Abstract
The most widely distributed blaCTX-M gene on a global scale is blaCTX-M-15. The dissemination has been associated with clonal spread and different types of mobile genetic elements. The objective of this review was to describe the genetic environments of the blaCTX-M-15 gene detected from Enterobacteriaceae in published literature from Africa. A literature search for relevant articles was performed through PubMed, AJOL, and Google Scholar electronic databases; 43 articles from 17 African countries were included in the review based on the eligibility criteria. Insertion sequences were reported as part of the genetic environment of blaCTX-M-15 gene in 32 studies, integrons in 13 studies, and plasmids in 23 studies. In this review, five insertion sequences including ISEcp1, IS26, orf447, IS903, and IS3 have been detected which are associated with the genetic environment of blaCTX-M-15 in Africa. Seven different genetic patterns were seen in the blaCTX-M-15 genetic environment. Insertion sequence ISEcp1 was commonly located upstream of the end of the blaCTX-M-15 gene, while the insertion sequence orf477 was located downstream. In some studies, ISEcp1 was truncated upstream of blaCTX-M-15 by insertion sequences IS26 and IS3. The class 1 integron (Intl1) was most commonly reported to be associated with blaCTX-M-15 (13 studies), with Intl1/dfrA17–aadA5 being the most common gene cassette array. IncFIA-FIB-FII multi-replicons and IncHI2 replicon types were the most common plasmid replicon types that horizontally transferred the blaCTX-M-15 gene. Aminoglycoside-modifying enzymes, and plasmid-mediated quinolone resistance genes were commonly collocated with the blaCTX-M-15 gene on plasmids. This review revealed the predominant role of ISEcp1, Intl1 and IncF plasmids in the mobilization and continental dissemination of the blaCTX-M-15 gene in Africa.
1. Introduction
The most widely distributed blaCTX-M gene on a global scale is blaCTX-M-15, especially in the enterobacterial species such as Escherichia coli, Klebsiella spp. and Salmonella enterica [1,2]. The global dissemination of the blaCTX-M-15 gene has been associated with the clonal spread of E. coli O25: H4-ST131 strains and different types of mobile genetic elements (MGEs) such as insertion sequences, transposons, integrons, phage elements, and conjugative plasmids [1,2,3]. Of these MGEs, insertion sequences (IS) are of special concern because this mobile element can facilitate the independent transposition with insertion mutation and genetic rearrangements in Enterobacteriaceae [4,5,6]. Several types of IS elements have been recognized; however, ISEcp1, IS26, orf447 and ISCR1 have been frequently found to be responsible for the mobilization and expression of different antimicrobial resistance genes [7]. ISEcp1 is the most frequently reported IS type [7]. ISEcp1 is a member of the IS1380 family and was first identified on the plasmid pST01 in E. coli strain 79 but has now been globally disseminated in association with different blaCTX-M phylogenetic clusters [8].
The roles of ISEcp1 and other MGEs in the genetic environments of blaCTX-M genes have been well described [7,9,10]. ISEcp1 is commonly located upstream of the blaCTX-M-15 gene and is responsible for the downstream mobilization and transposition of itself, adjacent genes, and the blaCTX-M-15 gene. IS26 has commonly been found to be located upstream of the blaCTX-M-15 alone or in association with ISEcp1 [1,7]. ISCR1, on the other hand, has been associated with class 1 integron, forming a transposition complex for the mobilization of blaCTX-M-15 and other beta-lactamase genes [8]. Integrons are site-specific recombination systems that capture various arrays of gene cassettes within the conserved regions and can integrate one or several non-functional gene cassettes and convert these into functional genes [6]. Molecular characterization and replicon typing of various plasmid groups have facilitated the recognition and location of blaCTX-M-15 genes co-existing with other AMR genes on both narrow host-range and, to a lesser extent, broad-host-range plasmids [11].
Genetic environments of blaCTX-M genes have been described and reported in enterobacterial species from different parts of the world, however variation in genetic patterns exists from region to region [10]. Additionally, analysis of genetic environments of blaCTX-M gene and associated MGEs on a continental scale may provide necessary information on the diversity and complexity of the genetic environments as well as provide opportunities for better understanding of the epidemiology of this globally disseminated resistance gene. This study aimed to review and describe the genetic environments of blaCTX-M-15 genes and associated MGEs in Enterobacteriaceae in published literature from Africa.
2. Materials and Methods
The literature search was conducted in the PubMed, AJOL and Google Scholar electronic databases between June 2018 and January 2019 for the purpose of this narrative and non-systematic review. The following terms were used for the literature search: blaCTX-M-15 gene AND Africa OR blaCTX-M-15 AND genetic environment AND Africa. A literature search was also conducted based on studies reporting the detection of blaCTX-M-15 from each African country, e.g., blaCTX-M-15 AND Nigeria, blaCTX-M-15 AND Egypt, blaCTX-M-15 and Kenya, etc. The reference lists of all eligible articles were further reviewed and used to carry out a supplementary literature search. The articles were further screened after the removal of duplicates by titles and abstracts for their relevance to the study objectives and purpose. The primary outcomes of interest were to describe the genetic environment of blaCTX-M-15 in Enterobacteriaceae from different African countries.
For studies to be included in the qualitative description, the studies must have reported the genetic environment of the blaCTXM-15 resistance gene with special reference to the associated insertion sequences. The data were abstracted onto an Excel (Microsoft Office Excel 2010) spreadsheet. For each eligible study, data extracted included: first author details, year of publication, country from which the study was conducted, sources of the samples (animal, human or environment), enterobacterial species in which the blaCTX-M-15 gene was detected, insertion sequences associated with the genetic environment, additional data on other mobile genetic elements including type of integron and associated gene cassette arrays, plasmid and associated replicon types, as well as additional antimicrobial resistance genes associated with the blaCTX-M-15 gene on different plasmids.
3. Results
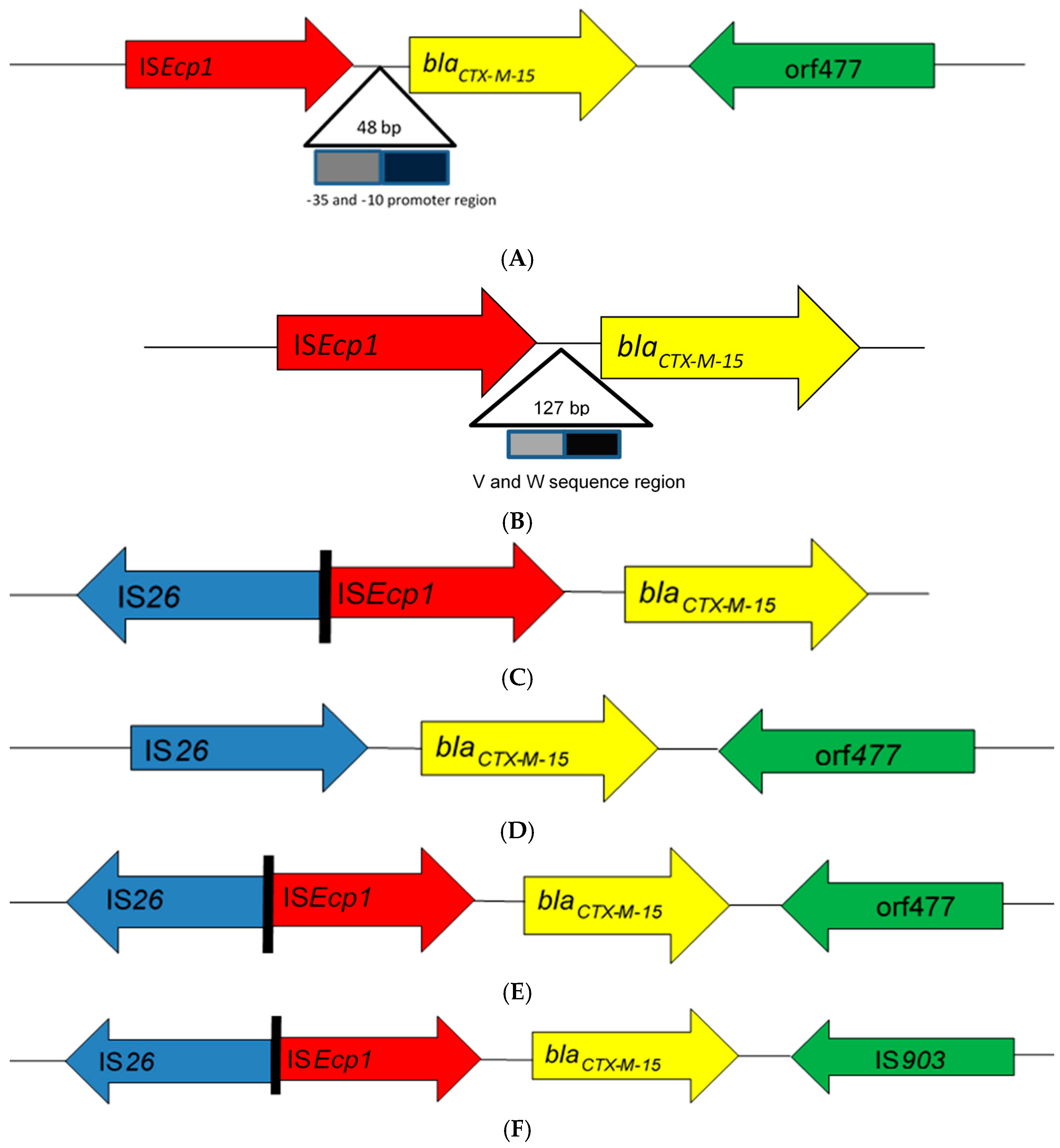
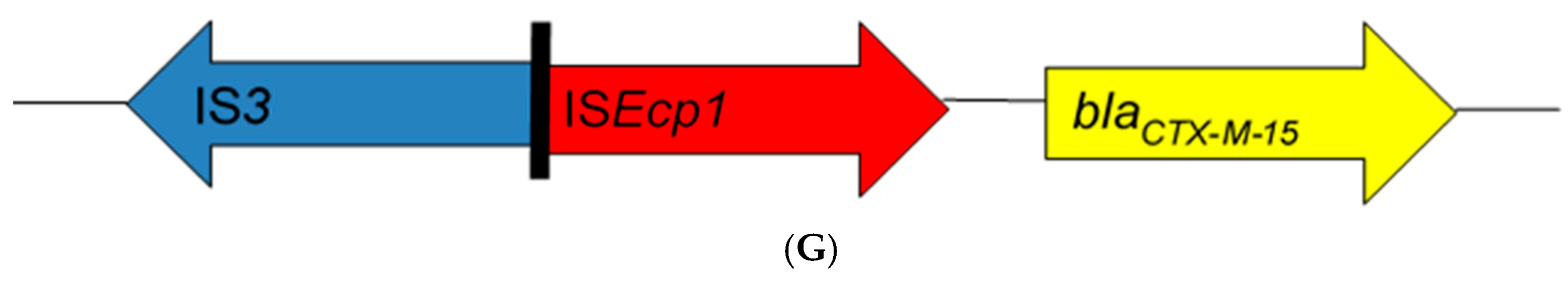
From the literature search, 43 articles from 17 African countries were included in the review based on the eligibility criteria (Table 1). Thirty-nine studies were based on blaCTX-M-15-producing Enterobacteriaceae isolated from human clinical cases, three studies from animals, and one study from the environment. Bacteria of Enterobacteriaceae reported were Escherichia coli alone (19 studies), Klebsiella spp. alone (8 studies), Salmonella enterica (6 studies), E. coli and Klebsiella spp. (4 studies), as well as combinations of other enterobacterial species (6 studies). Insertion sequences were reported in 32 of the 43 studies (Table 1). Seven different genetic patterns were observed among these studies (Figure 1). In eight studies [12,13,14,15,16,17,18,19], the insertion sequence ISEcp1 was located upstream of the end of the blaCTX-M-15 gene with insertion sequence orf477 located downstream (ISEcp1-blaCTX-M-15-orf477). Twenty-three studies [20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42] found ISEcp1 to be the only insertion sequence located upstream of the blaCTX-M-15 gene (ISEcp1-blaCTX-M-15). Additionally, two studies [16,42] reported the location of ISEcp1 upstream of blaCTX-M-15 truncated by IS26 without any downstream IS element (ISEcp1-IS26-blaCTX-M-15). In one these two studies [16], IS26 was located upstream of blaCTX-M-15 with orf447 located downstream in an enterobacterial isolate. In another two studies [12,43], ISEcp1 was truncated upstream of blaCTX-M-15 by IS26 (ISEcp1-IS26-blaCTX-M-15-orf477). In one study [42], ISEcp1 was truncated upstream of blaCTX-M-15 by IS26, with IS903 located downstream (ISEcp1-IS26-blaCTX-M-15-IS903); however, novel IS3 type [16] was reported in one study to truncate ISEcp1 upstream of the start of blaCTX-M-15 gene (ISEcp1-IS3-blaCTX-M-15). The promoter region (−35 and −10) of 48 bp [14,23,24,29,37], V and W promoter region of 127 bp [31], and other unspecified promoter regions of 400–1800 bp [17,19,26,35] of ISEcp1 were located upstream between the left end of ISEcp1 and the start codon of the blaCTX-M-15 gene.
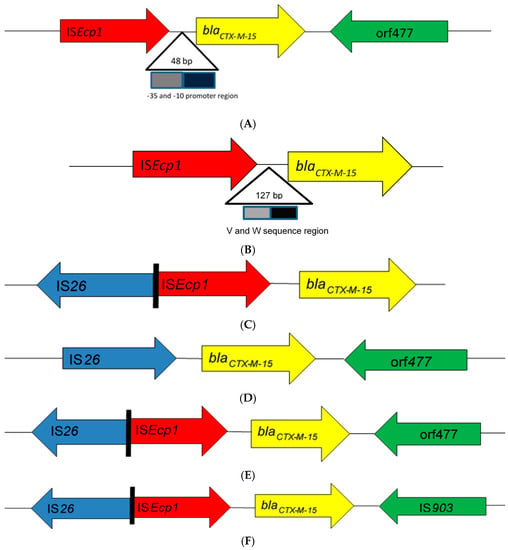
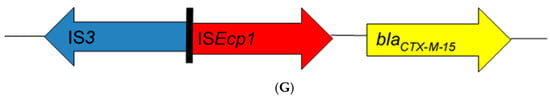
Figure 1.
Schematic representations of different genetic environments of blaCTX-M-15 reported in the literature from Africa. (A) The insertion sequence ISEcp1 was located upstream of the end of the blaCTX-M-15 gene with insertion sequence orf477 located downstream. (B) The ISEcp1 is the only insertion sequence located upstream of the blaCTX-M-15 gene. (C) ISEcp1 located upstream of blaCTX-M-15 truncated by IS26 without any downstream IS element. (D) IS26 was located upstream of blaCTX-M-15 with orf447 located downstream. (E) ISEcp1 was truncated upstream of blaCTX-M-15 by IS26 while orf477 was located downstream. (F) ISEcp1 was truncated upstream of blaCTX-M-15 by IS26, with IS903 located downstream. (G) Novel IS3 type truncated ISEcp1 upstream the start codon of blaCTX-M-15 gene.
Integrons were associated with blaCTX-M-15 genes in 13 studies [12,15,18,19,21,25,31,32,34,42,43,44,45]; a class 1 integron (Intl1) was reported in 13 studies, while a class 2 integron (Intl2) was reported in 3 studies together with a class 1 integron. A class 3 integron was not reported in all the studies reviewed, one gene cassette arrangement; Intl2/dfrA1-sat-aadA1 was detected in Intl2 in this review from only one study. However, with the exception of two studies, different gene cassette arrays were detected in variable regions of Intl1, with Intl1/dfrA17–aadA5 being the most reported gene cassette from 6 out of 43 studies reviewed (Table 1). Different types of plasmid incompatibility groups were reported to transfer blaCTX-M-15 gene horizontally. These plasmid groups include IncF, IncH, IncN, IncY, IncK, IncX, IncI, IncA, IncC, IncL, and IncM [14,16,25,30,32,33,34,35,36,38,39,42,43,45,46,47,48,49,50,51,52,53,54]. However, IncF plasmid was the most reported plasmid associated with blaCTX-M-15 gene from 17 out of the 43 studies reviewed (Table 1). Antimicrobial resistance genes including the narrow-spectrum blaOXA-1 and blaTEM-1 beta-lactamases, tetracycline resistance genes (tetA and tetB), sulfonamide resistance genes (sul2 and sul3) and plasmid-mediated quinolone resistance genes qnrA, qnrB, and qnrS and aminoglycoside-modifying enzyme encoding genes (aac-(6′)-lb-cr), were reportedly associated with blaCTX-M-15 genes on plasmids.

Table 1.
Genetic environment of CTXM-15 genes in enterobacterial species from Africa.
Table 1.
Genetic environment of CTXM-15 genes in enterobacterial species from Africa.
| References | Country | Sample Sources | Enterobacterial Species | Genetic Environment Pattern | Additional Resistance Genes | Mobile Genetic Elements | |
|---|---|---|---|---|---|---|---|
| Integron/Gene Cassettes | Plasmids | ||||||
| [12] | Nigeria | Environment | Escherichia coli | ISEcp1-IS26-orf477, ISEcp1-orf477 | Intl1/dfrA17-aadA5, Intl1/dfrA32-ereA-aadA2, Intl1/dfrA16-aadA2, Intl1/aadA1, Intl1/dfrA7, Int2 | ||
| [20] | Nigeria | Human | Escherichia coli | ISEcp1 | aac-(6′)-lb-cr, qnrB1, qnrA1 | ||
| [44] | Nigeria | Human | Enterobacter cloacae, Pantoea agglomerans | aadA1, aph, aac-(6′)-Ib), sul1, cat1, qnrB1, tet (A), tet (E) | Intl1, Intl2 | ||
| [21] | Nigeria | Human | Proteus mirabilis | ISEcp1 | aac (6′)-Ib-cr, qnrA, blaTEM-1 | Int1, Int2, aadA1, aadA1-qacH, aadB-aadA2, aadA5, dfrA7, dfrA15, dfrA17, dfrA17-aadA5 | |
| [47] | Nigeria | Human | Escherichia coli | aac (6′)-Ib-cr, qnrS1, qnrB1, qepA1, blaOXA-1, blaTEM-1, blaCMY-2 | IncFIA-FIB-FII, IncHI2, IncY, IncX, IncX2, IncI2 | ||
| [46] | Nigeria | Chicken, pig | Escherichia coli | qnrS1, blaTEM-1 | IncN | ||
| [42] | Nigeria | Human | Escherichia coli | ISEcp1, ISEcp1-IS26, ISEcp1-IS26-IS903 | qnrB, aac (6′)-lb-cr, blaTEM-1 | Int1/aadA1, int1/dfrA17-aadA5 | IncFIA, IncFIB, HI2 IncK |
| [13] | Nigeria | Human | Escherichia coli | ISEcp1-orf477 | aac (6′)-Ib-cr, blaOXA-1, blaSHV, blaTEM-1 | ||
| [48] | Nigeria | Human | Escherichia coli, Klebsiella spp. | blaCTXM-2, blaOXA-1, blaSHV, blaTEM-1, blaAmpC | IncF | ||
| [43] | Nigeria | Chicken | Escherichia coli | ISEcp1-IS26-orf477 | aac (3)-IIa, aac (6′)-Ib-cr, dfrA5, dfrA12, strA, strB, sul1, sul2, tet (A), tet (B), blaOXA-1, blaTEM-1 | Intl1/aadA2-orfF-dfrA12 | IncFIB, IncFIA-IncFIB-IncI1 |
| [22] | Nigeria | Human | Escherichia coli, Klebsiella spp., Proteus mirabilis | ISEcp1 | blaTEM, blaSHV | ||
| [23] | Nigeria | Human | Klebsiella spp. | ISEcp1 | tet(A), aac (3)-II, aac (6′)-Ib | ||
| [14] | Ghana | Human | Escherichia coli, Klebsiella spp. | ISEcp1- orf477 | blaTEM, aac (3)-II, blaOXA-30 | IncFII-FIA-FIB, IncFIIK | |
| [49] | Ghana | Human | Salmonella Poona | blaTEM-1B, blaOXA−1, qnrB1, aac (6′) Ib-cr, tet(A), dfrA15, sul2, catB3, strA, strB, aac (3)-Iia | TrfA-IncHI2-IncHI2A | ||
| [15] | Mauritania | Human | Escherichia coli | ISEcp1-orf477 | aac (6′)-Ib-cr, tet(A), sul2, sul3, strA, strB, blaOXA-1, blaTEM-1B | intI/dfrA17-aadA5 | |
| [24] | Niger | Human | Morganella morganii, Citrobacter freundii | ISEcp1 | blaDHA, blaCIT, blaTEM-1 | ||
| [50] | Niger | Human | Escherichia coli | blaCMY-2, blaSHV-44 | FII/FIA/FIB, FII/I1/Iγ, | ||
| [51] | Senegal | Human | Salmonella enterica | qnrB1, aac (6′)-Ib-cr | IncHI2, IncN, IncFII | ||
| [25] | Senegal | Human | Escherichia coli | ISEcp1 | blaTEM-1, blaOXA-1, aac (6)-Ib-cr, tet(A) | intI/dfrA17-aadA5 | IncFIA-FIB-FII |
| [26] | Senegal | Human | Salmonella Kentucky | ISEcp1 | blaTEM-1, blaOXA-30 | ||
| [52] | Sao Tome and Principle | Human | Escherichia coli | blaOXA-181, blaTEM-1, rmtB | IncX3 | ||
| [27] | DRC | Human | Salmonella Typhi | ISEcp1 | blaTEM-1D, sulI, dfrA7 | ||
| [53] | Central African Republic | Human | Escherichia coli, Enterobactercloacae | aac (6′)-Ib-cr, qnrB, qnrS | IncF | ||
| [28] | Cameroon | Human | Escherichia coli | ISEcp1 | blaOXA-181, blaTEM-1, aac (6′) Ib-cr | ||
| [54] | Cameroon | Human | Klebsiella spp. | sul1, fosA, oqxA, oqxB, blaTEM-1B, dfrA15, strA, strB | ColRNAI, IncFIB (K), IncFIA (HI1 | ||
| [29] | Egypt | Human | Escherichia coli | ISEcp1 | blaTEM-1 | ||
| [30] | Algeria | Human | Salmonella enterica ser Infantis | ISEcp1 | armA, blaTEM-1 | IncL, IncM | |
| [31] | Algeria | Human | Klebsiella spp. | ISEcp1 | blaTEM-1 | Int1 | |
| [16] | Angola | Human | Escherichia coli, Klebsiella spp. | ISEcp1-orf477, IS26-orf447, ISEcp1-IS3-orf477 | blaOXA-1, blaTEM-1, aac-6′-Ib-cr | IncFII, IncFIIK6, IncHI2 and IncY | |
| [45] | Angola | Dog | Escherichia coli | qepA, qnrS1, qnrB19, aac (6′)-Ib-cr | Intl1/dfrA17-aadA5, Intl1/dfrA1-aadA1, Intl2/dfrA1-sat-aadA1 | IncFIB, IncY, IncN, IncI1 | |
| [32] | Madagascar | Human | Escherichia coli, Klebsiella spp. | ISEcp1 | blaTEM-1, blaOXA-1, aac (6′)-Ib-cr, sul1-sul2, tet(A), qnrB | Intl1/ aadA1-aadA2-aadA4-aadA5-dfrA5-dfrA22 | IncFII-FIA-FIB, IncHI2 |
| [17] | Morocco | Human | Klebsiella spp. | ISEcp1-orf477 | blaTEM-1B , bla OXA-1 , aac (6′)- Ib-cr, qnrB1 | ||
| [33] | Morocco | Human | Klebsiella spp. | ISEcp1 | qnrB1, bla NDM-1 | IncH | |
| [34] | Kenya | Human | Salmonella Typhimurium | ISEcp1 | blaTEM-1, blaOXA-1, aac (6′)-Ib, sul1, sul2, aadA1 | Intl1/dfrA14-catA1 | IncFII, IncHI2 |
| [35] | Tanzania | Human | Escherichia coli | ISEcp1 | blaTEM-1 | IncFIA- FIB | |
| [41] | Tanzania | Human | Enterobacter spp. | ISEcp1 | |||
| [36] | Tanzania | Human | Klebsiella spp. | ISEcp1 | blaTEM-1, blaSHV-11 | IncFII, IncFIA | |
| [37] | Tunisia | Human | Escherichia coli | ISEcp1 | blaTEM-1, blaSHV-12 | ||
| [18] | Tunisia | Human | Escherichia coli | ISEcp1-orf477 | blaTEM-1, blaOXA-1, aac (3)-II, aac (6′)-Ib-cr, strA, strB, sul2, tet (B) | Intl1/ dfrA17–aadA5, Intl1/ dfrA12–orfF–aadA2, Intl1/aadA2 | |
| [38] | Tunisia | Human | Escherichia coli | ISEcp1 | blaTEM-52 | IncA, IncC | |
| [39] | Tunisia | Human | Klebsiella spp. | ISEcp1 | blaTEM-1, blaSHV-12 | IncFII, IncL, IncM | |
| [40] | Tunisia | Human | Escherichia coli | ISEcp1-IS26 | |||
| [19] | Tunisia | Human | Klebsiella spp. | ISEcp1- orf477 | blaTEM-1, blaOXA-1, blaSHV-1 | Intl1/ dfrA17–ereA2, Intl1/aadA | |
4. Discussion
This review was carried out to describe the genetic environments of the internationally disseminated blaCTX-M-15 gene in Enterobacteriaceae from Africa. Most of the studies in this review were from human clinical settings, which suggests that blaCTX-M-15-producing Enterobacteriaceae are a challenge to healthcare facilities in Africa. The blaCTX-M-15 gene has been associated with the pandemic-initiating E. coli O25: H4 ST131 clone that causes both community and human healthcare infections globally [2]. Review of the genetic environments of blaCTX-M-15 in Enterobacteriaceae revealed five ISs including ISEcp1, IS26, orf447, IS903, and IS3 which had been detected in Africa. With the exception of a novel IS3 type that was reported from Angola [16], all the other ISs have been reported from other parts of the world to be associated with the genetic environment of different AMR genes in general [7,8,55]. From all the studies reviewed, ISEcp1 was typically located upstream of blaCTX-M-15 gene. This IS often encodes a transposase that facilitates the mobilization of blaCTX-M-15 gene among integrons, transposons, plasmids, and chromosomes, as well as provides promoters that can activate the weakly expressed state of blaCTX-M-15 [56,57,58]. This IS has been reported to contribute to the global dissemination of blaCTX-M-15 genes in association with other MGEs [1,59,60]. The ISEcp1/ blaCTX-M-15 genetic association observed in this review has previously been reported from other parts of the world including India, France, Turkey, Poland, Canada, the United Kingdom, Spain, and China [8,9,10,55,56,58,61,62,63,64]. This IS element has also been commonly found associated with other blaCTX-M genes and other beta-lactamase resistance genes [10,59]. Of the genetic environments associated with blaCTX-M genes, ISEcp1 is one of the most commonly detected IS elements in the genetic environment of blaCTX-M genes, suggesting a possible co-evolutionary relationship between the ISEcp1 and blaCTX-M genes [9,52,61,65].
The insertion site of ISEcp1 was different from study to study in this review; this may be due to the variation in bacterial strains, IS promoter types, and other factors associated with genetic environments of the blaCTX-M-15 gene. Three studies provided information on the promoter regions in this review; the −35 and −10 putative promoter regions (48 bp) were reported in five studies, while V and W sequences (127 bp) were in one study. In all cases, these promoter regions are important in the transcription, mobilization, and expression of the blaCTX-M-15 gene as previously described [7,9,10]. IS26 was another IS described in Africa. However, this IS element was located upstream of blaCTX-M-15, disrupting ISEcp1 elements in all studies reporting the presence of IS26 and ISEcp1 in the genetic environment of blaCTX-M-15. IS26 has also been reported from other parts of the world to be associated with blaCTX-M genes alone without ISEcp1 [64] or associated with blaCTX-M genes together with and located upstream of ISEcp1 [55,56,66], or located truncating ISEcp1 [55,64] in genetic arrangements with blaCTX-M genes similar to the findings of this review. In all these genetic arrangements involving IS26, the IS was suggested to be associated with transposition and stabilization of the ISEcp1/ blaCTX-M-15 complex on plasmids [63,67].
The genetic environment downstream of the blaCTX-M-15 revealed flanking of the blaCTX-M-15 gene by two different types of insertion sequences, orf447 and IS903. Both IS elements are the major IS elements commonly reported downstream of blaCTX-M [8,68,69]. However, based on this review, orf447 is the major IS element downstream of blaCTX-M-15 gene in Africa. In this review, seven different genetic patterns were observed; four of the five genetic patterns have previously been reported. ISEcp1-blaCTX-M-15-orf477 genetic pattern has been reported from European and Indian strains of Enterobacteriaceae [55,61,66]; ISEcp1 blaCTX-M-15 has been reported from Spain, Canada, India, and Poland [64,70,71,72,73]; ISEcp1-IS26-blaCTX-M-15-orf447 has also been reported from France [55,74]; while the ISEcp1-IS3 blaCTX-M-15 pattern was reported to be novel from Angola [16]. Other genetic patterns have been reported in the genetic environments of other types of blaCTX-M and other beta-lactamase genes [8,61,75]. These genetic patterns from Africa reveal how the genetic environment of blaCTX-M-15 is consistent with what has been reported on global scales. Additionally, immigration, global migration, and traveling for tourism purposes could also contribute to these global genetic patterns of blaCTX-M-15. Similar genetic environments of blaCTX-M-15 reported in this review and other novel genetic patterns have previously been reported from travelers returning to the United Kingdom from the Middle East, Africa, and Asian countries, which suggests the possible overseas acquisition of these genetic patterns [66].
Class 1 integrons were more commonly associated with blaCTX-M-15 compared to class 2 integrons; this is consistent with previous reports elsewhere [8,76]. Class 1 integrons are often associated with IS elements such as ISEcp1 and ISCR1. These integrons are often located adjacent to ISEcp1 and ISCR1 and function in the mobilization and transposition of blaCTX-M-15 genes [8]. In addition, some AMR genes associated with blaCTX-M-15 are captured within the conserved regions of the class 1 integrons. AMR genes were harbored within the cassette arrays of class 1 integron in different studies in this review. Antimicrobial resistance genes including dfrA17, dfrA5, dfrA1, aadA5, aadA2, aadA1 and catA1 were observed within the conserved region of the class 1 integrons, and these genes often confer multi-drug resistance to trimethoprim, aminoglycosides, and chloramphenicol. Conjugative plasmids are essential for the evolution and global dissemination of the blaCTX-M-15 gene. Similar to this review, several studies have found that the narrow-host range plasmid IncF is the predominant plasmid group that harbors the blaCTX-M-15 gene [77]. The IncF plasmid is mainly restricted to Enterobacteriaceae with support mechanisms such as lower fitness cost, transferability properties, plasmid addiction, and stability systems that favor: (i) the higher prevalence of blaCTX-M-15 in Enterobacteriaceae compared to other Gram-negative bacteria; and (ii) global dissemination of blaCTX-M-15 in association with other mobile genetic elements [11,59,78]. The IncFII-FIA-FIB multi-replicon plasmids were more commonly associated with blaCTX-M-15 in this review and have been widely distributed in the Enterobacteriaceae, especially E. coli, globally [79,80]. This replicon group could be maintained and propagated between enterobacterial species and from host to host without antimicrobial selective pressure [59,77]. This may provide some explanation to the rapid and global spread of the blaCTX-M-15 gene. Another important finding of this review was the presence of other antimicrobial resistance associated with blaCTX-M-15 often co-located on the same plasmid. Different AMR genes commonly co-exist on plasmids, therefore facilitating the co-dissemination of resistance genes and greater survival fitness of bacteria under antimicrobial selective pressure [78]. Antimicrobial resistance genes including the narrow-spectrum blaOXA-1 and blaTEM-1 beta-lactamases, aminoglycoside-modifying enzymes (aac-(6′)-lb-cr), tetracycline resistance genes (tetA and tetB), sulfonamide resistance genes (sul2 and sul3) and plasmid-mediated quinolone resistance genes (qnrA, qnrB and qnrS) were found to be consistently associated with blaCTX-M-15 from different studies in the review. These AMR genes have previously been reported to be co-located on IncFII-FIA-FIB plasmid replicons in association with blaCTX-M-15-producing E. coli O25:H4-ST131 [81,82], conferring multi-drug resistance to different antimicrobial classes, complicating the genetic environments, and facilitating the global spread of blaCTX-M-15 in Enterobacteriaceae. In addition to the contribution of clonal spread of some bacteria of Enterobacteriaceae, especially E. coli and Klebsiella spp., the association of blaCTX-M-15 with mobile genetic elements such as insertion sequences, integrons, and conjugative plasmids may explain its global dominance and dissemination. This review has showed the diversity and the complexity of the genetic environments of blaCTX-M-15 beta-lactamase gene in Enterobacteriaceae from Africa. We recognize that a limited number of articles were included in this review, which was a limitation of this review. This is partly due to limited published articles on this subject matter in Africa. Our focus was to provide a narrative review that can serve as baseline literature for a future comprehensive and systematic review and indicate the need for more research on this internationally disseminated beta-lactamase resistance gene.
Author Contributions
Conceptualization, B.B.A.; Methodology, Formal Analysis and Literature Review, B.B.A., A.M.; Writing—Original Draft Preparation, B.B.A., A.M.; Writing—Review and Editing, B.B.A., A.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cantón, R.; González-Alba, J.M.; Galán, J.C. CTX-M enzymes: Origin and diffusion. Front. Microbiol. 2012, 3. [Google Scholar] [CrossRef]
- Peirano, G.; Pitout, J.D.D. Molecular epidemiology of Escherichia coli producing CTX-M β-lactamases: The worldwide emergence of clone ST131 O25:H4. Int. J. Antimicrob. Agents 2010, 35, 316–321. [Google Scholar] [CrossRef]
- Pitout, J.D.; Laupland, K.B. Extended-spectrum β-lactamase-producing Enterobacteriaceae: An emerging public-health concern. Lancet Infect. Dis. 2008, 8, 159–166. [Google Scholar] [CrossRef]
- Lee, H.; Doak, T.G.; Popodi, E.; Foster, P.L.; Tang, H. Insertion sequence-caused large-scale rearrangements in the genome of Escherichia coli. Nucleic Acids Res. 2016, 44, 7109–7119. [Google Scholar] [CrossRef] [PubMed]
- Nzabarushimana, E.; Tang, H. Insertion sequence elements-mediated structural variations in bacterial genomes. Mob. DNA 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Mahillon, J.; Chandler, M. Insertion sequences. Microbiol. Mol. Biol. Rev. 1998, 62, 725–774. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Naas, T.; Nordmann, P. Genetic support of extended-spectrum β-lactamases. Clin. Microbiol. Infect. 2008, 14, 75–81. [Google Scholar] [CrossRef]
- Zhao, W.-H.; Hu, Z.-Q. Epidemiology and genetics of CTX-M extended-spectrum β-lactamases in Gram-negative bacteria. Crit. Rev. Microbiol. 2013, 39, 79–101. [Google Scholar] [CrossRef]
- Karim, A.; Poirel, L.; Nagarajan, S.; Nordmann, P. Plasmid-mediated extended-spectrum beta-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol. Lett. 2001, 201, 237–241. [Google Scholar]
- Lartigue, M.-F.; Poirel, L.; Nordmann, P. Diversity of genetic environment of blaCTX-M genes. FEMS Microbiol. Lett. 2004, 234, 201–207. [Google Scholar] [CrossRef]
- Carattoli, A. Resistance plasmid families in enterobacteriaceae. Antimicrob. Agents Chemother. 2009, 53, 2227–2238. [Google Scholar] [CrossRef] [PubMed]
- Adelowo, O.O.; Caucci, S.; Banjo, O.A.; Nnanna, O.C.; Awotipe, E.O.; Peters, F.B.; Fagade, O.E.; Berendonk, T.U. Extended Spectrum Beta-Lactamase (ESBL)-producing bacteria isolated from hospital wastewaters, rivers and aquaculture sources in Nigeria. Environ. Sci. Pollut. Res. 2018, 25, 2744–2755. [Google Scholar] [CrossRef]
- Iroha, I.R.; Esimone, C.O.; Neumann, S.; Marlinghaus, L.; Korte, M.; Szabados, F.; Gatermann, S.; Kaase, M. First description of Escherichia coli producing CTX-M-15- extended spectrum beta lactamase (ESBL) in out-patients from south eastern Nigeria. Ann. Clin. Microbiol. Antimicrob. 2012, 11, 19. [Google Scholar] [CrossRef] [PubMed]
- Agyekum, A.; Fajardo-Lubián, A.; Ansong, D.; Partridge, S.R.; Agbenyega, T.; Iredell, J.R. blaCTX-M-15 carried by IncF-type plasmids is the dominant ESBL gene in Escherichia coli and Klebsiella pneumoniae at a hospital in Ghana. Diagn. Microbiol. Infect. Dis. 2016, 84, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Sallem, R.B.; Slama, K.B.; Estepa, V.; Cheikhna, E.O.; Mohamed, A.M.; Chairat, S.; Ruiz-Larrea, F.; Boudabous, A.; Torres, C. Detection of CTX-M-15-producing Escherichia coli isolates of lineages ST410-A, ST617-A and ST354-D in faecal samples of hospitalized patients in a Mauritanian hospital. J. Chemother. 2015, 27, 114–116. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, T.G.; Novais, Â.; Peixe, L.; Machado, E. Atypical epidemiology of CTX-M-15 among Enterobacteriaceae from a high diversity of non-clinical niches in Angola: Table 1. J. Antimicrob. Chemother. 2016, 71, 1169–1173. [Google Scholar] [CrossRef]
- Barguigua, A.; El Otmani, F.; Talmi, M.; Reguig, A.; Jamali, L.; Zerouali, K.; Timinouni, M. Prevalence and genotypic analysis of plasmid-mediated β-lactamases among urinary Klebsiella pneumoniae isolates in Moroccan community. J. Antibiot. 2013, 66, 11–16. [Google Scholar] [CrossRef]
- Ben Slama, K.; Ben Sallem, R.; Jouini, A.; Rachid, S.; Moussa, L.; Sáenz, Y.; Estepa, V.; Somalo, S.; Boudabous, A.; Torres, C. Diversity of genetic lineages among CTX-M-15 and CTX-M-14 producing Escherichia coli strains in a Tunisian hospital. Curr. Microbiol. 2011, 62, 1794–1801. [Google Scholar] [CrossRef]
- Abbassi, M.S.; Torres, C.; Achour, W.; Vinué, L.; Sáenz, Y.; Costa, D.; Bouchami, O.; Hassen, A.B. Genetic characterisation of CTX-M-15-producing Klebsiella pneumoniae and Escherichia coli strains isolated from stem cell transplant patients in Tunisia. Int. J. Antimicrob. Agents 2008, 32, 308–314. [Google Scholar] [CrossRef]
- Aibinu, I.; Odugbemi, T.; Koenig, W.; Ghebremedhin, B. Sequence type ST131 and ST10 complex (ST617) predominant among CTX-M-15-producing Escherichia coli isolates from Nigeria. Clin. Microbiol. Infect. 2012, 18, E49–E51. [Google Scholar] [CrossRef][Green Version]
- Alabi, O.S.; Mendonça, N.; Adeleke, O.E.; Da Silva, G.J. Molecular screening of antibiotic-resistant determinants among multidrug-resistant clinical isolates of Proteus mirabilis from South West Nigeria. Afr. Health Sci. 2017, 17, 356. [Google Scholar] [CrossRef]
- Raji, M.A.; Jamal, W.; Ojemeh, O.; Rotimi, V.O. Sequence analysis of genes mediating extended-spectrum beta-lactamase (ESBL) production in isolates of Enterobacteriaceae in a Lagos Teaching Hospital, Nigeria. BMC Infect. Dis. 2015, 15. [Google Scholar] [CrossRef]
- Soge, O.O.; Queenan, A.M.; Ojo, K.K.; Adeniyi, B.A.; Roberts, M.C. CTX-M-15 extended-spectrum β-lactamase from Nigerian Klebsiella pneumoniae. J. Antimicrob. Chemother. 2006, 57, 24–30. [Google Scholar] [CrossRef]
- Soleimanian, S.; Gordon, N.C.; Wareham, D.W. Polymicrobial necrotizing fasciitis involving enterobacteria producing CTX-M-15 extended-spectrum -lactamases. J. Med. Microbiol. 2011, 60, 135–137. [Google Scholar] [CrossRef]
- Ruppe, E.; Woerther, P.-L.; Diop, A.; Sene, A.-M.; Da Costa, A.; Arlet, G.; Andremont, A.; Rouveix, B. Carriage of CTX-M-15-producing Escherichia coli isolates among children living in a remote village in Senegal. Antimicrob. Agents Chemother. 2009, 53, 3135–3137. [Google Scholar] [CrossRef]
- Weill, F.-X.; Perrier-Gros-Claude, J.-D.; Demartin, M.; Coignard, S.; Grimont, P.A.D. Characterization of extended-spectrum-Î2-lactamase (CTX-M-15)-producing strains of Salmonella enterica isolated in France and Senegal. FEMS Microbiol. Lett. 2004, 238, 353–358. [Google Scholar]
- Phoba, M.-F.; Barbé, B.; Lunguya, O.; Masendu, L.; Lulengwa, D.; Dougan, G.; Wong, V.K.; Bertrand, S.; Ceyssens, P.J.; Jacobs, J.; et al. Salmonella enterica serovar typhi producing CTX-M-15 extended spectrum β-lactamase in the democratic Republic of the Congo. Clin. Infect. Dis. 2017, 65, 1229–1231. [Google Scholar] [CrossRef] [PubMed]
- Lonchel Magoué, C.; Melin, P.; Gangoué-Piéboji, J.; Assoumou, M.-C.O.; Boreux, R.; De Mol, P. Prevalence and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae in Ngaoundere, Cameroon. Clin. Microbiol. Infect. 2013, 19, E416–E420. [Google Scholar] [CrossRef] [PubMed]
- Khalaf, N.G.; Eletreby, M.M.; Hanson, N.D. Characterization of CTX-M ESBLs in Enterobacter cloacae, Escherichia coli and Klebsiella pneumoniae clinical isolates from Cairo, Egypt. BMC Infect. Dis. 2009, 9. [Google Scholar] [CrossRef]
- Naas, T.; Bentchouala, C.; Cuzon, G.; Yaou, S.; Lezzar, A.; Smati, F.; Nordmann, P. Outbreak of Salmonella enterica serotype Infantis producing ArmA 16S RNA methylase and CTX-M-15 extended-spectrum β-lactamase in a neonatology ward in Constantine, Algeria. Int. J. Antimicrob. Agents 2011, 38, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Messai, Y.; Iabadene, H.; Benhassine, T.; Alouache, S.; Tazir, M.; Gautier, V.; Arlet, G.; Bakour, R. Prevalence and characterization of extended-spectrum β-lactamases in Klebsiella pneumoniae in Algiers hospitals (Algeria). Pathol. Biol. 2008, 56, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Rakotonirina, H.C.; Garin, B.; Randrianirina, F.; Richard, V.; Talarmin, A.; Arlet, G. Molecular characterization of multidrug-resistant extended-spectrum β-lactamase-producing Enterobacteriaceae isolated in Antananarivo, Madagascar. BMC Microbiol. 2013, 13, 85. [Google Scholar] [CrossRef]
- Villa, L.; Poirel, L.; Nordmann, P.; Carta, C.; Carattoli, A. Complete sequencing of an IncH plasmid carrying the blaNDM-1, blaCTX-M-15 and qnrB1 genes. J. Antimicrob. Chemother. 2012, 67, 1645–1650. [Google Scholar] [CrossRef]
- Kariuki, S.; Okoro, C.; Kiiru, J.; Njoroge, S.; Omuse, G.; Langridge, G.; Kingsley, R.A.; Dougan, G.; Revathi, G. Ceftriaxone-resistant Salmonella enterica serotype typhimurium sequence type 313 from Kenyan patients is associated with the bla CTX-M-15 gene on a novel IncHI2 plasmid. Antimicrob. Agents Chemother. 2015, 59, 3133–3139. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mshana, S.E.; Imirzalioglu, C.; Hain, T.; Domann, E.; Lyamuya, E.F.; Chakraborty, T. Multiple ST clonal complexes, with a predominance of ST131, of Escherichia coli harbouring blaCTX-M-15 in a tertiary hospital in Tanzania. Clin. Microbiol. Infect. 2011, 17, 1279–1282. [Google Scholar] [CrossRef] [PubMed]
- Mshana, S.E.; Hain, T.; Domann, E.; Lyamuya, E.F.; Chakraborty, T.; Imirzalioglu, C. Predominance of Klebsiella pneumoniaeST14 carrying CTX-M-15 causing neonatal sepsis in Tanzania. BMC Infect. Dis. 2013, 13. [Google Scholar] [CrossRef] [PubMed]
- Réjiba, S.; Mercuri, P.S.; Power, P.; Kechrid, A. Emergence and dominance of CTX-M-15 extended spectrum beta-lactamase among Escherichia coli isolates from children. Microb. Drug Resist. 2011, 17, 135–140. [Google Scholar] [CrossRef]
- Chouchani, C.; Salabi, A.; Marrakchi, R.; Ferchichi, L.; Walsh, T.R. Characterization of IncA/C conjugative plasmid harboring bla TEM-52 and bla CTX-M-15 extended-spectrum β-lactamases in clinical isolates of Escherichia coli in Tunisia. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 1081–1087. [Google Scholar] [CrossRef]
- Elhani, D.; Bakir, L.; Aouni, M.; Passet, V.; Arlet, G.; Brisse, S.; Weill, F.-X. Molecular epidemiology of extended-spectrum β-lactamase-producing Klebsiella pneumoniae strains in a university hospital in Tunis, Tunisia, 1999–2005. Clin. Microbiol. Infect. 2010, 16, 157–164. [Google Scholar] [CrossRef]
- Ayari, K.; Bourouis, A.; Chihi, H.; Mahrouki, S.; Naas, T.; Belhadj, O. Dissemination and genetic support of broad-spectrum beta-lactam-resistant Escherichia coli strain isolated from two Tunisian hospitals during 2004–2012. Afr. Health Sci. 2017, 17, 346. [Google Scholar] [CrossRef]
- Mshana, S.E.; Gerwing, L.; Minde, M.; Hain, T.; Domann, E.; Lyamuya, E.; Chakraborty, T.; Imirzalioglu, C. Outbreak of a novel Enterobacter sp. carrying blaCTX-M-15 in a neonatal unit of a tertiary care hospital in Tanzania. Int. J. Antimicrob. Agents 2011. [Google Scholar] [CrossRef]
- Inwezerua, C.; Mendonça, N.; Calhau, V.; Domingues, S.; Adeleke, O.E.; Da Silva, G.J. Occurrence of extended-spectrum beta-lactamases in human and bovine isolates of Escherichia coli from Oyo state, Nigeria. J. Infect. Dev. Ctries. 2014, 8. [Google Scholar] [CrossRef]
- Ojo, O.E.; Schwarz, S.; Michael, G.B. Detection and characterization of extended-spectrum β-lactamase-producing Escherichia coli from chicken production chains in Nigeria. Vet. Microbiol. 2016, 194, 62–68. [Google Scholar] [CrossRef]
- Aibinu, I.; Pfeifer, Y.; Peters, F.; Ogunsola, F.; Adenipekun, E.; Odugbemi, T.; Keonig, W. Emergence of blaCTX-M-15, qnrB1 and aac(6′)-Ib-cr resistance genes in Pantoea agglomerans and Enterobacter cloacae from Nigeria (sub-Saharan Africa). J. Med. Microbiol. 2012, 61, 165–167. [Google Scholar] [CrossRef][Green Version]
- Albrechtova, K.; Kubelova, M.; Mazancova, J.; Dolejska, M.; Literak, I.; Cizek, A. High prevalence and variability of CTX-M-15-producing and fluoroquinolone-resistant Escherichia coli observed in stray dogs in rural angola. Microb. Drug Resist. 2014, 20, 372–375. [Google Scholar] [CrossRef]
- Fortini, D.; Fashae, K.; Garcia-Fernandez, A.; Villa, L.; Carattoli, A. Plasmid-mediated quinolone resistance and -lactamases in Escherichia coli from healthy animals from Nigeria. J. Antimicrob. Chemother. 2011, 66, 1269–1272. [Google Scholar] [CrossRef]
- Fortini, D.; Fashae, K.; Villa, L.; Feudi, C.; García-Fernández, A.; Carattoli, A. A novel plasmid carrying blaCTX-M-15 identified in commensal Escherichia coli from healthy pregnant women in Ibadan, Nigeria. J. Glob. Antimicrob. Resist. 2015, 3, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Ogbolu, D.O.; Alli, O.A.T.; Olanipekun, L.B.; Ojo, O.I.; Makinde, O.O. Faecal carriage of extended-spectrum beta-lactamase (ESBL)-producing commensal Klebsiella pneumoniae and Escherichia coli from hospital out-patients in Southern Nigeria. Int. J. Med. Med. Sci. 2013, 9, 97–105. [Google Scholar] [CrossRef]
- Kudirkiene, E.; Andoh, L.A.; Ahmed, S.; Herrero-Fresno, A.; Dalsgaard, A.; Obiri-Danso, K.; Olsen, J.E. The use of a combined bioinformatics approach to locate antibiotic resistance genes on plasmids from whole genome sequences of Salmonella enterica serovars from humans in Ghana. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Woerther, P.-L.; Angebault, C.; Jacquier, H.; Hugede, H.-C.; Janssens, A.-C.; Sayadi, S.; El Mniai, A.; Armand-Lefevre, L.; Ruppe, E.; Barbier, F.; et al. Massive increase, spread, and exchange of extended spectrum β-lactamase-encoding genes among intestinal enterobacteriaceae in hospitalized children with severe acute malnutrition in Niger. Clin. Infect. Dis. 2011, 53, 677–685. [Google Scholar] [CrossRef]
- Harrois, D.; Breurec, S.; Seck, A.; Delauné, A.; Hello, S.L.; de la Gándara, M.P.; Sontag, L.; Perrier-Gros-Claude, J.-D.; Sire, J.-M.; Garin, B.; et al. Prevalence and characterization of extended-spectrum β-lactamase-producing clinical Salmonella enterica isolates in Dakar, Senegal, from 1999 to 2009. Clin. Microbiol. Infect. 2014, 20, O109–O116. [Google Scholar] [CrossRef]
- Poirel, L.; Aires-de-Sousa, M.; Kudyba, P.; Kieffer, N.; Nordmann, P. Screening and characterization of multidrug-resistant gram-negative bacteria from a remote African Area, São Tomé and Príncipe. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [PubMed]
- Rafaï, C.; Frank, T.; Manirakiza, A.; Gaudeuille, A.; Mbecko, J.-R.; Nghario, L.; Serdouma, E.; Tekpa, B.; Garin, B.; Breurec, S. Dissemination of IncF-type plasmids in multiresistant CTX-M-15-producing Enterobacteriaceae isolates from surgical-site infections in Bangui, Central African Republic. BMC Microbiol. 2015, 15, 15. [Google Scholar] [CrossRef] [PubMed]
- Founou, L.L.; Founou, R.C.; Allam, M.; Ismail, A.; Djoko, C.F.; Essack, S.Y. Genome sequencing of extended-spectrum β-lactamase (esbl)-producing Klebsiella pneumoniae isolated from pigs and abattoir workers in cameroon. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Eckert, C.; Gautier, V.; Arlet, G. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 2006, 57, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Ensor, V.M.; Shahid, M.; Evans, J.T.; Hawkey, P.M. Occurrence, prevalence and genetic environment of CTX-M β-lactamases in Enterobacteriaceae from Indian hospitals. J. Antimicrob. Chemother. 2006, 58, 1260–1263. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gourbeyre, E.; Siguier, P.; Chandler, M. Bacterial insertion sequences: Their genomic impact and diversity. FEMS Microbiol. Rev. 2014, 38, 865–891. [Google Scholar]
- Lartigue, M.-F.; Poirel, L.; Aubert, D.; Nordmann, P. In vitro analysis of ISEcp1B-mediated mobilization of naturally occurring β-lactamase gene blaCTX-M of Kluyvera ascorbata. Antimicrob. Agents Chemother. 2006, 50, 1282–1286. [Google Scholar] [CrossRef]
- Bevan, E.R.; Jones, A.M.; Hawkey, P.M. Global epidemiology of CTX-M β-lactamases: Temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 2017, 72, 2145–2155. [Google Scholar] [CrossRef]
- Cantón, R.; Coque, T.M. The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 2006, 9, 466–475. [Google Scholar] [CrossRef]
- Singh, N.S.; Singhal, N.; Virdi, J.S. Genetic environment of blaTEM-1, blaCTX-M-15, blaCMY-42 and characterization of ontegrons of Escherichia coli isolated from an Indian urban aquatic environment. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Shahid, M.; Sobia, F.; Singh, A.; Khan, H.M. Concurrent occurrence of blaampC families and blaCTX-M genogroups and association with mobile genetic elements ISEcp1, IS26, ISCR1, and sul1-type class 1 integrons in Escherichia coli and Klebsiella pneumoniae isolates originating from India. J. Clin. Microbiol. 2012, 50, 1779–1782. [Google Scholar] [CrossRef]
- Smet, A.; Van Nieuwerburgh, F.; Vandekerckhove, T.T.M.; Martel, A.; Deforce, D.; Butaye, P.; Haesebrouck, F. Complete nucleotide sequence of CTX-M-15-plasmids from clinical Escherichia coli isolates: Insertional events of transposons and insertion sequences. PLoS ONE 2010, 5, e11202. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Etayo, L.; Berzosa, M.; González, D.; Vitas, A. Prevalence of integrons and insertion sequences in ESBL-producing, E. coli isolated from different sources in Navarra, Spain. Int. J. Environ. Res. Public Health 2018, 15, 2308. [Google Scholar] [CrossRef] [PubMed]
- Saladin, M. Diversity of CTX-M β-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 2002, 209, 161–168. [Google Scholar] [CrossRef]
- Dhanji, H.; Patel, R.; Wall, R.; Doumith, M.; Patel, B.; Hope, R.; Livermore, D.M.; Woodford, N. Variation in the genetic environments of blaCTX-M-15 in Escherichia coli from the faeces of travellers returning to the United Kingdom. J. Antimicrob. Chemother. 2011, 66, 1005–1012. [Google Scholar] [CrossRef]
- Johnson, T.J.; Danzeisen, J.L.; Youmans, B.; Case, K.; Llop, K.; Munoz-Aguayo, J.; Flores-Figueroa, C.; Aziz, M.; Stoesser, N.; Sokurenko, E.; et al. Separate F-type plasmids have shaped the evolution of the H 30 Subclone of Escherichia coli sequence type 131. mSphere 2016, 1. [Google Scholar] [CrossRef]
- Poirel, L.; Decousser, J.-W.; Nordmann, P. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob. Agents Chemother. 2003, 47, 2938–2945. [Google Scholar] [CrossRef]
- Poirel, L.; Lartigue, M.-F.; Decousser, J.-W.; Nordmann, P. ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob. Agents Chemother. 2005, 49, 447–450. [Google Scholar] [CrossRef]
- Gómez-Garcés, J.L.; Saéz, D.; Almagro, M.; Fernández-Romero, S.; Merino, F.; Campos, J.; Oteo, J. Osteomyelitis associated to CTX-M-15-producing Aeromonas hydrophila: First description in the literature. Diagn. Microbiol. Infect. Dis. 2011, 70, 420–422. [Google Scholar] [CrossRef] [PubMed]
- Boyd, D.A.; Tyler, S.; Christianson, S.; McGeer, A.; Muller, M.P.; Willey, B.M.; Bryce, E.; Gardam, M.; Nordmann, P.; Mulvey, M.R. Complete nucleotide sequence of a 92-Kilobase plasmid harboring the CTX-M-15 extended-spectrum beta-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob. Agents Chemother. 2004, 48, 3758–3764. [Google Scholar] [CrossRef]
- Baraniak, A. Ceftazidime-hydrolysing CTX-M-15 extended-spectrum beta-lactamase (ESBL) in Poland. J. Antimicrob. Chemother. 2002, 50, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.; Hussain, A.; Mishra, S.; Maurya, A.P.; Bhattacharjee, A.; Joshi, S.R. Genetic environment of plasmid mediated CTX-M-15 extended spectrum beta-lactamases from clinical and food borne bacteria in North-Eastern India. PLoS ONE 2015, 10, e0138056. [Google Scholar] [CrossRef]
- Fabre, L.; Delauné, A.; Espié, E.; Nygard, K.; Pardos de la Gandara, M.; Polomack, L.; Guesnier, F.; Galimand, M.; Lassen, J.; Weill, F.X. Chromosomal integration of the extended-spectrum beta-lactamase gene blaCTX-M-15 in Salmonella enterica serotype Concord isolates from internationally adopted children. Antimicrob. Agents Chemother. 2009, 53, 1808–1816. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, J.; Stephan, R.; Zurfluh, K.; Hachler, H.; Fanning, S. Characterization of the genetic environment of blaESBL genes, integrons and toxin-antitoxin systems identified on large transferrable plasmids in multi-drug resistant Escherichia coli. Front. Microbiol. 2015, 5. [Google Scholar] [CrossRef]
- Kaushik, M.; Kumar, S.; Kapoor, R.K.; Virdi, J.S.; Gulati, P. Integrons in Enterobacteriaceae: Diversity, distribution and epidemiology. Int. J. Antimicrob. Agents 2018, 51, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Lahlaoui, H.; Ben Haj Khalifa, A.; Ben Moussa, M. Epidemiology of Enterobacteriaceae producing CTX-M type extended spectrum β-lactamase (ESBL). Med. Mal. Infect. 2014, 44, 400–404. [Google Scholar] [CrossRef]
- Woodford, N.; Carattoli, A.; Karisik, E.; Underwood, A.; Ellington, M.J.; Livermore, D.M. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob. Agents Chemother. 2009, 53, 4472–4482. [Google Scholar] [CrossRef]
- Nicolas-Chanoine, M.-H.; Blanco, J.; Leflon-Guibout, V.; Demarty, R.; Alonso, M.P.; Caniça, M.M.; Park, Y.J.; Lavigne, J.P.; Pitout, J.; Johnson, J.R. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 2007, 61, 273–281. [Google Scholar] [CrossRef]
- Rogers, B.A.; Sidjabat, H.E.; Paterson, D.L. Escherichia coli O25b-ST131: A pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 2010, 66, 1–14. [Google Scholar] [CrossRef]
- Marcadé, G.; Deschamps, C.; Boyd, A.; Gautier, V.; Picard, B.; Branger, C.; Denamur, E.; Arlet, G. Replicon typing of plasmids in Escherichia coli producing extended-spectrum β-lactamases. J. Antimicrob. Chemother. 2009, 63, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Zurfluh, K.; Glier, M.; Hächler, H.; Stephan, R. Replicon typing of plasmids carrying blaCTX-M-15 among Enterobacteriaceae isolated at the environment, livestock and human interface. Sci. Total Environ. 2015, 521–522, 75–78. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).