Abstract
Enteric fermentation of methane by ruminant animals represents a major source of anthropogenic methane. Significantly less information is available on the existence of methanotrophs in the gut of ruminants. Therefore, detailed strain descriptions of methanotrophs isolated from ruminant faeces or gut are rare. We present a first report on the enrichment and isolation of a methanotroph, strain BlB1, from the faeces of an Indian antelope (blackbuck). The 16S rRNA gene sequence of strain BlB1 showed the highest identity (98.40% identity) to Methylobacter marinus A45T and Methylobacter luteus NCIMB 11914T. Strain BlB1 showed coccoidal cells (1.5–2 µm in diameter), which formed chains or aggregates of 3–4 cells of light yellow-coloured colonies on agarose when incubated with methane in the gas phase. The draft genome of BlB1 (JADMKV01) is 4.87 Mbp in size, with a G + C content of 51.3%. The draft genome showed 27.4% digital DNA-DNA hybridization (DDH) and 83.07% average nucleotide identity (ANIb) values with that of its closest phylogenetic neighbour, Methylobacter marinus A45T. Due to the lower values of DDH and ANIb with the nearest species, and <98.7% 16S rRNA gene sequence identity, we propose that strain BlB1 belongs to a novel species of Methylobacter. However, as the culture has to be maintained live and resisted cryopreservation, deposition in culture collections was not possible and hence we propose a Candidatus species name, ‘Ca. Methylobacter coli’ BlB1. ‘Ca. Methylobacter coli’ BlB1 would be the first described methanotroph from ruminants worldwide, with a sequenced draft genome. This strain could be used as a model for studies concerning methane mitigation from ruminants.
Keywords:
methanotrophs; ruminants; Methylobacter; cultivation; novel species; Indian antelope; blackbuck 1. Introduction
Ruminants are significant sources of anthropogenic methane emissions and contribute to around 12–37% of global methane emissions [1]. Methane is a by-product of plant biomass digestion by microbial communities present in their rumen or gastrointestinal tract. Methane is the second most important greenhouse gas and is not assimilated by the ruminant animals and hence released into the atmosphere contributing to global warming [2]. As the only producers of enteric methane, methanogens are responsible for the contribution of livestock industries to climate change [3]. They have thus become the focus of research towards developing mitigation strategies.
In other systems, like wetlands, rice fields, and landfills, methanotrophs or methane-oxidizing bacteria have been the only and the most efficient contributors to methane mitigation [4]. The classical methanotrophs or methane-oxidizing bacteria belong to Gammaproteobacteria (Type I) and Alphaproteobacteria (Type II). Recently, methanotrophs have also been isolated from the phylum Verrucomicrobia. The gammaproteobacterial methanotrophs are further classified as Type Ia or Type Ib (Type X methanotrophs) [5].
Very few researchers have tried to investigate the presence of methanotrophs in ruminants. The rumen is classically thought to be completely anoxic; however, there are indications of minor amounts of oxygen ([6] and references therein). Methanotrophs require oxygen; however, they also exist in places where significantly reduced oxygen quantities are present [7].
A research group in Australia studied the cow rumen for the presence of methanotrophs [6]. According to that report, a Methylocystis species was consistently detected in the rumen epithelium of the cow. They could also enrich a Methylobacter species from one of the cow rumen samples. Two older examples of methanotrophs isolation from cow rumen or mouth exist where Methylobacter species were isolated or enriched [8,9]. Significantly less is known about the presence of methanotrophs in the gut of other ruminants or non-ruminant herbivores. Usually, it is commonly observed that methanogens and methanotrophs co-exist in habitats.
The Indian antelope (Indian blackbuck) is one of the few small ruminants which is considered to be a strict grazer and has approximately 80% grass in their diet [10]. The digestive tract shows similarity to cattle-type rumen [10]. Considering this background, we attempted enrichment, cultivation, and isolation of methanotrophs from blackbuck faeces.
2. Materials and Methods
2.1. Sampling
A faecal sample of an Indian blackbuck was collected from Rajiv Gandhi Zoological Park and Wildlife Research Centre, Pune, in January 2017. The sample was transferred to the laboratory within 24 h of collection. The sample was handled under aseptic conditions for enrichment in a methane environment, and aliquots were stored at 8 °C.
2.2. Enrichment and Isolation of a Methanotroph
On a clean bench, ~0.1 g of a sample was taken with a sterile spatula and diluted to 1:10. Further, a 1:20 dilution was carried out by adding 0.5 mL of the diluted sample to 19.5 mL of sterile dilute nitrate mineral salts (NMS) medium in a serum bottle [11], resulting in a 5 × 10−3 dilution. Twenty-five percent volume of methane gas was added to the 65 mL capacity serum bottle’s headspace. The bottles were incubated at 39 °C. The decrease in methane was followed every two days, and the gas phase was refreshed every week. A control was kept with medium and added methane with no sample.
The bottles were incubated for 20–60 days, and fresh methane was added periodically, or the gas phase was replaced. The decrease in methane was documented by injecting the headspace gas in a Chemito Gas Chromatograph, as described before [12]. Turbidity or pellicle formation was considered to be indicative of growth. Liquid from this actively growing methane enrichment was streaked on dilute NMS medium. The plates were incubated in a 25% methane atmosphere in a glass desiccator at 39 °C.
Single colonies were picked using a sterilized toothpick and streaked on a fresh medium plate and incubated in a methane-containing atmosphere. This process was repeated several times to get the axenic culture. The purity of the methanotrophic isolate was checked by streaking on dilute nutrient agar and nutrient agar plates, where no heterotrophic growth was observed. We performed 16S rRNA-based identification for the axenic culture.
2.3. Morphological Characterization
Live cells of a methanotroph strain were observed under a phase-contrast microscope (Nikon 80i, Japan microscope with a camera) under 100× magnifications with oil emulsion. The Gram-character of heat-fixed culture was determined using a standard Gram-staining protocol and observed under a bright-field microscope. Bacterial culture was fixed and processed to observe under Scanning Electron Microscopy (SEM) (Zeiss model EVO-MA-15 SEM). Sample preparation for SEM has been described before [12].
2.4. DNA Extraction and PCR Amplification
The amplification of particulate methane monooxygenase β subunit gene (pmoA) and 16S rRNA gene was carried out using A189f-mb661r primers and 27f and 1492r primers, respectively using genomic DNA as the template. The primers and PCR protocols were used as described before [12]. The amplified product was sequenced by First Base Laboratories, Malaysia. The obtained sequence was subjected to NCBI-BLAST analysis and aligned using the MAFFT alignment server (https://mafft.cbrc.jp/alignment/server/ (accessed on 1 March 2021)). The 16S rRNA-based evolutionary tree was constructed using the maximum likelihood method and Tamura-Nei model [13] using MEGA X [14].
The evolutionary history of the pmoA gene of BlB1 with its closest members was conducted using the maximum likelihood method and Poisson correction model [15] in MEGA X [14].
Based on cut off values of 98.65% and 87% for the 16S rRNA gene [16] and pmoA gene nucleotide sequence [17], respectively, strains were considered as a putative novel species. The whole genome was sequenced for further understanding the novelty and salient features of the BlB1 strain, which was a putative novel species of Methylobacter.
The genomic DNA was extracted from larger biomass obtained from a liquid culture of strain BlB1 and sequenced. The whole-genome sequencing was done using the Illumina HiSeqX platform (150*2) at Med genome laboratory, Bangalore, India. De novo assembly of the sequenced reads was done by using SPADES de novo Assembler (v3.13.0). RAST, (http://rast.nmpdr.org/ (accessed on 1 March 2021)) and the NCBI prokaryotic genome annotation pipeline PGAP (https://www.ncbi.nlm.nih.gov/genome/annotation_prok/ (accessed on 1 March 2021)) were used for further annotation. The details of whole genome analysis are provided in the Supplementary Material.
2.5. Biochemical Characterization
Strain BlB1 was characterized for details pertaining to its novelty. For determination of different carbon sources utilized by Methylobacter sp. BlB1, dilute NMS medium [9] was supplemented with one of the following filter-sterilized or autoclaved (at 10psi) substrates (0.1%, w/v): formate, lactose, fructose, formaldehyde, maltose, glucose, sucrose, pyruvate, acetate, and mannose in 35-mL serum bottles. The ability of strain BlB1 to grow on methanol was tested by adding 5–2000 mM methanol in a liquid medium in serum bottles. The range of nitrogen sources utilized by strain BlB1 was tested by replacing KNO3 from the dilute NMS medium with one of the following compounds: urea, glycine, serine, valine, asparagine, aspartate, L-glutamic acid, glutamate, peptone, and yeast extract (0.25 g/L). Growth without nitrogen source was tested under micro-oxic conditions (~10% air). The optimum pH (buffered and non-buffered) and temperature ranges were evaluated in dilute NMS liquid medium [12].
2.6. Strain Maintenance
The culture was difficult to maintain and preserve using the cryopreservation techniques used for preserving methanotrophs. Therefore, the live culture was maintained by using a combination of methods such as subculturing from solid to solid, solid to liquid and then again to solid, etc.
3. Results
3.1. Enrichment, Isolation, and Identification
We observed decline in methane concentration indicating methane oxidation due to the inoculated faecal sample of blackbuck, as compared to the control. The methanotroph enrichment from the blackbuck formed a thick yellowish pellet after eight to ten days of incubation. The growth was further incubated, followed by streaking the growth on solid agarose plates, and was incubated in a methane environment. After incubation of 10–14 days, dark yellow-coloured colonies intermingled with dark grey-blackish colonies of actinomycetes were observed on dilute NMS agarose medium, indicating mixed colonies. After repeated subcultures, only yellow-coloured colonies grew in the methane containing atmosphere. The colony size shrunk down after repeated subculture and purification. The actinomycete was isolated on dilute nutrient agar plates and later identified as a Streptomyces species. The 16S rRNA gene of strain BlB1 showed 98.4% identity with Methylobacter marinus A45T and Methylobacter luteus strain 11914T.
3.2. Microscopic Identification
Strain BlB1 shows coccoidal cells of 1–2 µm diameter and showed chains or aggregates of 3–4 cells (Figure 1A,B). The cells are Gram-negative (Figure 1C). The pure culture of BlB1 formed yellow-coloured colonies that were 2–3 mm in diameter (Figure 1D) and grew exclusively with methane in the gas phase. BlB1 forms a turbid suspension with a yellow tinge in liquid dilute NMS medium with methane and air (20:80) in the headspace (Figure 1E). BlB1 grew well on plates and did not grow to high turbidity in the liquid medium.
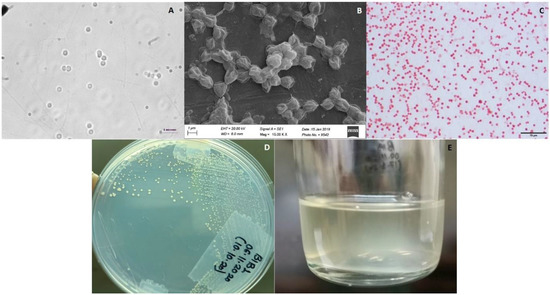
Figure 1.
Morphology of strain BlB1: (A) live cells were observed under a phase-contrast microscope (Nikon 80i, Japan microscope with a camera) under 100X magnification with oil emulsion; (B) fixed and processed culture was observed under Scanning Electron Microscopy (SEM) (Zeiss model EVO-MA-15 SEM); (C) Gram-staining under light microscopy; (D) colony morphology on solid NMS medium plate; (E) BlB1 forms a turbid suspension with a yellow tinge in liquid dilute NMS medium.
3.3. Phylogenetic and Phylogenomic Affiliation
We obtained the nearly complete 16S rRNA gene based on the sequencing of the PCR product, deposited in GenBank as MH424899.1. The entire 16S rRNA gene sequence (1496 bp) of strain BlB1 was determined from the draft genome and deposited as MH424899.2. BLAST analysis with the type material showed that the 16S rRNA gene was 98.4% identical to both Methylobacter marinus A45T and Methylobacter luteus strain NCIMB 11914. In a maximum likelihood tree, the nearest neighbour of BlB1 is Methylobacter marinus A45T and Methylobacter luteus strain NCIMB 11914T (Figure 2).
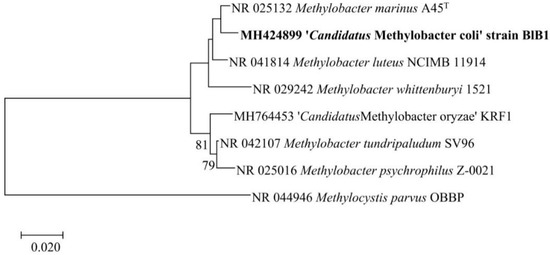
Figure 2.
Maximum likelihood bootstrap tree of the 16S rRNA gene sequence of strain BlB1 in comparison with the 16S rRNA genes of closest valid species. The evolutionary history was inferred by using the maximum likelihood method and Tamura-Nei model and 1000 bootstraps (values >50 shown). The bar represents 2% divergence. The phylogenetic tree was constructed using MEGA X.
The complete pMMO B protein (genome-based) showed 98.38% similarity to that of Methylobacter luteus IMV-B-3098. The maximum likelihood-based tree based on pMMO B subunit from type material determined that Methylobacter BlB1 clustered with Methylobacter marinus A45, Methylobacter luteus IMV-B-3098, and Methylobacter whittenburyi ACM 3310 (Figure 3).
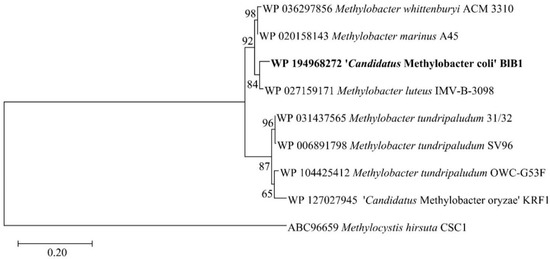
Figure 3.
Maximum likelihood bootstrap tree of the pMMO B subunit protein sequences of strain BlB1 in comparison with the protein from type strains of valid species. The evolutionary history was completed using the maximum likelihood method based on the Poisson correction model and 1000 bootstraps. The bar represents 2% divergence. The phylogenetic tree was constructed using MEGA X.
3.4. Draft Genome Features and Salient Genome Characteristics
The draft genome was sequenced to determine the phylogenetic position of strain BlB1. A total of ~240 contigs were generated. The blastx analysis of all the contigs showed similarity to members of Methylobacter. Based on RAST annotation, the BlB1 draft genome is 4.87 Mbp in size, with a G + C content of 51.3% genes and has 306 subsystems, while as per the PGAP annotation (https://www.ncbi.nlm.nih.gov/genome/14391?genome_assembly_id=1491660 (accessed on 1 March 2021)), 4430 genes and 4319 proteins with three rRNA genes and 38 tRNA were found. The phylogenomic relation of strain BlB1 with the genome of other Methylobacter genus was determined using the PATRIC database (https://patricbrc.org/ (accessed on 1 March 2021)) and shows a similar affiliation with the two Methylobacter species, Methylobacter marinus A45T and Methylobacter luteus strain NCIMB 11914T (Figure 4).
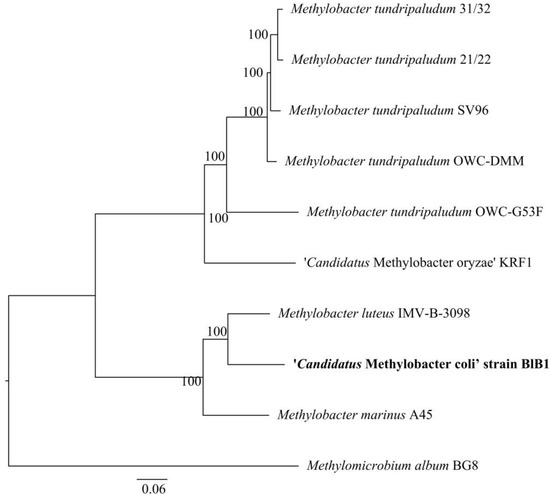
Figure 4.
Phylogenomic tree constructed using the PATRIC database showing the relationship of ‘Ca. Methylobacter coli’ strain BlB1 with other species of Methylobacter with an outgroup.
The general characteristics of the draft genome of strain BlB1 are listed in Supplementary Table S1. The NCBI accession number and the Bioproject of BlB1 draft genome are JADMKV01 and PRJNA675424, respectively (https://www.ncbi.nlm.nih.gov/genome/14391?genome_assembly_id=1491660 (accessed on 1 March 2021)).
3.5. Carbon Metabolism Genes
The strain BlB1 genome contains a complete set of genes for methane oxidation, including one copy of the particulate methane monooxygenase pmoCAB gene cluster for methane to methanol oxidation reaction. Methanol is dehydrogenated to formaldehyde by two copies of each pyrroloquinoline quinone (PQQ) and zinc dependent and one copy of NAD(P)-dependent methanol dehydrogenase. The BlB1 genome also contains genes encoding quinol-dependent methanol dehydrogenase (moxF) with its system protein MoxJ. The tetrahydromethanopterin (H4MPT) pathway for formaldehyde oxidation and formate dehydrogenase were also found in the genome (Supplementary Table S2). Formaldehyde assimilation can be done via both the RuMP pathway as well as the serine pathway. However, the type-I methanotrophs usually assimilate formaldehyde via the RuMP pathway only [4]. The BlB1 genome showed both serine and RuMP pathways for formaldehyde assimilation and encodes a complete glycolytic pathway, pentose phosphate pathway, Entner‒Doudoroff pathway, and citrate cycle.
3.6. Nitrogen Metabolism Genes
The draft genome of BlB1 contains cyanase (cynS) (also known as cyanate hydratase enzyme) genes. Cyanate hydratase is used to decompose cyanate, a salt or ester of cyanic acid, which can be used as the sole source of nitrogen [18,19]. This enzyme catalyses the reaction to produce ammonia and CO2 using cyanate and bicarbonate [19].
The BlB1 genome contains a complete set of dissimilatory nitrate reduction genes such as nasAB and nirBD clusters for nitrate to nitrite to ammonia dissimilation. The complete set of genes for the classical nitrogen fixation pathway were detected. Within the denitrification pathway, nitrite reductase nirBD genes and nitric oxide reductase norB and norC genes are present. Additionally, in the draft genome of BlB1, genes qnorB encoding for quinol-dependent nitric oxide reductase subunit B (qNorB) were found along with its anaerobic nitric oxide reductase transcriptional regulator (norR). The qNorB protein encoded in the BlB1 genome shows 72.85% similarity with Rugosibacter sp. (Supplementary Table S2). Some of the strains of the genus Methylomonas also possess qnorB genes.
The Methylobacter BlB1 genome also contains genes for bacteriohemerythrin synthesis, which is reported to be responsible for oxygen scavenging and transport to particulate methane monooxygenase [20,21].
3.7. DDH, ANIb, and AAI Indicate That Strain BlB1 Represents a New Species of Methylobacter
Most of the protein sequences derived from the BlB1 genome showed high similarity to those of Methylobacter marinus A45 and Methylobacter luteus (blast analysis of the proteins, Supplementary Table S2). The genome-based comparison of BlB1 with Methylobacter marinus A45, Methylobacter oryzae KRF1, and Methylobacter tundripaludum (strains SV96T, OWC-DMM, OWC-G53F, 21/22 and 31/32) were used to calculate the average nucleotide identity (ANIb-G, http://jspecies.ribohost.com/jspeciesws/#analyse (accessed on 1 March 2021)), digital DNA–DNA hybridization (dDDH, http://ggdc.dsmz.de/ (accessed on 1 March 2021)) and the average amino-acid identity (AAI, http://enve-omics.ce.gatech.edu/aai/ (accessed on 1 March 2021)). The ANIb, AAI, and DDH values between BlB1 genome and Methylobacter marinus A45, Methylobacter oryzae KRF1, and Methylobacter tundripaludum strains were 73–83%, 70–83%, and 20–27%, respectively (Table 1), which were lower than the threshold values (95% for ANI or AAI and 70% for DDH). Hence, strain BlB1 is proposed to represent a novel species of Methylobacter genus within the family Methylococccaceae.

Table 1.
ANIb, AAI, and DDH calculation of strain BlB1 with its closely related members.
3.8. Biochemical Characterisation
Strain BlB1 can only utilise methane or methanol as carbon and energy sources. Strain BlB1 can utilise aspartic acid, glutamate, and yeast extract as a nitrogen source (Table 2). Initially, we used 39 °C for the isolation of BlB1, but later we observed that the strain could grow better at 30 °C. Strain BlB1 was found to be a mesophilic methanotroph and could grow optimally at 30 °C in the 15–39 °C temperature range. It was found to grow best at neutral pH in the range of 4–9 and best at pH 6.8, under buffered condition (Table 2). The strain could tolerate NaCl up to 0.5% and up to 5% methanol. However, it grew best with no additional NaCl. The optimum methanol concentration for the strain was 0.2%. Strain BlB1 oxidised methane and grew under micro-oxic conditions with 5–20% air and 20% methane in the headspace, with the remainder nitrogen, however, the optimum conditions were full oxic, with 80% air and 20% methane in the gas phase. The culture could also grow without addition of nitrogen sources indicating that it could fix nitrogen. The complete pathway for nitrogen fixation was encoded in the genome, supporting this observation (Supplementary Table S2).

Table 2.
Biochemical characterization of strain BlB1.
The major morphological and biochemical features are compared between BlB1 and other Methylobacter sp. in Table 3. Strain BlB1 showed unique cell morphology. Strain BlB1 could not be cryo-preserved using published methods [12] and has to be maintained via serial transfers or storage for a maximum of ~3 months at 4–8 °C, similar to Ca. Methylobacter KRF1 [22,23]. The same difficulties were faced for ‘Ca. Methylobacter oryzae’ strain KRF1 reported earlier [22] and other micro-aerophilic methanotrophs such as Methyloglobulus morosus [24], Methylosoma difficile [25,26], and Ca. Methylospira [27].

Table 3.
Differential morphological and biochemical characteristics of Methylobacter species in comparison with strain BlB1.
3.9. Description of ‘Candidatus Methylobacter coli’ Strain BlB1 Isolated from an Indian Blackbuck
Strain BlB1 has to be maintained live (transfer from solid to liquid to solid) and could not be cryo-preserved and hence could not be deposited in international culture collections. The culture is being preserved live in our laboratory. Based on the recent prokaryotic taxonomy for the proposal of Candidatus status [29], we have proposed a Candidatus name for the novel species: ‘Ca. Methylobacter coli’ strain BlB1.
‘Ca. Methylobacter coli’ strain BlB1 was isolated from the faecal sample of an Indian blackbuck. Live cells are coccoid or diplococci or aggregates or chains of 3–4 cells (1.5–2 µm in diameter). The cells are non-motile and show Gram-negative character. It forms opaque yellow-coloured colonies of 2–3 mm in size under methane and air rich environment. BlB1 can utilise methane as well as methanol as a carbon source and aspartic acid, glutamate, and yeast extract as a nitrogen source. The % GC content is 51.3%, determined from the draft genome. The whole-genome shotgun project was deposited at DDBJ/ENA/GenBank under the accession JADMKV000000000.1.
4. Discussion
Ruminants such as cows, sheep, and goats ferment plant material, anaerobically generating methane. The digestive system of ruminants typically has four compartments: the rumen, reticulum, omasum (act as channels), and the abomasum. The ruminant’s digestive system is characterized by anatomical adjustments that allow them to digest cellulose and other complex carbohydrates [30]. In the rumen, methane production has been reported via the activity of methanogenic Archaea, which reside primarily in the rumen and the lower segment of the intestines of ruminants [31]. While researchers know a great deal about methanogenesis in the rumen, the possibility that methane oxidation occurs with methanogenesis has received little attention [6].
Methanotrophs require oxygen; however, they also exist in places where significantly reduced oxygen quantities are present, e.g., anoxic parts of the lakes [7]. Earlier studies have indicated that although the rumen of the cow is generally considered to be anoxic, oxygen concentrations of up to 0.3 μM L−1 and 6.7 μM L−1 have been measured in cattle rumen fluid and gas, respectively [32]. In cattle the rumen epithelium is also highly oxygenated, with blood flow to the rumen epithelial tissue rising to 3.1 mL min−1 g−1 after feeding [32]. Methanotrophs have been enriched from the mouth of cattle [9] and the rumen of fistulated cattle previously [33].
The cow rumen was suspected to harbour methanotrophs, and one of the first detailed studies was done as a part of an Australian project [6]. The researchers detected and enriched rumen methanotrophs from cow rumen for their potential as a biocontrol in rumen methane mitigation [6]. They found sequences of Methylocystis in the rumen epithelium of four of the cow rumen samples and could enrich a Methylobacter species from one rumen [6].
Methanotrophs from both the families Methylococcaceae and Methylocystaceae have been detected using metagenomics of the cow rumen. A cow rumen shotgun approach yielded 21804 hits from Methylococcales and methanotrophs including Methylobacter tundripaludum, Methylococcus capsulatus, Methylocystis, and Methylocella. Methanotrophs were detected in all four sets of metagenome samples [34]. Thus, we had hypothesized that there is a possibility of the presence of methanotrophs in ruminants or non-ruminant herbivores.
Whittenbury and colleagues cultivated Methylobacter bovis (synonymous to Methylobacter luteus) from cow’s mouth [9]. Methylobacter species was enriched from a cow rumen sample in an Australian study [6]. Strain BlB1 is, to the best of our knowledge, the first pure culture of a methanotroph isolated from ruminants in recent times. The present study also highlights the salient genome characteristics which could be useful for understanding the properties of a ruminant methanotroph and could act as a model for methane mitigation studies. Future studies focusing on the physiological properties of strain BlB1 would help us in understanding the role of methanotrophs in rumen methane mitigation.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microbiolres12020035/s1, Table S1: Genomic features of Methylobacter BlB1, Table S2: Blastp analysis of the annotated proteins of the Methylobacter BlB1 genome. Supplementary Material: Whole genome sequencing details and data analysis.
Author Contributions
Formal analysis, R.A.B.; funding acquisition, M.C.R.; investigation, K.K. and M.C.R.; methodology, K.K., J.M. and P.P.; visualization, M.C.R.; writing—original draft, K.K. and M.C.R., writing—review and editing, M.C.R. and R.A.B. All authors have read and agreed to the published version of the manuscript.
Funding
MCR acknowledges the Department of Science and Technology, DST, SERB (EMR/2017/002817) for the funds. KK acknowledges CSIR for the research fellowship. JM acknowledges SERB, Department of Science and Technology, for the research fellowship. PSP acknowledges the Department of Science and Technology for providing a women scientist (WOS-A) fellowship and funds (SR/WOS-A/LS-410/2017). We are also grateful to Agharkar Research Institute for the basic funding provided.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The whole-genome shotgun project is available in the GenBank under the accession JADMKV000000000.1.
Acknowledgments
We are thankful to Rajkumar Jadhav, Zoo Director, Rajiv Gandhi Zoological Park and Wildlife Research Centre, Pune, for helping in the faecal sample collection. We are thankful to Akshay Joshi and Sumit Singh Dagar for the faecal sample collection and important discussions.
Conflicts of Interest
We declare no conflict of interests.
References
- Broucek, J. Production of methane emissions from ruminant husbandry: A review. J. Environ. Prot. 2014, 5, 1482–1493. [Google Scholar] [CrossRef]
- Conrad, R. The global methane cycle: Recent advances in understanding the microbial processes involved. Environ. Microbiol. Rep. 2009, 1, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, A. Enteric fermentation and ruminant eructation: The role (and control?) of methane in the climate change debate. Clim. Chang. 2009, 93, 407–431. [Google Scholar] [CrossRef]
- Hanson, R.S.; Hanson, T.E. Methanotrophic bacteria. Microbiol. Rev. 1996, 60, 439–471. [Google Scholar] [CrossRef] [PubMed]
- Dedysh, S.N.; Knief, C. Diversity and Phylogeny of Described Aerobic Methanotrophs. In Methane Biocatalysis: Paving the Way to Sustainability; Springer: Berlin/Heidelberg, Germany, 2018; pp. 17–42. [Google Scholar]
- Finn, D.; Ouwerkerk, D.; Klieve, A. Methanotrophs from Natural Ecosystems as Biocontrol Agents for Ruminant Methane Emissions; Project Report; Meat & Livestock Australia Limited: North Sydney, Australia, February 2012. [Google Scholar]
- Oswald, K.; Milucka, J.; Brand, A.; Hach, P.; Littmann, S.; Wehrli, B.; Kuypers, M.M.; Schubert, C.J. Aerobic gammaproteobacterial methanotrophs mitigate methane emissions from oxic and anoxic lake waters. Limnol. Oceanogr. 2016, 61, S101–S118. [Google Scholar] [CrossRef]
- Stocks, P.K.; McCleskey, C. Morphology and physiology of Methanomonas methanooxidans. J. Bacteriol. 1964, 88, 1071–1077. [Google Scholar] [CrossRef] [PubMed]
- Whittenbury, R.; Phillips, K.; Wilkinson, J. Enrichment, isolation and some properties of methane-utilizing bacteria. Microbiology 1970, 61, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Sauer, C.; Bertelsen, M.F.; Hammer, S.; Lund, P.; Weisbjerg, M.R.; Clauss, M. Macroscopic digestive tract anatomy of two small antelopes, the blackbuck (Antilope cervicapra) and the Arabian sand gazelle (Gazella subgutturosa marica). Anat. Histol. Embryol. 2016, 45, 392–398. [Google Scholar] [CrossRef]
- Pandit, P.S.; Rahalkar, M.C.; Dhakephalkar, P.K.; Ranade, D.R.; Pore, S.; Arora, P.; Kapse, N. Deciphering community structure of methanotrophs dwelling in rice rhizospheres of an Indian rice field using cultivation and cultivation-independent approaches. Microb. Ecol. 2016, 71, 634–644. [Google Scholar] [CrossRef]
- Pandit, P.S.; Hoppert, M.; Rahalkar, M.C. Description of ‘Candidatus Methylocucumis oryzae’, a novel Type I methanotroph with large cells and pale pink colour, isolated from an Indian rice field. Antonie Van Leeuwenhoek 2018, 111, 2473–2484. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Zuckerkandl, E.; Pauling, L. Evolutionary Divergence and Convergence in Proteins. In Evolving Genes and Proteins; Elsevier: Amsterdam, The Netherlands, 1965; pp. 97–166. [Google Scholar]
- Rossi-Tamisier, M.; Benamar, S.; Raoult, D.; Fournier, P.-E. Cautionary tale of using 16S rRNA gene sequence similarity values in identification of human-associated bacterial species. Int. J. Syst. Evol. Microbiol. 2015, 65, 1929–1934. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Yang, S.; Liebner, S. Evaluation and update of cutoff values for methanotrophic pmoA gene sequences. Arch. Microbiol. 2016, 198, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Kozliak, E.I.; Fuchs, J.A.; Guilloton, M.B.; Anderson, P.M. Role of bicarbonate/CO2 in the inhibition of Escherichia coli growth by cyanate. J. Bacteriol. 1995, 177, 3213–3219. [Google Scholar] [CrossRef] [PubMed]
- Walsh, M.A.; Otwinowski, Z.; Perrakis, A.; Anderson, P.M.; Joachimiak, A. Structure of cyanase reveals that a novel dimeric and decameric arrangement of subunits is required for formation of the enzyme active site. Structure 2000, 8, 505–514. [Google Scholar] [CrossRef][Green Version]
- Kao, W.-C.; Wang, V.C.-C.; Huang, Y.-C.; Yu, S.S.-F.; Chang, T.-C.; Chan, S.I. Isolation, purification and characterization of hemerythrin from Methylococcus capsulatus (Bath). J. Inorg. Biochem. 2008, 102, 1607–1614. [Google Scholar] [CrossRef] [PubMed]
- Ross, M.O.; Rosenzweig, A.C. A tale of two methane monooxygenases. J. Biol. Inorg. Chem. 2017, 22, 307–319. [Google Scholar] [CrossRef]
- Khatri, K.; Mohite, J.A.; Pandit, P.S.; Bahulikar, R.; Rahalkar, M.C. Description of ‘Ca. Methylobacter oryzae’ KRF1, a novel species from the environmentally important Methylobacter clade 2. Antonie Van Leeuwenhoek 2020, 113, 729–735. [Google Scholar] [CrossRef]
- Rahalkar, M.C.; Khatri, K.; Pandit, P.S.; Dhakephalkar, P.K. A putative novel Methylobacter member (KRF1) from the globally important Methylobacter clade 2: Cultivation and salient draft genome features. Antonie Van Leeuwenhoek 2019, 112, 1399–1408. [Google Scholar] [CrossRef]
- Deutzmann, J.S.; Hoppert, M.; Schink, B. Characterization and phylogeny of a novel methanotroph, Methyloglobulus morosus gen. nov., spec. nov. Syst. Appl. Microbiol. 2014, 37, 165–169. [Google Scholar] [CrossRef]
- Rahalkar, M.; Bussmann, I.; Schink, B. Methylosoma difficile gen. nov., sp. nov., a novel methanotroph enriched by gradient cultivation from littoral sediment of Lake Constance. Int. J. Syst. Evol. Microbiol. 2007, 57, 1073–1080. [Google Scholar] [CrossRef]
- Schink, B.; Rahalkar, M. Genus Methylosoma. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley Publications: Hoboken, NJ, USA, 2016; pp. 1–4. [Google Scholar]
- Oshkin, I.Y.; Miroshnikov, K.K.; Danilova, O.V.; Hakobyan, A.; Liesack, W.; Dedysh, S.N. Thriving in wetlands: Ecophysiology of the spiral-shaped methanotroph Methylospira mobilis as revealed by the complete genome sequence. Microorganisms 2019, 7, 683. [Google Scholar] [CrossRef]
- Collins, D.A.; Akberdin, I.R.; Kalyuzhnaya, M. Genus Methylobacter. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley Publications: Hoboken, NJ, USA, 2017. [Google Scholar]
- Whitman, W.B.; Sutcliffe, I.C.; Rossello-Mora, R. Proposal for changes in the International Code of Nomenclature of Prokaryotes: Granting priority to Candidatus names. Int. J. Syst. Evol. Microbiol. 2019, 69, 2174–2175. [Google Scholar] [CrossRef]
- Niwiska, B. Digestion in Ruminants. In Carbohydrates-Comprehensive Studies on Glycobiology and Glycotechnology; Chang, C.-F., Ed.; InTech: Rijeka, Croatia, 2012; pp. 245–258. [Google Scholar]
- Patra, A.; Park, T.; Kim, M.; Yu, Z. Rumen methanogens and mitigation of methane emission by anti-methanogenic compounds and substances. J. Anim. Sci. Biotechnol. 2017, 8, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.; Yarlett, N.; Hillman, K.; Williams, A.; Lloyd, D.; Williams, T. The presence of oxygen in rumen liquor and its effects on methanogenesis. J. Appl. Bacteriol. 1983, 55, 143–149. [Google Scholar] [CrossRef]
- Dobson, A. Blood flow and absorption from the rumen. Q. J. Exp. Physiol. Transl. Integr. 1984, 69, 599–606. [Google Scholar] [CrossRef]
- Wallace, R.J.; Rooke, J.A.; McKain, N.; Duthie, C.-A.; Hyslop, J.J.; Ross, D.W.; Waterhouse, A.; Watson, M.; Roehe, R. The rumen microbial metagenome associated with high methane production in cattle. BMC Genom. 2015, 16, 452. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).