Analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS ONE 2012, 7, e38544. [Google Scholar] [CrossRef]
- Russell, J.A. The ants (Hymenoptera: Formicidae) are unique and enigmatic hosts of prevalent Wolbachia (Alphaproteobacteria) symbionts. Myrmecol. News 2012, 16, 7–23. [Google Scholar]
- Russell, J.A.; Funaro, C.F.; Giraldo, Y.M.; Goldman-Huertas, B.; Suh, D.; Kronauer, D.J.C.; Moreau, C.S.; Pierce, N.E. A veritable menagerie of heritable bacteria from ants, butterflies, and beyond: Broad molecular surveys and a systematic review. PLoS ONE 2012, 7, e51027. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Chen, S.; Huang, Y.; Pierce, N.E.; Riegler, M.; Yang, F.; Zeng, L.; Lu, Y.; Liang, G.; Xu, Y. Symbiotic microbiota may reflect host adaptation by resident to invasive ant species. PLoS Pathog. 2019, 15, e1007942. [Google Scholar] [CrossRef]
- Singh, R.; Linksvayer, T.A. Wolbachia-infected ant colonies have increased reproductive investment and an accelerated life cycle. J. Exp. Biol. 2020, 223, jeb220079. [Google Scholar] [CrossRef]
- Kautz, S.; Rubin, B.E.R.; Moreau, C.S. Bacterial infections across the ants: Frequency and prevalence of Wolbachia, Spiroplasma, and Asaia. Psyche (Stuttg.) 2013. [Google Scholar] [CrossRef]
- Ramalho, M.O.; Kim, Z.; Wang, S.; Moreau, C.S. Wolbachia across social insects: Patterns and implications. Ann. Entomol. Soc. Am. 2021. [Google Scholar] [CrossRef]
- Baldo, L.; Lo, N.; Werren, J.H. Mosaic nature of the Wolbachia surface protein. J. Bacteriol. 2005, 187, 5406–5418. [Google Scholar] [CrossRef] [PubMed]
- Baldo, L.; Dunning Hotopp, J.C.; Jolley, K.A.; Bordenstein, S.R.; Biber, S.A.; Choudhury, R.R.; Hayashi, C.; Maiden, M.C.J.; Tettelin, H.; Werren, J.H. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl. Environ. Microbiol. 2006, 72, 7098–7110. [Google Scholar] [CrossRef]
- Frost, C.L.; FernÁndez-MarÍn, H.; Smith, J.E.; Hughes, W.O.H. Multiple gains and losses of Wolbachia symbionts across a tribe of fungus-growing ants. Mol. Ecol. 2010, 19, 4077–4085. [Google Scholar] [CrossRef]
- Kelly, M.; Price, S.L.; Ramalho, M.O.; Moreau, C.S. Diversity of Wolbachia associated with the giant turtle ant, Cephalotes atratus. Curr. Microbiol. 2019, 76, 1330–1337. [Google Scholar] [CrossRef]
- Ramalho, M.O.; Martins, C.; Silva, L.M.R.; Martins, V.G.; Bueno, O.C. Intracellular symbiotic bacteria of Camponotus textor, Forel (Hymenoptera, Formicidae). Curr. Microbiol. 2017, 74, 589–597. [Google Scholar] [CrossRef]
- Reeves, D.D.; Price, S.L.; Ramalho, M.O.; Moreau, C.S. The diversity and distribution of Wolbachia, Rhizobiales, and Ophiocordyceps within the widespread neotropical turtle ant, Cephalotes atratus (Hymenoptera: Formicidae). Neotrop. Entomol. 2020, 49, 52–60. [Google Scholar] [CrossRef]
- Russell, J.A.; Goldman-Huertas, B.; Moreau, C.S.; Baldo, L.; Stahlhut, J.K.; Werren, J.H.; Pierce, N.E. Specialization and geographic isolation among Wolbachia symbionts from ants and lycaenid butterflies. Evolution (N. Y.) 2009, 63, 624–640. [Google Scholar] [CrossRef]
- Bolton, B. An Online Catalog of the Ants of the World. Available online: http://antcat.org (accessed on 3 April 2021).
- Pitts, J.P.; Camacho, G.P.; Gotzek, D.; Mchugh, J.V.; Ross, K.G. Revision of the fire ants of the Solenopsis saevissima species-group (Hymenoptera: Formicidae). Proc. Entomol. Soc. Wash. 2018, 120, 308–411. [Google Scholar] [CrossRef]
- Ahrens, M.E.; Ross, K.G.; Shoemaker, D.D. Phylogenographic structure of the fire ant Solenopsis invicta in its native South American range: Roles of natural barriers and habitat connectivity. Evolution (N. Y.) 2005, 59, 1733–1743. [Google Scholar] [CrossRef]
- Dedeine, F.; Ahrens, M.; Calcaterra, L.; Shoemaker, D.D. Social parasitism in fire ants (Solenopsis spp.): A potential mechanism for interspecies transfer of Wolbachia. Mol. Ecol. 2005, 14, 1543–1548. [Google Scholar] [CrossRef] [PubMed]
- Souza, R.F.; Ramalho, J.D.S.; Morini, M.S.C.; Wolff, J.L.C.; Araújo, R.C.; Mascara, D. Identification and characterization of Wolbachia in Solenopsis saevissima fire ants (Hymenoptera: Formicidae) in southeastern Brazil. Curr. Microbiol. 2009, 58, 189–194. [Google Scholar] [CrossRef]
- Martins, C.; Souza, R.F.; Bueno, O.C. Presence and distribution of the endosymbiont Wolbachia among Solenopsis spp. (Hymenoptera: Formicidae) from Brazil and its evolutionary history. J. Invertebr. Pathol. 2012, 109, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, D.D.; Ahrens, M.; Sheill, L.; Mescher, M.; Keller, L.; Ross, K.G. Distribution and prevalence of Wolbachia infections in native populations of the fire ant Solenopsis invicta (Hymenoptera: Formicidae). Environ. Entomol. 2003, 32, 1329–1336. [Google Scholar] [CrossRef][Green Version]
- Shoemaker, D.D.; Ross, K.G.; Keller, L.; Vargo, E.L.; Werren, J.H. Wolbachia infections in native and introduced populations of fire ants (Solenopsis spp.). Insect Mol. Biol. 2000, 9, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Souza, R.F.; Martins, C.; Pereira, R.; Bueno, O.C. Analysis of the hypervariable regions (HVRs) of the wsp gene of Wolbachia from Solenopsis invicta ants in southeastern Brazil. Adv. Entomol. 2014, 2, 135–143. [Google Scholar] [CrossRef][Green Version]
- Bleidorn, C.; Gerth, M. A critical re-evaluation of multilocus sequence typing (MLST) efforts in Wolbachia. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, T.M.; Bruzzese, D.J.; Klasson, L.; Corretto, E.; Lečić, S.; Stauffer, C.; Feder, J.L.; Schuler, H. Comparative genome sequencing reveals insights into the dynamics of Wolbachia in native and invasive cherry fruit flies. Mol. Ecol. 2021. [Google Scholar] [CrossRef]
- Scholz, M.; Albanese, D.; Tuohy, K.; Donati, C.; Segata, N.; Rota-Stabelli, O. Large scale genome reconstructions illuminate Wolbachia evolution. Nat. Commun. 2020, 11, 5235. [Google Scholar] [CrossRef] [PubMed]
- Dhaygude, K.; Nair, A.; Johansson, H.; Wurm, Y.; Sundström, L. The first draft genomes of the ant Formica exsecta, and its Wolbachia endosymbiont reveal extensive gene transfer from endosymbiont to host. BMC Genom. 2019, 20, 301. [Google Scholar] [CrossRef] [PubMed]
- Braig, H.R.; Zhou, W.; Dobson, S.L.; O’Neill, S.L. Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont Wolbachia pipientis. J. Bacteriol. 1998, 180, 2373–2378. [Google Scholar] [CrossRef]
- Pitts, J.P.; McHugh, J.V.; Ross, K.G. Cladistic analysis of the fire ants of the Solenopsis saevissima species-group (Hymenoptera: Formicidae). Zool. Scr. 2005, 34, 493–505. [Google Scholar] [CrossRef]
- Trager, J.C. A Revision of the fire ants, Solenopsis geminata group (Hymenoptera: Formicidae: Myrmicinae). J. N. Y. Entomol. Soc. 1991, 99, 141–198. [Google Scholar]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Higgins, D.G.; Bleasby, A.J.; Fuchs, R. Clustal V: Improved software for multiple sequence alignment. Comput. Appl. Biosci. 1992, 8, 189–191. [Google Scholar] [CrossRef]
- Martins, C.; Souza, R.F.; Bueno, O.C. Molecular characterization of fire ants, Solenopsis spp., from Brazil based on analysis of mtDNA gene cytochrome oxidase I. J. Insect Sci. 2014, 14, 50. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. The CIPRES Portals. Available online: http://www.phylo.org/ (accessed on 15 January 2020).
- Ramalho, M.O.; Moreau, C.S. The evolution and biogeography of Wolbachia in ants (Hymenoptera: Formicidae). Diversity 2020, 12, 426. [Google Scholar] [CrossRef]
- R Development Core Team R: A Language and Environment for Statistical Computing. 2017. Available online: https://www.R-project.org/ (accessed on 16 October 2017).
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Bivand, R.; Keitt, T.; Rowlingson, B. rgdal: Bindings for the Geospatial Data Abstraction Library. 2013. Available online: https://CRAN.R-project.org/package=rgdal (accessed on 16 October 2017).
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. vegan: Community Ecology Package. R Package Version 2.4-5 2017. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 16 October 2017).
- Species Mapper. Available online: http://splink.cria.org.br/mapper?criaLANG=en (accessed on 5 May 2017).
- Baldo, L.; Bordenstein, S.; Wernegreen, J.J.; Werren, J.H. Widespread recombination throughout Wolbachia genomes. Mol. Biol. Evol. 2006, 23, 437–449. [Google Scholar] [CrossRef]
- Reuter, M.; Keller, L. High levels of multiple Wolbachia infection and recombination in the ant Formica exsecta. Mol. Biol. Evol. 2003, 20, 748–753. [Google Scholar] [CrossRef][Green Version]
- Van Borm, S.; Wenseleers, T.; Billen, J.; Boomsma, J.J. Cloning and sequencing of wsp encoding gene fragments reveals a diversity of co-infecting Wolbachia strains in Acromyrmex leafcutter ants. Mol. Phylogenet. Evol. 2003, 26, 102–109. [Google Scholar] [CrossRef]
- Ahrens, M.E.; Shoemaker, D. Evolutionary history of Wolbachia infections in the fire ant Solenopsis invicta. BMC Evol. Biol. 2005, 5, 35. [Google Scholar] [CrossRef][Green Version]
- Viljakainen, L.; Reuter, M.; Pamilo, P. Wolbachia transmission dynamics in Formica wood ants. BMC Evol. Biol. 2008, 8, 55. [Google Scholar] [CrossRef]
- Ramalho, M.O.; Bueno, O.C.; Moreau, C.S. Species-specific signatures of the microbiome from Camponotus and Colobopsis ants across developmental stages. PLoS ONE 2017, 12, e0187461. [Google Scholar] [CrossRef]
- Ramalho, M.O.; Bueno, O.C.; Moreau, C.S. Microbial composition of spiny ants (Hymenoptera: Formicidae: Polyrhachis) across their geographic range. BMC Evol. Biol. 2017. [Google Scholar] [CrossRef]
- Zhang, Y.-K.; Zhang, K.-J.; Sun, J.-T.; Yang, X.-M.; Ge, C.; Hong, X.-Y. Diversity of Wolbachia in natural populations of spider mites (genus Tetranychus): Evidence for complex infection history and disequilibrium distribution. Microb. Ecol. 2013, 65, 731–739. [Google Scholar] [CrossRef]
- Ramalho, M.O.; Martins, C.; Campos, T.; Nondillo, A.; Botton, M.; Bueno, O.C. Occurrence of 15 haplotypes of Linepithema micans (Hymenoptera: Formicidae) in southern Brazil. J. Econ. Entomol. 2017, 110, 1841–1846. [Google Scholar] [CrossRef]
- Yun, Y.; Lei, C.; Peng, Y.; Liu, F.; Chen, J.; Chen, L. Wolbachia strains typing in different geographic population spider, Hylyphantes graminicola (Linyphiidae). Curr. Microbiol. 2011, 62, 139–145. [Google Scholar] [CrossRef]
- Baldo, L.; Ayoub, N.A.; Hayashi, C.Y.; Russell, J.A.; Stahlhut, J.K.; Werren, J.H. Insight into the routes of Wolbachia invasion: High levels of horizontal transfer in the spider genus Agelenopsis revealed by Wolbachia strain and mitochondrial DNA diversity. Mol. Ecol. 2008, 17, 557–569. [Google Scholar] [CrossRef]
- Souza, R.F. Aspectos Bioecológicos e Genéticos de Solenopsis spp. No Mosaico de Paisagens do Alto Tietê Cabeceiras, São Paulo. Ph.D. Thesis, Universidade de Mogi das Cruzes, São Paulo, Brazil, 2019. [Google Scholar]
- Ramalho, M.O.; Vieira, A.S.; Pereira, M.C.; Moreau, C.S.; Bueno, O.C. Transovarian transmission of Blochmannia and Wolbachia endosymbionts in the neotropical weaver ant Camponotus textor (Hymenoptera, Formicidae). Curr. Microbiol. 2018, 75, 866–873. [Google Scholar] [CrossRef]
- Werren, J.H. Biology of Wolbachia. Annu. Rev. Entomol. 1997, 42, 587–609. [Google Scholar] [CrossRef]
- Schilthuizen, M.O.; Stouthamer, R. Horizontal transmission of parthenogenesis–inducing microbes in Trichogramma wasps. Proc. R. Soc. Lond. Ser. B Biol. Sci. 1997, 264, 361–366. [Google Scholar] [CrossRef]
- Tolley, S.J.A.; Nonacs, P.; Sapountzis, P. Wolbachia horizontal transmission events in ants: What do we know and what can we learn? Front. Microbiol. 2019, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Raychoudhury, R.; Baldo, L.; Oliveira, D.C.S.G.; Werren, J.H. Modes of acquisition of Wolbachia: Horizontal transfer, hybrid introgression, and codivergence in the Nasonia species complex. Evolution (N. Y.) 2009, 63, 165–183. [Google Scholar] [CrossRef]
- Watanabe, M.; Tagami, Y.; Miura, K.; Kageyama, D.; Stouthamer, R. Distribution patterns of Wolbachia endosymbionts in the closely related flower bugs of the genus Orius: Implications for coevolution and horizontal transfer. Microb. Ecol. 2012, 64, 537–545. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).