Metabacillus schmidteae sp. nov., Cultivated from Planarian Schmidtea mediterranea Microbiota
Abstract
1. Introduction
2. Materials and Methods
2.1. Planarian Schmidtea Mediterranea Culture
2.2. Culture Conditions from Schmidtea mediterranea and Strain Isolation
2.3. DNA Extraction, Sequencing, Assembly, and Annotation
2.4. Phylogenetic Analysis
2.5. Genomic Comparison
2.6. Phenotypic and Biochemical Features
2.7. Antibiotic Susceptibility
2.8. Chemotaxonomic Analysis
3. Results
3.1. Phenotypic and Biochemical Characteristics
3.2. Phylogenetic Analysis
| Property | Term |
|---|---|
| Current classification | Kingdom: Bacteria [69] |
| Phylum: Firmicutes [16] | |
| Class: Bacilli [70] | |
| Order: Bacillales [71] | |
| Family: Bacillaceae [15] | |
| Genus: Metabacillus [11] | |
| Species name: Schmidteae | |
| Specific epithet: Metabacillus schmidteae | |
| Type strain: Marseille-P9898 | |
| Species status | sp. nov. |
| Gram stain | Negative |
| Cell shape | Rod-shape |
| Motility | Motile |
| Sporulation | Spore-forming |
| Temperature range for growth | 19 to 50 |
| Temperature optimum | 28 |
| pH range for growth | 7.5 to 10 |
| pH optimum | 8 |
| pH category | alkaline |
| Lowest NaCl concentration for growth | 0 |
| Highest NaCl concentration for growth | 12 g/L |
| Salinity optimum | 10 g/L |
| O2 conditions for strain testing | Strict aerobic |
| Catalase | Positive |
| Oxidase | Negative |
| Habitat | Planarians |
| Biotic relationship | Microbiota planarian |
| Property | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Gram-staining | − | + | + | − |
| Mobility | Motile | Motile | Motile | Motile |
| Sporulation | + | + | + | + |
| Growth temperature range (°C) | 19–50 | 15–40 | 20–45 | 4–45 |
| Aerobic growth | Strict aerobic | Aerobic | Strict aerobic | Strict aerobic |
| Source | Planarians | Cotton waste | Lake | Tidal flat |
| Colony color | White | Yellowish white | Cream | Yellowish white |
| Catalase | + | + | + | + |
| Oxidase | − | − | + | + |
| ENZYMATIC ACTIVITIES | ||||
| 2-nitrophenyl-βD-galactopyranoside (ONPG) | − | + | NA | NA |
| 4-Nitrophenyl-βD-Galactopyranoside | + | + | NA | NA |
| Acid phosphatase | − | − | NA | + |
| Alkaline phosphatase | + | − | NA | + |
| Cystine arylamidase | − | − | − | − |
| Esculin ferric citrate (hydolyse) | + | + | + | NA |
| Esterase (C4) | + | + | NA | + |
| Esterase lipase (C8) | + | + | NA | + |
| Gelatin | − | − | + | + |
| Indole production | − | − | − | − |
| L-arginin | − | − | − | − |
| Leucine arylamidase | − | − | − | − |
| Lipase (C14) | − | − | NA | − |
| L-lysin | − | − | − | − |
| L-ormithin | − | − | − | − |
| N-acetyl-β-glucosaminidase | − | − | NA | − |
| Naphtol-AS-BI-phosphohydrolase | + | + | NA | + |
| Natriumpyruvat | + | − | NA | − |
| Natriumthiosulfat | − | − | − | − |
| Trinatriumcitrat | − | − | NA | NA |
| Trypsin | − | − | NA | − |
| Urea | − | − | − | − |
| Valine arylamidase | − | − | − | − |
| α-chymotrypsin | − | − | NA | − |
| α-fucosidase | − | − | NA | − |
| α-galactosidase | − | − | NA | − |
| α-glucosidase | + | − | NA | − |
| α-mannosidase | − | − | NA | − |
| β-glucosidase | − | + | NA | − |
| β-glucuronidase | + | − | NA | − |
| CARBOHYDRATE ASSIMILATION | ||||
| Adipic acid | − | − | NA | NA |
| Amygdalin | + | + | NA | NA |
| Arbutin | − | + | NA | NA |
| Capric acid | − | − | NA | NA |
| D-adonitol | − | − | NA | − |
| D-arabinose | − | − | + | NA |
| D-arabitol | + | − | NA | NA |
| D-cellobiose | + | + | + | + |
| D-fructose | + | + | + | + |
| D-fucose | − | − | NA | NA |
| D-galactose | + | + | + | + |
| D-glucose | + | + | + | + |
| D-lactose | + | + | + | + |
| D-lyxose | + | − | NA | NA |
| D-maltose | + | − | + | + |
| D-mannitol | + | − | + | − |
| D-mannose | + | + | + | − |
| D-melezitose | + | − | NA | + |
| D-melibiose | + | + | + | + |
| D-raffinose | + | + | + | + |
| D-ribose | − | − | + | + |
| D-saccharose | + | + | + | + |
| D-sorbitol | − | + | NA | − |
| D-tagatose | − | − | NA | NA |
| D-trehalose | + | + | + | + |
| D-turanose | − | − | NA | NA |
| Dulcitol | − | − | NA | NA |
| D-xylose | + | + | + | + |
| Erythritol | − | − | NA | NA |
| Gentiobiose | + | + | NA | NA |
| Glycerol | + | + | + | NA |
| Glycogen | − | + | NA | NA |
| Inositol | − | + | NA | − |
| Inulin | + | + | NA | NA |
| L-arabinose | + | - | + | + |
| L-arabitol | − | − | NA | NA |
| L-fucose | − | − | NA | NA |
| L-rhamnose | − | + | NA | + |
| L-sorbose | − | − | NA | NA |
| L-xylose | − | − | + | NA |
| Malic acid | + | − | NA | + |
| Methyl-αD-glucopyranoside | + | − | − | NA |
| Methyl-αD-mannopyranoside | − | − | − | NA |
| Methyl-βD-xylopyranoside | + | + | NA | NA |
| N-Acetyl-glucosamine | + | − | NA | NA |
| Phenylacetic acid | − | − | NA | NA |
| Potassium 2-ketoGluconate | − | − | NA | NA |
| Potassium 5-ketogluconate | − | − | NA | NA |
| Potassium gluconate | + | − | NA | NA |
| Salicin | + | + | + | NA |
| Starch | + | + | + | + |
| Trisodium citrate | + | − | NA | − |
| Xylitol | − | − | NA | NA |
| SUBSTRAT REDUCTION | ||||
| L-tryptophan | + | + | NA | − |
| Potassium nitrate | − | + | − | NA |
| Name | Size (bp) | GC% | Contigs | Refseq |
|---|---|---|---|---|
| Bacillus flexus | 3,906,163 | 37.6 | 259 | BCVD01000001.1 |
| Bacillus acidicola | 5,137,992 | 39.4 | 10 | LWJG01000001.1 |
| Mesobacillus foraminis | 5,730,823 | 43.0 | 35 | SLVV01000001.1 |
| Metabacillus halosaccharovorans | 5,399,327 | 36.1 | 8 | MTIR01000001.1 |
| Metabacillus litoralis | 5,230,624 | 35.9 | 1 | NZ_CP033043.1 |
| Metabacillus niabensis | 4,987,608 | 35.5 | 462 | NZ_CADEPK010000000 |
| Neobacillus niacini | 2,201,253 | 38.3 | 143 | JRYQ01000001.1 |
| Metabacillus schmidteae | 5,499,502 | 35.8 | 3 | CAESCH000000000.1 |
| Mesobacillus subterraneus | 4,571,170 | 43.9 | 42 | RSFW01000001.1 |
| Bacillus frigoritolerans | 5,475,560 | 43.9 | 1 | NZ_CP030063.1 |
3.3. Genomic Comparison
3.4. Chemotaxonomic Analysis
3.5. Antibiotic Susceptibility
4. Discussion
5. Conclusions
5.1. Protologue
5.2. Nucleotide Sequence Accession Number
5.3. Deposit in Culture Collections
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DSMZ | Deutsche Sammlung von Mikroorganismen und Zellkulturen |
| CSUR | Collection de Souches de l′Unité des Rickettsies |
| dDDH | digital DNA-DNA hybridization |
| GGDC | Genome-to-Genome Distance Calculator |
| MALDI-TOF-MS | Matrix-Assisted Laser Desorption Ionization Time-Of-Flight Mass Spectrometry |
| MEGA | Molecular Evolutionary Genetics Analysis |
| FAME | Fatty Acid Methyl Ester |
References
- Ramasamy, D.; Mishra, A.K.; Lagier, J.-C.; Padhmanabhan, R.; Rossi, M.; Sentausa, E.; Raoult, D.; Fournier, P.-E. A Polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int. J. Syst. Evol. Microbiol. 2014, 64 Pt 2, 384–391. [Google Scholar] [CrossRef]
- Fournier, P.-E.; Lagier, J.-C.; Dubourg, G.; Raoult, D. From Culturomics to Taxonomogenomics: A need to change the taxonomy of prokaryotes in clinical microbiology. Anaerobe 2015, 36, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Morel, A.-S.; Dubourg, G.; Prudent, E.; Edouard, S.; Gouriet, F.; Casalta, J.-P.; Fenollar, F.; Fournier, P.E.; Drancourt, M.; Raoult, D. Complementarity between targeted real-time specific PCR and conventional Broad-Range 16S rDNA PCR in the syndrome-driven diagnosis of infectious diseases. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Tindall, B.J.; Rosselló-Móra, R.; Busse, H.J.; Ludwig, W.; Kämpfer, P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 2010, 60 Pt 1, 249–266. [Google Scholar] [CrossRef]
- Drancourt, M.; Berger, P.; Raoult, D. Systematic 16S rRNA gene sequencing of atypical clinical isolates identified 27 new bacterial species associated with Humans. J. Clin. Microbiol. 2004, 42, 2197–2202. [Google Scholar] [CrossRef] [PubMed]
- Wayne, L.G. International committee on systematic bacteriology: Announcement of the report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Zent. Bakteriol. Mikrobiol. Hyg. Ser. A Med. Microbiol. Infect. Dis. Virol. Parasitol. 1988, 268, 433–434. [Google Scholar] [CrossRef]
- Abnave, P.; Mottola, G.; Gimenez, G.; Boucherit, N.; Trouplin, V.; Torre, C.; Conti, F.; Ben Amara, A.; Lepolard, C.; Djian, B.; et al. Screening in planarians identifies Morn2 as a key component in lc3-associated phagocytosis and resistance to bacterial infection. Cell Host. Microbe 2014, 16, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Maciel, E.I.; Jiang, C.; Barghouth, P.G.; Nobile, C.J.; Oviedo, N.J. The planarian schmidtea mediterranea is a new model to study host-pathogen interactions during fungal infections. Dev. Comp. Immunol. 2019, 93, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Torre, C.; Ghigo, É. Planaria: An immortal worm to clarify human immune response. Med. Sci. 2015, 31, 20–22. [Google Scholar] [CrossRef][Green Version]
- Seng, P.; Abat, C.; Rolain, J.M.; Colson, P.; Lagier, J.-C.; Gouriet, F.; Fournier, P.E.; Drancourt, M.; La Scola, B.; Raoult, D. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: Impact of Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry. J. Clin. Microbiol. 2013, 51, 2182–2194. [Google Scholar] [CrossRef]
- Patel, S.; Gupta, R.S. A phylogenomic and comparative genomic framework for resolving the polyphyly of the genus Bacillus: Proposal for six new genera of Bacillus species, Peribacillus gen. nov., Cytobacillus gen. nov., Mesobacillus gen. nov., Neobacillus gen. nov., Metabacillus gen. nov. and Alkalihalobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 2020, 70, 406–438. [Google Scholar] [CrossRef] [PubMed]
- Fritze, D. Taxonomy of the Genus Bacillus and Related Genera: The aerobic endospore-forming bacteria. Phytopathology 2004, 94, 1245–1248. [Google Scholar] [CrossRef] [PubMed]
- Cohn, F. Untersuchungen über Bacterien. Beiträge Biologie Pflanzen 1872, 1, 127–224. [Google Scholar]
- Fritze, D.; Pukall, R. Reclassification of bioindicator strains Bacillus subtilis DSM 675 and Bacillus subtilis DSM 2277 as Bacillus Atrophaeus. Int. J. Syst. Evol. Microbiol. 2001, 51, 35–37. [Google Scholar] [CrossRef]
- Fischer, A. Untersuchungen über bakterien. Jahrbücher Wissenschaftliche Botanik 1895, 27, 1–163. [Google Scholar]
- GIBBONS, N.E.; MURRAY, R.G.E. Proposals concerning the higher taxa of bacteria. Int. J. Syst. Evol. Microbiol. 1978, 28, 1–6. [Google Scholar] [CrossRef]
- Arahal, D.R.; Ventosa, A. Moderately halophilic and halotolerant species of bacillus and related genera. In Applications and Systematics of Bacillus and Relatives; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2002; pp. 83–99. [Google Scholar] [CrossRef]
- Márquez, M.C.; Sánchez-Porro, C.; Ventosa, A. halophilic and haloalkaliphilic, aerobic endospore-forming bacteria in soil. In Endospore-Forming Soil Bacteria; Logan, N.A., Vos, P., Eds.; Soil Biology; Springer: Berlin/Heidelberg, Germany, 2011; pp. 309–339. [Google Scholar] [CrossRef]
- Ventosa, A. Unusual micro-organisms from unusual habitats: Hypersaline environments. In Prokaryotic Diversity: Mechanisms and Significance; Lappin-Scott, H.M., Logan, N.A., Oyston, P.C.F., Eds.; Society for General Microbiology Symposia; Cambridge University Press: Cambridge, UK, 2006; pp. 223–254. [Google Scholar] [CrossRef]
- Ventosa, A.; Nieto, J.J.; Oren, A. Biology of Moderately Halophilic Aerobic Bacteria. Microbiol Mol. Biol. Rev 1998, 62, 504–544. [Google Scholar] [CrossRef]
- De la Haba, R.R.; Sánchez-Porro, C.; Marquez, M.C.; Ventosa, A. Taxonomy of Halophiles. In Extremophiles Handbook; Horikoshi, K., Ed.; Springer: Tokyo, Japan, 2011; pp. 255–308. [Google Scholar] [CrossRef]
- Seng, P.; Drancourt, M.; Gouriet, F.; La Scola, B.; Fournier, P.-E.; Rolain, J.M.; Raoult, D. Ongoing Revolution in Bacteriology: Routine Identification of Bacteria by Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry. Clin. Infect. Dis. 2009, 49, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Ravi, R.K.; Walton, K.; Khosroheidari, M. MiSeq: A Next Generation Sequencing platform for genomic analysis. Methods Mol. Biol. 2018, 1706, 223–232. [Google Scholar] [CrossRef]
- Karamitros, T.; Magiorkinis, G. Multiplexed targeted sequencing for Oxford Nanopore MinION: A detailed library preparation procedure. Methods Mol. Biol. 2018, 1712, 43–51. [Google Scholar] [CrossRef]
- Jain, M.; Olsen, H.E.; Paten, B.; Akeson, M. The Oxford Nanopore MinION: Delivery of Nanopore sequencing to the genomics community. Genome Biol. 2016, 17. [Google Scholar] [CrossRef]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Res 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M.; Spröer, C.; Klenk, H.-P. When should a DDH experiment be mandatory in microbial taxonomy? Arch. Microbiol. 2013, 195, 413–418. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial dna in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Auch, A.F.; von Jan, M.; Klenk, H.-P.; Göker, M. Digital DNA-DNA Hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genom. Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Ouk Kim, Y.; Park, S.-C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Unaogu, I.C.; Gugnani, H.C.; Boiron, P. The enzymatic profile of some pathogenic aerobic actinomycetes as determined by api-zym method. J. Med. Mycol. 2008, 9, 235. [Google Scholar]
- Gruner, E.; von Graevenitz, A.; Altwegg, M. The API ZYM System: A Tabulated Review from 1977 to Date. J. Microbiol. Methods 1992, 16, 101–118. [Google Scholar] [CrossRef]
- Humble, M.W.; King, A.; Phillips, I. API ZYM: A Simple rapid system for the detection of bacterial enzymes. J. Clin. Pathol. 1977, 30, 275–277. [Google Scholar] [CrossRef] [PubMed]
- Søgaard, P.; Gahrn-Hansen, B.; Zhou, H.P.; Frederiksen, W. An Investigation of three commercial methods for rapid identification of non-enteric gram-negative rods. application on pseudomonas paucimobilis and some other pseudomonas species. Acta Pathol. Microbiol. Immunol. Scand. B 1986, 94, 357–363. [Google Scholar] [CrossRef] [PubMed]
- MK, B.; DA, B.; GL, C.; JG, G. Comparison of five commercial methods for the identification of non- fermentative and oxidase positive fermentative gram-negative Bacilli. N. Z. J. Med. Lab. Technol. 1988, 42, 8–12. [Google Scholar]
- Swanson, E.C.; Collins, M.T. Use of the API 20E System to identify veterinary enterobacteriaceae. J. Clin. Microbiol. 1980, 12, 10–14. [Google Scholar] [CrossRef]
- Smith, P.B.; Tomfohrde, K.M.; Rhoden, D.L.; Balows, A. API System: A multitube micromethod for identification of enterobacteriaceae. Appl. Microbiol. 1972, 24, 449–452. [Google Scholar] [CrossRef]
- Véron, M.; Le Minor, L. Nutrition and taxonomy of “enterobacteriaceae” and related bacteria. III. Nutritional characters and differentiation of the taxonomic groups. Ann. Microbiol. 1975, 126, 125–147. [Google Scholar]
- Bergey, D.H.; Krieg, N.R.; Holt, J.G. Bergey’s Manual of Systematic Bacteriology; Williams & Wilkins: Baltimore, MD, USA, 1984. [Google Scholar]
- Rogosa, M.; Sharpe, M.E. An Approach to the Classification of the Lactobacilli. J. Appl. Bacteriol. 1960, 22, 329–340. [Google Scholar]
- Sharpe, M.E.; Hill, L.R.; Lapage, S.P. Pathogenic Lactobacilli. J. Med. Microbiol. 1973, 6, 281–286. [Google Scholar] [CrossRef]
- Kwon, S.-W.; Lee, S.-Y.; Kim, B.-Y.; Weon, H.-Y.; Kim, J.-B.; Go, S.-J.; Lee, G.-B. Bacillus niabensis sp. nov., isolated from cotton-waste composts for mushroom cultivation. Int. J. Syst. Evol. Microbiol. 2007, 57, 1909–1913. [Google Scholar] [CrossRef] [PubMed]
- Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids. MIDI 1990, 20, 1–6. [Google Scholar]
- Dione, N.; Sankar, S.A.; Lagier, J.-C.; Khelaifia, S.; Michele, C.; Armstrong, N.; Richez, M.; Abrahão, J.; Raoult, D.; Fournier, P.-E. Genome Sequence and Description of Anaerosalibacter massiliensis Sp. Nov. New Microbes New Infect. 2016, 10, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Mehrshad, M.; Amoozegar, M.A.; Didari, M.; Bagheri, M.; Fazeli, S.A.S.; Schumann, P.; Spröer, C.; Sánchez-Porro, C.; Ventosa, A. Bacillus halosaccharovorans sp. nov., a moderately halophilic bacterium from a hypersaline lake. Int. J. Syst. Evol. Microbiol. 2013, 63 Pt 8, 2776–2781. [Google Scholar] [CrossRef] [PubMed]
- Abbas, S.; Ahmed, I.; Kudo, T.; Iqbal, M.; Lee, Y.-J.; Fujiwara, T.; Ohkuma, M. A heavy metal tolerant novel bacterium, Bacillus malikii sp. nov., isolated from tannery effluent wastewater. Antonie Van Leeuwenhoek 2015, 108, 1319–1330. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.-H.; Oh, T.-K. Bacillus litoralis sp. nov., isolated from a tidal flat of the yellow sea in korea. Int. J. Syst. Evol. Microbiol. 2005, 55 Pt 5, 1945–1948. [Google Scholar] [CrossRef] [PubMed]
- Delaporte, B.; Sasson, A. Study of bacteria from arid soils of Morocco: Brevibacterium haloterans n. sp. and Brevibacterium frigoritolerans n. sp. Comptes Rendus Hebd. Seances Acad. Sci. Ser. D Sci. Nat. 1967, 264, 2257–2260. [Google Scholar]
- Liu, G.H.; Liu, B.; Wang, J.P.; Che, J.M.; Li, P.F. Reclassification of Brevibacterium frigoritolerans DSM 8801T as Bacillus frigoritolerans comb. nov. Based on Genome Analysis. Curr. Microbiol. 2020, 77, 1916–1923. [Google Scholar] [CrossRef]
- Tiago, I.; Pires, C.; Mendes, V.; Morais, P.V.; da Costa, M.S.; Veríssimo, A. Bacillus foraminis sp. nov., isolated from a non-saline alkaline groundwater. Int. J. Syst. Evol. Microbiol. 2006, 56 Pt 11, 2571. [Google Scholar] [CrossRef]
- Kanso, S.; Greene, A.C.; Patel, B.K.C. Bacillus subterraneus sp. nov., an iron- and manganese-reducing bacterium from a deep subsurface australian thermal aquifer. Int. J. Syst. Evol. Microbiol. 2002, 52 Pt 3, 869. [Google Scholar] [CrossRef]
- Wieser, M.; Worliczek, H.; Kämpfer, P.; Busse, H.-J. Bacillus herbersteinensis sp. nov. Int. J. Syst. Evol. Microbiol. 2005, 55 Pt 5, 2119–2123. [Google Scholar] [CrossRef]
- NAGEL, M.; ANDREESEN, J.R. Bacillus niacini sp. nov., a nicotinate-metabolizing mesophile isolated from soil. Int. J. Syst. Evol. Microbiol. 1991, 41, 134–139. [Google Scholar] [CrossRef][Green Version]
- Albert, R.A.; Archambault, J.; Rosselló-Mora, R.; Tindall, B.J.; Matheny, M. Bacillus acidicola sp. nov., a novel mesophilic, acidophilic species isolated from acidic sphagnum peat bogs in wisconsin. Int. J. Syst. Evol. Microbiol. 2005, 55 Pt 5, 2125–2130. [Google Scholar] [CrossRef]
- Zhang, X.-X.; Gao, J.-S.; Zhang, L.; Zhang, C.-W.; Ma, X.-T.; Zhang, J. Bacillus oryzisoli sp. nov., isolated from rice rhizosphere. Int. J. Syst. Evol. Microbiol. 2016, 66, 3432–3436. [Google Scholar] [CrossRef]
- Priest, F.G.; Goodfellow, M.; Todd, C. A numerical classification of the genus bacillus. J. Gen. Microbiol. 1988, 134, 1847–1882. [Google Scholar] [CrossRef]
- Suresh, K.; Prabagaran, S.R.; Sengupta, S.; Shivaji, S. Bacillus indicus sp. nov., an arsenic-resistant bacterium isolated from an aquifer in west bengal, india. Int. J. Syst. Evol. Microbiol. 2004, 54 Pt 4, 1369–1375. [Google Scholar] [CrossRef] [PubMed]
- Seiler, H.; Wenning, M.; Schmidt, V.; Scherer, S. Bacillus gottheilii sp. nov., isolated from a pharmaceutical manufacturing site. Int. J. Syst. Evol. Microbiol. 2013, 63 Pt 3, 867–872. [Google Scholar] [CrossRef]
- Nielsen, P.; Fritze, D.; Priest, F.G. Phenetic diversity of alkaliphilic bacillus strains: Proposal for nine new species. Microbiology 1995, 141, 1745–1761. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhang, D.-F.; Khieu, T.-N.; Son, C.K.; Zhang, X.-M.; Cheng, J.; Tian, X.-P.; Zhang, S.; Li, W.-J. Bacillus tianshenii sp. nov., isolated from a marine sediment sample. Int. J. Syst. Evol. Microbiol. 2014, 64 Pt 6, 1998–2002. [Google Scholar] [CrossRef]
- Shivaji, S.; Chaturvedi, P.; Begum, Z.; Pindi, P.K.; Manorama, R.; Padmanaban, D.A.; Shouche, Y.S.; Pawar, S.; Vaishampayan, P.; Dutt, C.B.S.; et al. Janibacter hoylei sp. nov., Bacillus isronensis sp. nov. and Bacillus aryabhattai sp. nov., isolated from cryotubes used for collecting air from the upper atmosphere. Int. J. Syst. Evol. Microbiol. 2009, 59 Pt 12, 2977–2986. [Google Scholar] [CrossRef]
- Parag, B.; Sasikala, C.; Ramana, C.V. Bacillus endolithicus sp. nov., isolated from pebbles. Int. J. Syst. Evol. Microbiol. 2015, 65, 4568–4573. [Google Scholar] [CrossRef]
- Heyrman, J.; Vanparys, B.; Logan, N.A.; Balcaen, A.; Rodríguez-Díaz, M.; Felske, A.; De Vos, P. Bacillus novalis sp. nov., Bacillus vireti sp. nov., Bacillus soli sp. nov., Bacillus bataviensis sp. nov. and Bacillus drentensis sp. nov., from the drentse a grassland. Int. J. Syst. Evol. Microbiol. 2004, 54 Pt 1, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Balcázar, J.L.; Pintado, J.; Planas, M. Bacillus galliciensis sp. nov., isolated from faeces of wild seahorses (Hippocampus Guttulatus). Int. J. Syst. Evol. Microbiol. 2010, 60 Pt 4, 892–895. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-H.; Tian, X.-R.; Ruan, Y.; Yang, L.-L.; He, Z.-Q.; Tang, S.-K.; Li, W.-J.; Shi, H.; Chen, Y.-G. Bacillus crassostreae sp. nov., isolated from an oyster (Crassostrea Hongkongensis). Int. J. Syst. Evol. Microbiol. 2015, 65 Pt 5, 1561–1566. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R.; Kandler, O.; Wheelis, M.L. Towards a Natural System of Organisms: Proposal for the domains archaea, bacteria, and eucarya. Proc. Natl. Acad. Sci. USA 1990, 87, 4576–4579. [Google Scholar] [CrossRef]
- Ludwig, W.; Schleifer, K.-H.; Whitman, W.B. Bacilli class. Nov. In Bergey’s Manual of Systematics of Archaea and Bacteria; American Cancer Society: Atlanta, GA, USA, 2015. [Google Scholar] [CrossRef]
- Vos, P.D. Bacillales. In Bergey’s Manual of Systematics of Archaea and Bacteria; American Cancer Society: Atlanta, GA, USA, 2015. [Google Scholar] [CrossRef]
- Reddien, P.W. The Cellular and Molecular Basis for Planarian Regeneration. Cell 2018, 175, 327–345. [Google Scholar] [CrossRef]
- Kangale, L.J.; Raoult, D.; Ghigo, E.; Fournier, P.E. Pedobacter schmidteae sp. nov., a new bacterium isolated from the microbiota of the planarian Schmidtea mediterranea. Sci. Rep. 2020, 10, 6113. [Google Scholar] [CrossRef]
- Pei, A.Y.; Oberdorf, W.E.; Nossa, C.W.; Agarwal, A.; Chokshi, P.; Gerz, E.A.; Jin, Z.; Lee, P.; Yang, L.; Poles, M.; et al. Diversity of 16S rRNA genes within individual prokaryotic genomes. Appl. Environ. Microbiol. 2010, 76, 3886–3897. [Google Scholar] [CrossRef]
- Ochman, H.; Elwyn, S.; Moran, N.A. Calibrating bacterial evolution. Proc. Natl. Acad. Sci. USA 1999, 96, 12638–12643. [Google Scholar] [CrossRef]
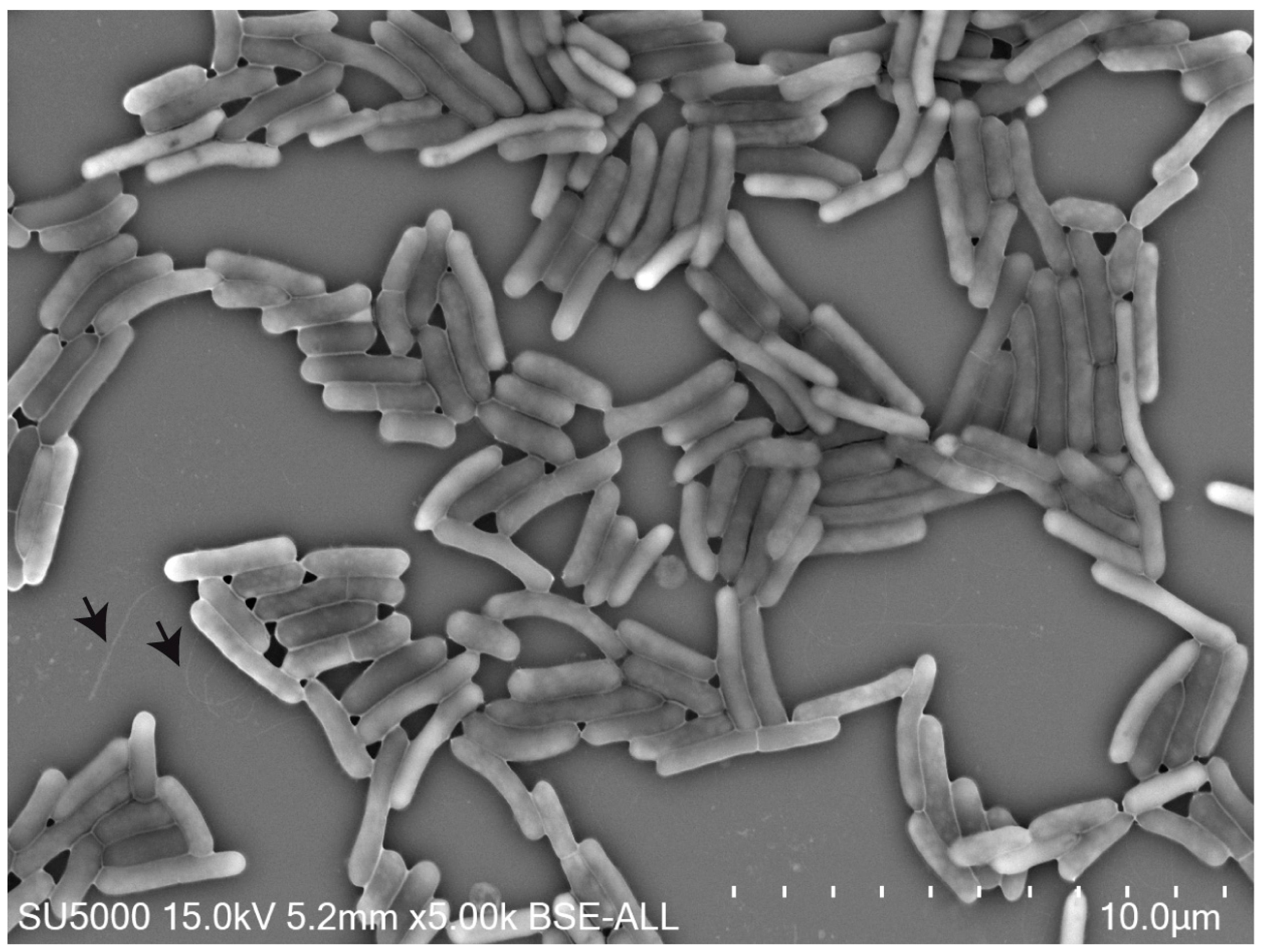
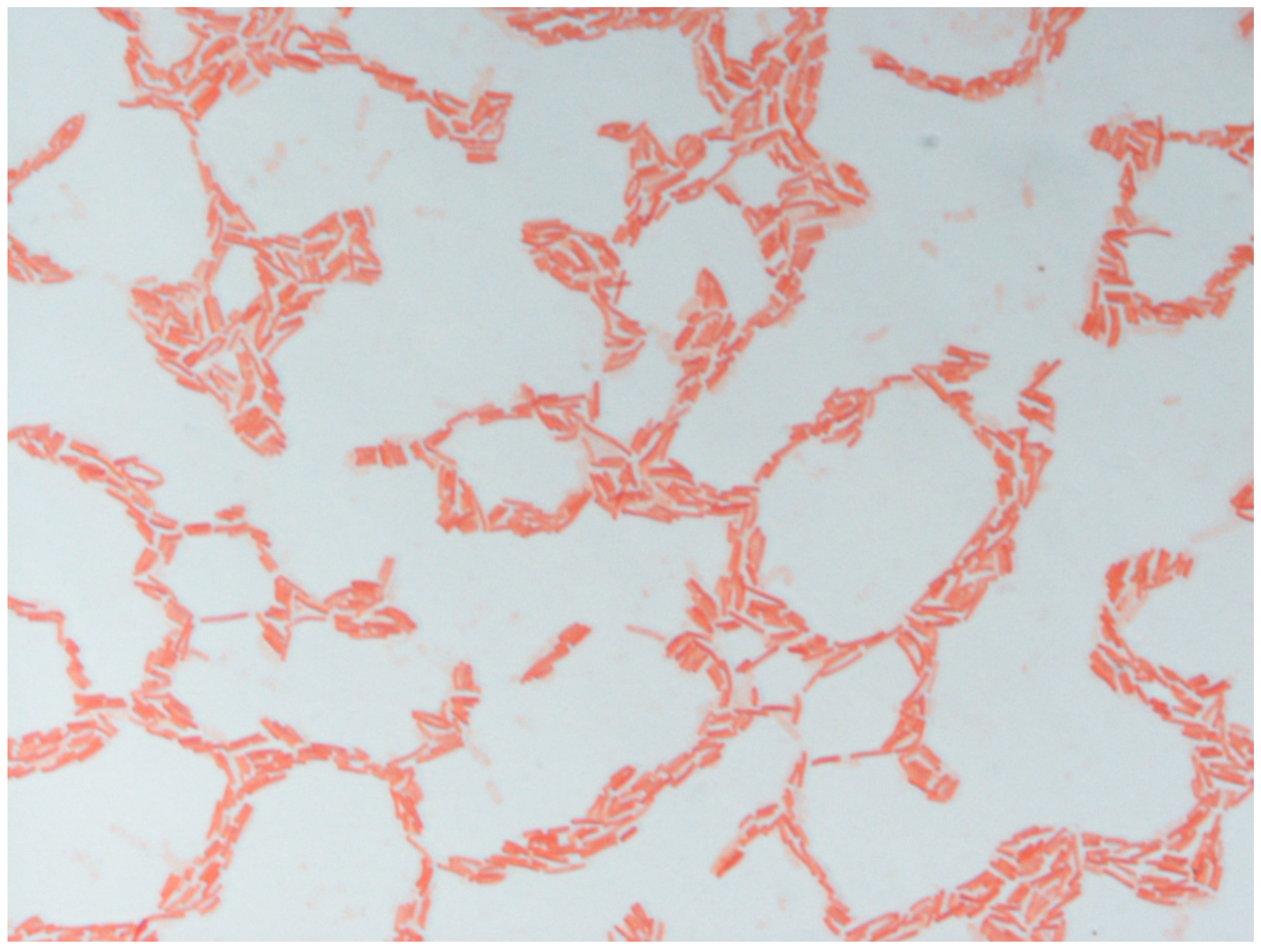

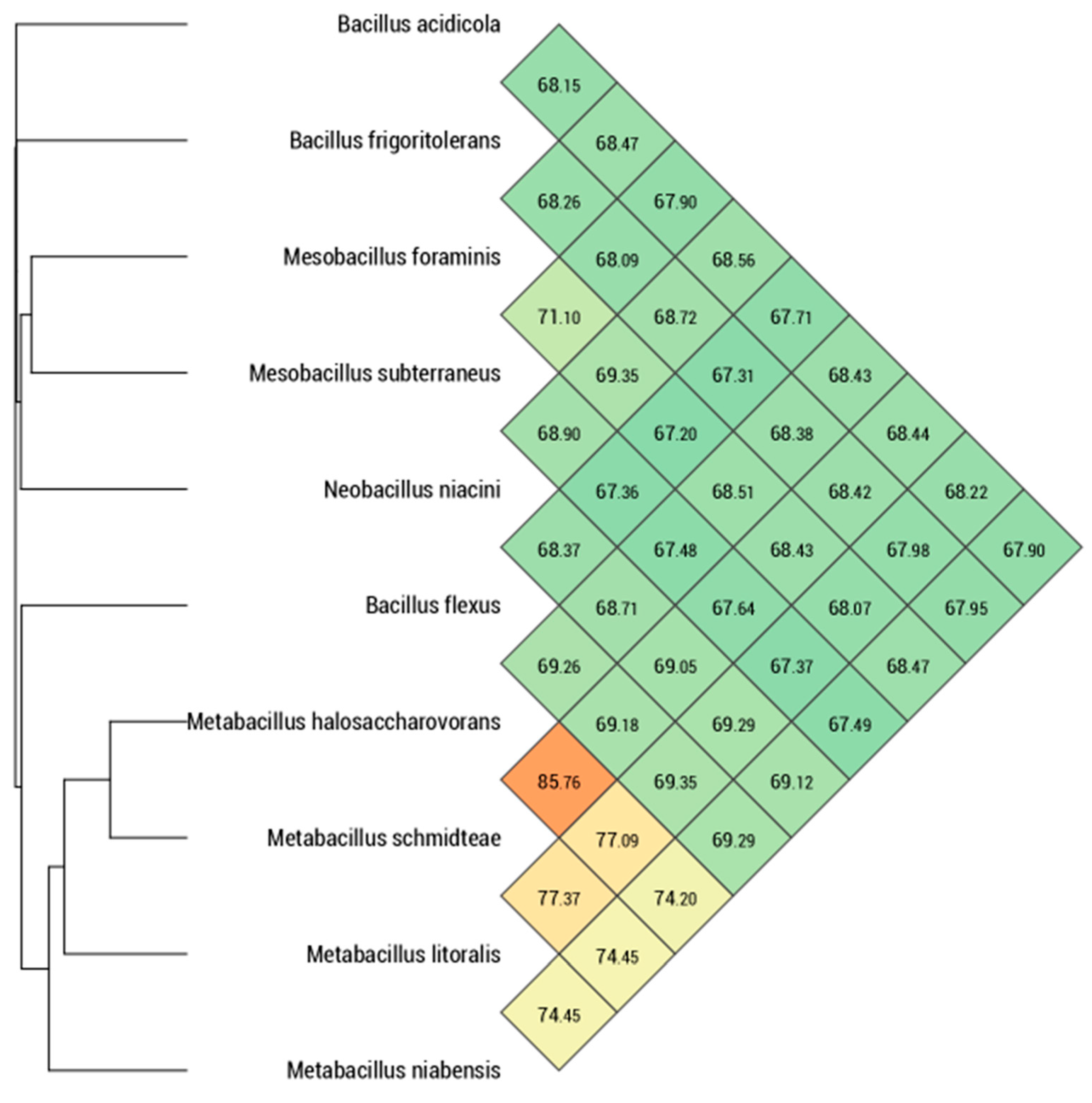

| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|
| Bacillus flexus | 28.20 | ||||||||
| Mesobacillus foraminis | 27.80 | 19.80 | |||||||
| Metabacillus halosaccharovorans | 31.80 | 23.70 | 27.90 | ||||||
| Metabacillus litoralis | 39.10 | 23.70 | 27.00 | 22.70 | |||||
| Metabacillus niabensis | 27.00 | 20.10 | 22.00 | 22.00 | 22.60 | ||||
| Neobacillus niacini | 25.90 | 19.90 | 21.50 | 25.50 | 29.70 | 23.10 | |||
| Metabacillus schmidteae | 34.20 | 24.40 | 27.10 | 31.10 | 23.00 | 22.30 | 27.70 | ||
| Mesobacillus subterraneus | 25.30 | 21.70 | 18.80 | 29.90 | 31.30 | 20.70 | 21.30 | 30.10 | |
| Bacillus frigoritolerans | 32.50 | 26.70 | 25.60 | 31.30 | 34.10 | 28.70 | 24.70 | 32.80 | 28.20 |
| CODE | VALUE | DESCRIPTION |
|---|---|---|
| Information storage and processing | ||
| [J] | 169 | Translation, ribosomal structure, and biogenesis |
| [A] | 0 | RNA processing and modification |
| [K] | 347 | Transcription |
| [L] | 221 | Replication, recombination, and repair |
| [B] | 1 | Chromatin structure and dynamics |
| Cellular processes and signaling | ||
| [D] | 36 | Cell cycle control, cell division, chromosome partitioning |
| [V] | 0 | Defense mechanisms |
| [T] | 81 | Signal transduction mechanisms |
| [M] | 190 | Cell wall/membrane/envelope biogenesis |
| [N] | 197 | Cell motility |
| [Z] | 77 | Cytoskeleton |
| [W] | 0 | Extracellular structures |
| [U] | 0 | Intracellular trafficking, secretion, and vesicular transport |
| [O] | 30 | Posttranslational modification, protein turnover, chaperones |
| [X] | 125 | Mobilome: prophages, transposons |
| Metabolism | ||
| [C] | 181 | Energy production and conversion |
| [G] | 444 | Carbohydrate transport and metabolism |
| [E] | 308 | Amino acid transport and metabolism |
| [F] | 92 | Nucleotide transport and metabolism |
| [H] | 120 | Coenzyme transport and metabolism |
| [I] | 137 | Lipid transport and metabolism |
| [P] | 205 | Inorganic ion transport and metabolism |
| [Q] | 47 | Secondary metabolites biosynthesis, transport, and catabolism |
| Poorly characterized | ||
| [R] | 469 | General function prediction only |
| [S] | 403 | Function unknown |
| Fatty Acids | Name | 1 | 2 | 3 | 4 |
|---|---|---|---|---|---|
| Straight-chain saturated | |||||
| 15:0 | Pentadecanoic acid | 2.7 | tr | − | − |
| 16:0 | Hexadecanoic acid | 7.1 | 15.7 | − | 5.5 |
| Branched saturated | |||||
| 13:0 iso | 11-methyl-Dodecanoic acid | 1.1 | tr | − | − |
| 14:0 iso | 12-methyl-Tridecanoic acid | 3.3 | 5.8 | − | 10.0 |
| 15:0 iso | 13-methyl-tetradecanoic acid | 12.5 | 7.2 | 21.0 | 15.6 |
| 16:0 iso | 12-methyl-Hexadecanoic acid | 2.3 | 6.0 | − | 12.5 |
| 17:0 iso | 15-methyl-Hexadecanoic acid | 1.6 | 1.7 | − | − |
| 15:0 anteiso | 12-methyl-tetradecanoic acid | 62.6 | 45.0 | 43.5 | 34.8 |
| 17:0 anteiso | 15-methyl-Hexadecanoic acid | 3.8 | 11.4 | 9.5 | 6.4 |
| Monounsaturated | |||||
| 16:1ω9 | 7-Hexadecenoic acid | 3.1 | 2.1 | − | − |
| 18:1n9 | 9-Octadecenoic acid | − | 1.1 | − | − |
| Drug (Antibiotics) | CC µg/mL | Marseille-P9898 MIC | 4T19T MIC |
|---|---|---|---|
| amikacin | 0.016–256 | 50 | 1.5 |
| fosfomycin | 0.064–1024 | 16 | >1024 |
| benzylpenicelin | 0.016–256 | 0.016 | 0.016 |
| ciprofloxacin | 0.002–32 | 19 | 0.38 |
| amoxicillin | 0.016–256 | 0.016 | 0.016 |
| ceftriaxone | 0.016–256 | 0.064 | 0.50 |
| daptomycin | 0.016–256 | 25 | 1 |
| doxycyclin | 0.016–256 | 0.016 | 0.016 |
| vancomycin | 0.016–256 | 0.094 | 0.50 |
| ampicillin | 0.016–256 | 0.016 | 0.016 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kangale, L.J.; Raoult, D.A.; Ghigo, E.; Fournier, P.-E. Metabacillus schmidteae sp. nov., Cultivated from Planarian Schmidtea mediterranea Microbiota. Microbiol. Res. 2021, 12, 299-316. https://doi.org/10.3390/microbiolres12020021
Kangale LJ, Raoult DA, Ghigo E, Fournier P-E. Metabacillus schmidteae sp. nov., Cultivated from Planarian Schmidtea mediterranea Microbiota. Microbiology Research. 2021; 12(2):299-316. https://doi.org/10.3390/microbiolres12020021
Chicago/Turabian StyleKangale, Luis Johnson, Didier A. Raoult, Eric Ghigo, and Pierre-Edouard Fournier. 2021. "Metabacillus schmidteae sp. nov., Cultivated from Planarian Schmidtea mediterranea Microbiota" Microbiology Research 12, no. 2: 299-316. https://doi.org/10.3390/microbiolres12020021
APA StyleKangale, L. J., Raoult, D. A., Ghigo, E., & Fournier, P.-E. (2021). Metabacillus schmidteae sp. nov., Cultivated from Planarian Schmidtea mediterranea Microbiota. Microbiology Research, 12(2), 299-316. https://doi.org/10.3390/microbiolres12020021








