Dolutegravir Resistance in Mozambique: Insights from a Programmatic HIV Resistance Testing Intervention in a Highly Antiretroviral Therapy-Experienced Cohort
Abstract
1. Introduction
2. Materials and Methods
2.1. Setting
2.2. Study Design and Population
2.2.1. Eligible Participants
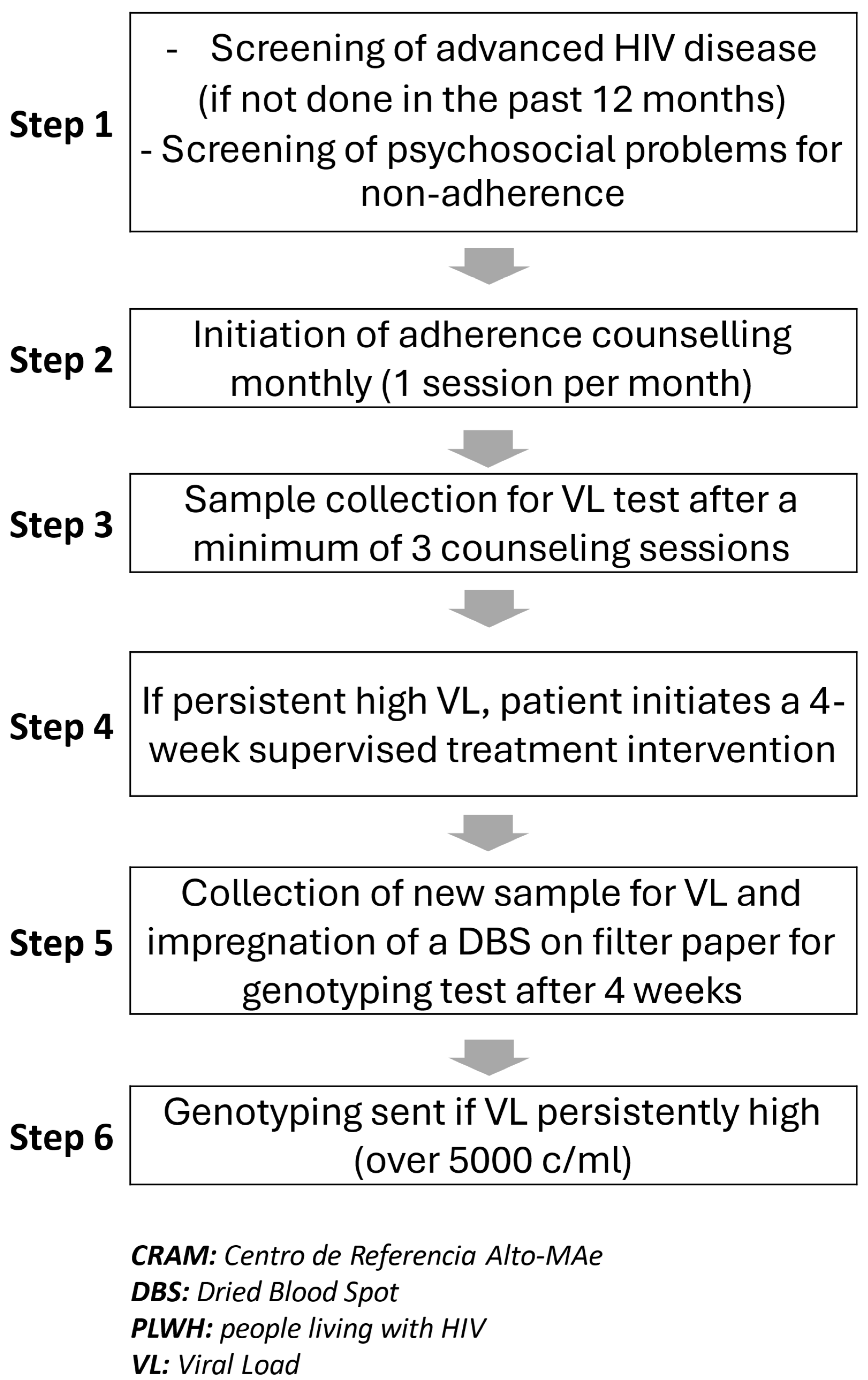
Algorithm for Selecting PLWH for Resistance Testing
Positive Predictive Value (PPV)
Genotypic HIV Drug-Resistance (HIVDR) Testing
2.2.2. Sampling and Sample Size of Program Participants
2.2.3. Program Data Collection
2.2.4. Program Data Analysis
3. Results
3.1. Cohort Main Characteristics
3.2. INSTI Mutations and Resistance
4. Discussion
4.1. Main Findings
4.2. Other Relevant Findings
4.3. Limitations of the Study
5. Conclusions and Recommendations
What Are the Bullet Points of This Study?
- High Detection of Dolutegravir Resistance: The study reveals a strikingly high detection of dolutegravir (DTG) resistance (89.5%) among individuals with virologic failure (VF) who had extensive prior exposure to antiretroviral therapy (ART). This underscores the risk of resistance in heavily ART-experienced populations and challenges the assumption that DTG resistance is still rare.
- Effective Algorithm for Resistance Identification: The study outlines a structured algorithm combining viral load monitoring, adherence reinforcement, and supervised ART support to identify individuals at high risk of resistance. The add-on approach demonstrated a high positive predictive value (89.5%), enabling targeted use of genotypic resistance testing where resources are limited.
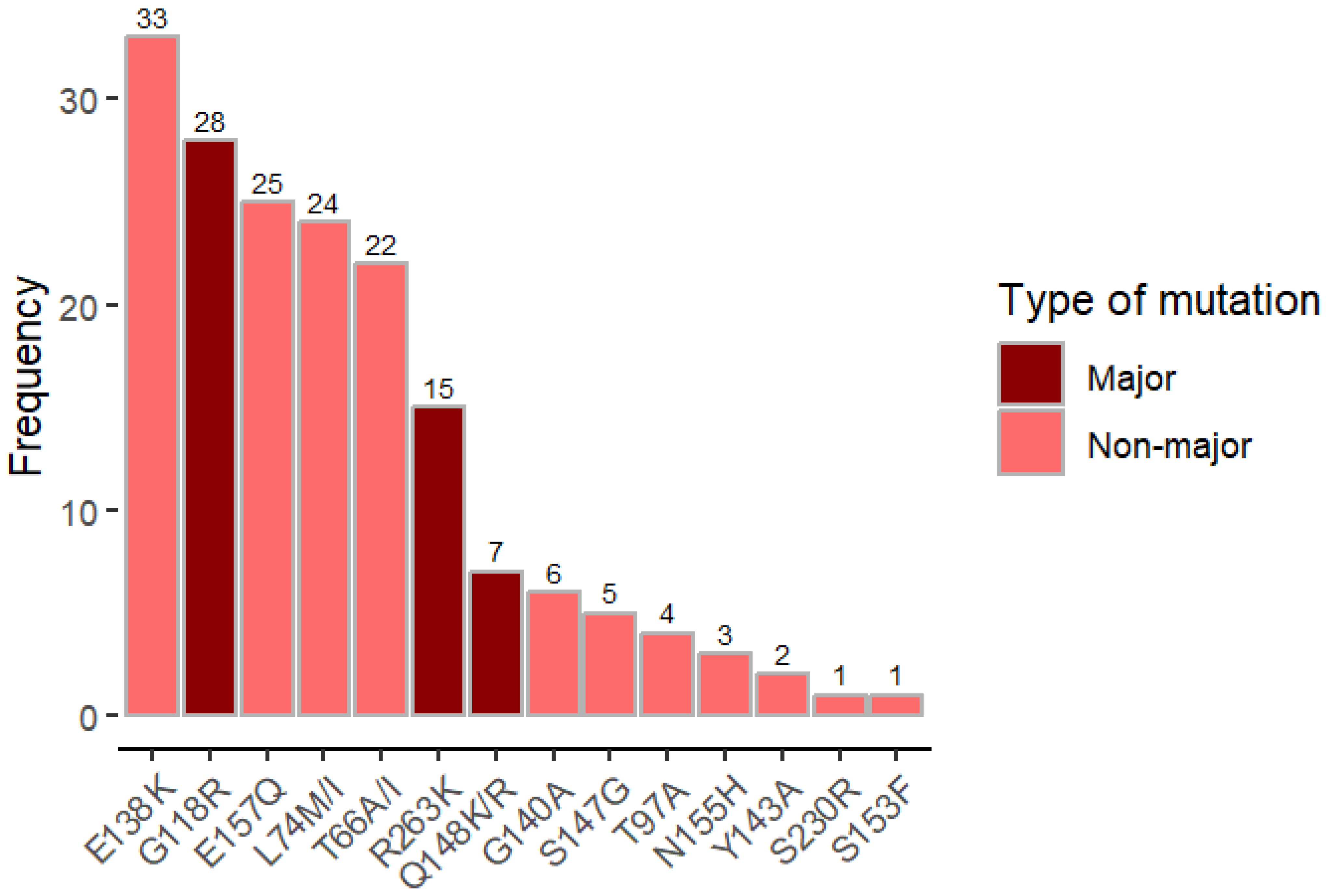
- Mutational Patterns and Resistance Scores: The study documents the most common DTG-associated mutations (e.g., G118R, R263K, Q148RK) and provides detailed resistance scores, offering insights into the genetic basis of DTG resistance in this cohort.
- Programmatic Implications: The findings highlight the need for integrating resistance testing into ART programs in resource-limited settings, particularly for individuals with prolonged ART exposure and virologic failure. The study substantiates the need for adherence support and supervised interventions before considering regimen switches, which can help preserve effective treatment options like DTG. It also highlights the importance of elucidating the role of regular HIVDR surveillance vs. clinically driven individual resistance testing in settings where the affordability and feasibility of HIVDR testing are perceived as limited.
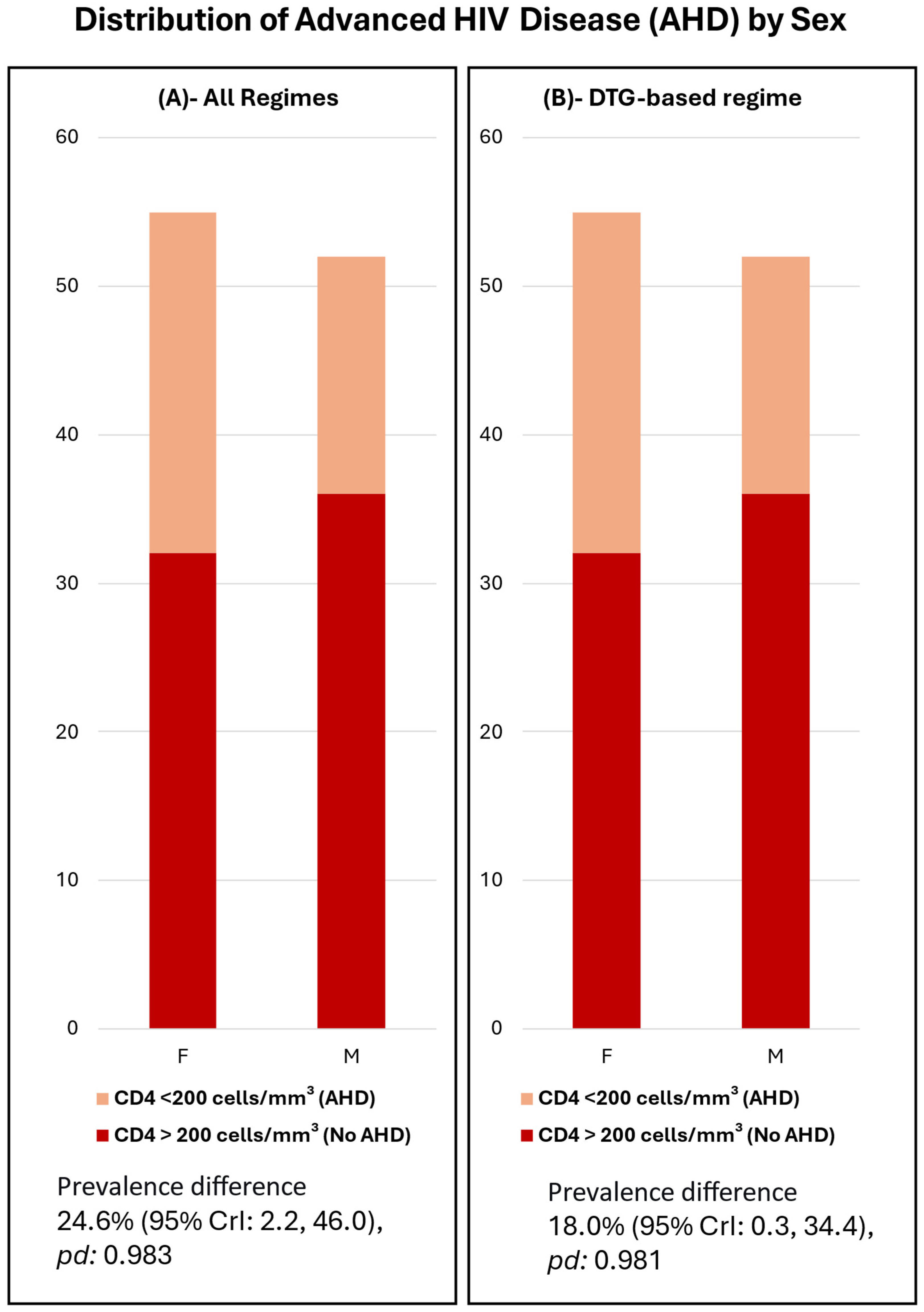
- Sex Disparities in Advanced HIV Disease: The study notes significant immunological differences between men and women in the cohort, with men exhibiting lower CD4 counts and higher rates of advanced HIV disease. This aligns with broader trends in sub-Saharan Africa and emphasizes the need for targeted interventions for male populations.
- Limitations and Future Directions: The study acknowledges the challenges of using dried blood spot (DBS) samples for resistance testing, particularly their lower sensitivity for detecting resistance in cases of low-level viremia. It calls for further research to validate the algorithm in different subpopulations and to explore plasma-based testing for improved sensitivity.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABC | Abacavir |
| AHD | Advanced HIV disease |
| ART | Antiretroviral therapy |
| ATV | Atazanavir |
| AZT | Zidovudine |
| CRAM | Centro de Referência de Alto-Mae |
| CDC | Centers for Disease Control and Prevention |
| D4T | Stavudine |
| DBS | Dried blood spot |
| DRV | Darunavir |
| DTG | Dolutegravir |
| DTG-R | Dolutegravir resistance testing |
| HRSA | Health Resources and Services Administration |
| HIV-GT | HIV genotypic resistance testing |
| IQR | Interquartile range |
| INSTI | Integrase strand transfer inhibitor |
| ITECH | International Training and Education Center for Health |
| pd | Probability of direction (pd) |
| PIs | Protease inhibitors |
| PLWH | People living with HIV |
| LPV | Lopinavir |
| MSF | Médecins Sans Frontières |
| NRTI | Nucleotide reverse transcriptase inhibitors |
| RTV | Ritonavir |
| RAL | Raltegravir |
| TDF | Tenofovir |
| VF | Virologic failure |
| WHO | World Health Organization |
| 95% Crl | 95% Bayesian credible interval |
References
- WHO. WHO Recommends Dolutegravir as Preferred HIV Treatment Option in all Populations; World Health Organization (WHO): Geneva, Switzerland, 2019; Available online: https://www.who.int/news/item/22-07-2019-who-recommends-dolutegravir-as-preferred-hiv-treatment-option-in-all-populations (accessed on 20 November 2024).
- Borry, P.; Bentzen, H.B.; Budin-Ljøsne, I.; Cornel, M.C.; Howard, H.C.; Feeney, O.; Jackson, L.; Mascalzoni, D.; Mendes, Á.; Peterlin, B.; et al. The challenges of the expanded availability of genomic information: An agenda-setting paper. J. Community Genet. 2018, 9, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Fusco, P.; Nasta, P.; Quiros-Roldan, E.; Tondinelli, A.; Costa, C.; Fornabaio, C.; Mazzini, N.; Prosperi, M.; Torti, C.; Carosi, G. Efficacy, Convenience, Safety and Durability of DTG-Based Antiretroviral Therapies: Evidence from a Prospective Study by the Italian MaSTER Cohort. Viruses 2023, 15, 924. [Google Scholar] [CrossRef]
- Tschumi, N.; Lukau, B.; Tlali, K.; Motaboli, L.; Kao, M.; Kopo, M.; Haenggi, K.; Mokebe, M.; Naegele, K.; Ayakaka, I.; et al. Emergence of Acquired Dolutegravir Resistance in Treatment-experienced People With HIV in Lesotho. Clin. Infect. Dis. 2024, 79, 1208–1222. [Google Scholar] [CrossRef] [PubMed]
- Giovanetti, M.; Ciccozzi, M.; Parolin, C.; Borsetti, A. Molecular Epidemiology of HIV-1 in African Countries: A Comprehensive Overview. Pathogens 2020, 9, 1072. [Google Scholar] [CrossRef]
- DNAM—Direcção Nacional de Assistência Médica. COMITÉ TARV; Ministério da Saúde de Moçambique; MISAU: Maputo, Mozambique, 2021; Available online: https://comitetarvmisau.co.mz (accessed on 20 April 2022).
- Nacarapa, E.; Verdu, M.E.; Nacarapa, J.; Macuacua, A.; Chongo, B.; Osorio, D.; Munyangaju, I.; Mugabe, D.; Paredes, R.; Chamarro, A.; et al. Predictors of attrition among adults in a rural HIV clinic in southern Mozambique: 18-year retrospective study. Sci. Rep. 2021, 11, 17897. [Google Scholar] [CrossRef]
- Fox. A. I-TECH Supported Reference Center Serves as Critical Lifeline for People with Advanced HIV Disease; I-TECH, University of Washington—Global Health Department: Seattle, WA, USA, 2023; Available online: https://www.go2itech.org/2023/02/i-tech-supported-reference-center-serves-as-critical-lifeline-for-people-with-advanced-hiv-disease/ (accessed on 24 February 2023).
- KHuik, K.; Hill, S.; George, J.; Pau, A.; Kuriakose, S.; Lange, C.M.; Dee, N.; Stoll, P.; Khan, M.; Rehman, T.; et al. High-level dolutegravir resistance can emerge rapidly from few variants and spread by recombination: Implications for integrase strand transfer inhibitor salvage therapy. AIDS 2022, 36, 1835–1840. [Google Scholar] [CrossRef]
- INS–Instituto Nacional de Saude. INSIDA 2021: Divulgados Resultados do Inquérito sobre o Impacto do HIV e SIDA em Moçambique; INS: Maputo, Mozambique, 2021. Available online: https://ins.gov.mz/wp-content/uploads/2022/12/53059_14_INSIDA_Summary-sheet_POR.pdf (accessed on 1 June 2023).
- SISMA—MoH Mozambique. O Sistema de Informação de Saúde para Monitoria e Avaliação (SIS-MA); SISMA—MoH Mozambique: Maputo, Mozambique, 2025. Available online: https://sisma.misau.gov.mz/ (accessed on 17 February 2025).
- I-TECH. I-TECH International Training and Education Center for Health, Mozambique Partnerships. Available online: https://www.go2itech.org/2017/08/mozambique-partnerships/ (accessed on 20 April 2022).
- ITECH Mozambique 2017. Annual Progress Report Narrative COP16. September 2017. Available online: https://www.go2itech.org/where-we-work/mozambique/ (accessed on 8 September 2023).
- HIV/AIDS (HIV). WHO Manual for HIV Drug Resistance Testing Using Dried Blood Spot Specimens; World Health Organization (WHO): Geneva, Switzerland, 2020; Available online: https://www.who.int/publications/i/item/9789240009424 (accessed on 24 June 2025).
- MReust, M.J.; Lee, M.H.; Xiang, J.; Zhang, W.; Xu, D.; Batson, T.; Zhang, T.; Downs, J.A.; Dupnik, K.M. Dried Blood Spot RNA Transcriptomes Correlate with Transcriptomes Derived from Whole Blood RNA. Am. J. Trop. Med. Hyg. 2018, 98, 1541–1546. [Google Scholar] [CrossRef] [PubMed]
- Crossley, B.M.; Bai, J.; Glaser, A.; Maes, R.; Porter, E.; Killian, M.L.; Clement, T.; Toohey-Kurth, K. Guidelines for Sanger sequencing and molecular assay monitoring. J. Vet. Diagn. Investig. 2020, 32, 767–775. [Google Scholar] [CrossRef]
- Yerly, S.; Kaiser, L.; Race, E.; Bru, J.P.; Clavel, F.; Perrin, L. Transmission of antiretroviral-drug-resistant HIV-1 variants. Lancet 1999, 354, 729–733. [Google Scholar] [CrossRef]
- Yerly, S.; Rakik, A.; De Loes, S.K.; Hirschel, B.; Descamps, D.; Brun-Vézinet, F.; Perrin, L. Switch to unusual amino acids at codon 215 of the human immunodeficiency virus type 1 reverse transcriptase gene in seroconvertors infected with zidovudine-resistant variants. J. Virol. 1998, 72, 3520–3523. [Google Scholar] [CrossRef] [PubMed]
- Stanford University. Stanford Hivdb Algorithm Updates. Available online: https://hivdb.stanford.edu/page/algorithm-updates/#version.9.4.update.2022-12-07 (accessed on 6 May 2023).
- Kanise, H.; van Oosterhout, J.J.; Bisani, P.; Songo, J.; Matola, B.W.; Chipungu, C.; Simon, K.; Cox, C.; Hosseinipour, M.C.; Sagno, J.-B.; et al. Virological Findings and Treatment Outcomes of Cases That Developed Dolutegravir Resistance in Malawi’s National HIV Treatment Program. Viruses 2023, 16, 29. [Google Scholar] [CrossRef] [PubMed]
- Kamori, D.; Barabona, G.; Rugemalila, J.; Maokola, W.; Masoud, S.S.; Mizinduko, M.; Sabasaba, A.; Ruhago, G.; Sambu, V.; Mushi, J.; et al. Emerging integrase strand transfer inhibitor drug resistance mutations among children and adults on ART in Tanzania: Findings from a national representative HIV drug resistance survey. J. Antimicrob. Chemother. 2023, 78, 779–787. [Google Scholar] [CrossRef]
- Loosli, T.; Hossmann, S.; Ingle, S.M.; Okhai, H.; Kusejko, K.; Mouton, J.; Bellecave, P.; van Sighem, A.; Stecher, M.; Monforte, A.D.; et al. HIV-1 drug resistance in people on dolutegravir-based antiretroviral therapy: A collaborative cohort analysis. Lancet HIV 2023, 10, e733–e741. [Google Scholar] [CrossRef] [PubMed]
- Kitenge, M.K.; Fatti, G.; Eshun-Wilson, I.; Aluko, O.; Nyasulu, P. Prevalence and trends of advanced HIV disease among antiretroviral therapy-naïve and antiretroviral therapy-experienced patients in South Africa between 2010-2021: A systematic review and meta-analysis. BMC Infect. Dis. 2023, 23, 549. [Google Scholar] [CrossRef]
| All | DTG-Based | PI-Based | |
|---|---|---|---|
| N | 106 | 62 | 44 |
| Sex Women, N (%) | 57 (53.8) | 30 (48.4) | 27 (61.4) |
| Men, N (%) | 49 (46.2) | 35 (53.8) | 17 (38.6) |
| Age, median (IQR) | 42 (38, 48) | 42 (38, 48) | 42 (37, 50) |
| Time on ART (years), median (IQR) | 11.6 (9.1, 14.5) | 11.2 (6.5, 14.4) | 12.1 (10.4, 14.6) |
| Time on current ART regimen (years), median (IQR) | 3.2 (2.2, 4.5) | 2.5 (1.9, 3.3) | 4.8 (3.7, 6.0) |
| TDF backbone, N (%) | 80 (75.5) | 49 (79.0) | 31 (70.5) |
| Non-first-line ART, N (%) | 102 (96.2) | 60 (96.8) | 42 (95.5) |
| Viral load (log10), median (IQR) | 4.6 (4.2, 5.2) | 4.7 (4.3, 5.2) | 4.6 (4.2, 4.9) |
| CD4, median (IQR) | 134 (58, 234) | 128 (56, 233) | 146 (59, 258) |
| Advanced HIV, N (%) 1 | 72 (67.9) | 43 (69.4) | 29 (65.9) |
| Subtype C, N (%) | 99 (93.4) | 58 (93.5) | 41 (93.2) |
| All | DTG-Based | PI-Based | |
|---|---|---|---|
| Number of NRTI mutations, median (IQR) 1 | 3 (2, 5) | 4 (2, 5) | 3 (2, 5) |
| TDF-R score, median (IQR) | 20 (5, 35) | 20 (13.8, 36.2) | 15 (0, 28.8) |
| TDF-R score ≥ 5, N (%) | 78 (76.5) | 48 (80.0) | 30 (71.4) |
| AZT-R score, median (IQR) | 57.5 (0, 90) | 60 (0, 90) | 55 (5, 88.8) |
| AZT-R score ≥ 5, N (%) | 73 (71.5) | 41 (68.3) | 32 (76.2) |
| Number of PI mutations, median (IQR) 2 | 2 (0, 5) | 1 (0, 2) | 5 (4, 7) |
| ATV-R score, median (IQR) | 0 (0, 70) | 0 (0, 0) | 65 (17.5, 110) |
| ATV-R score ≥ 5, N (%) | 51 (49.5) | 12 (20.0) | 39 (90.7) |
| DRV-R score, median (IQR) | 0 (0, 0) | 0 (0, 0) | 0 (0, 0) |
| DRV-R score ≥ 5, N (%) | 14 (13.6) | 6 (10.0) | 8 (18.6) |
| Number of INSTI mutations, median (IQR) 3 | 0.5 (0, 3) | 3 (1, 4) | 0 (0, 0) |
| DTG-R score, median (IQR) | 30 (0, 80) | 75 (40, 81.2) | 0 (0, 0) |
| DTG-R score ≥ 5, N (%) | 51 (53.6%) | 51 (91.1%) | 0 (0%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruano, M.; Flores, A.; Couto, A.; Gaspar, I.; Yerly, S.; Zamudio, A.G.G.; Bene, R.; Maiela, A.; Macuacua, H.; Lane, J.; et al. Dolutegravir Resistance in Mozambique: Insights from a Programmatic HIV Resistance Testing Intervention in a Highly Antiretroviral Therapy-Experienced Cohort. Infect. Dis. Rep. 2025, 17, 123. https://doi.org/10.3390/idr17050123
Ruano M, Flores A, Couto A, Gaspar I, Yerly S, Zamudio AGG, Bene R, Maiela A, Macuacua H, Lane J, et al. Dolutegravir Resistance in Mozambique: Insights from a Programmatic HIV Resistance Testing Intervention in a Highly Antiretroviral Therapy-Experienced Cohort. Infectious Disease Reports. 2025; 17(5):123. https://doi.org/10.3390/idr17050123
Chicago/Turabian StyleRuano, Maria, Antonio Flores, Aleny Couto, Irénio Gaspar, Sabine Yerly, Ana Gabriela Gutierrez Zamudio, Rosa Bene, Adelina Maiela, Helder Macuacua, Jeff Lane, and et al. 2025. "Dolutegravir Resistance in Mozambique: Insights from a Programmatic HIV Resistance Testing Intervention in a Highly Antiretroviral Therapy-Experienced Cohort" Infectious Disease Reports 17, no. 5: 123. https://doi.org/10.3390/idr17050123
APA StyleRuano, M., Flores, A., Couto, A., Gaspar, I., Yerly, S., Zamudio, A. G. G., Bene, R., Maiela, A., Macuacua, H., Lane, J., Mudender, F., & Nacarapa, E. (2025). Dolutegravir Resistance in Mozambique: Insights from a Programmatic HIV Resistance Testing Intervention in a Highly Antiretroviral Therapy-Experienced Cohort. Infectious Disease Reports, 17(5), 123. https://doi.org/10.3390/idr17050123







