Circulation of SARS-CoV-2 and Co-Infection with Plasmodium falciparum in Equatorial Guinea
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Malaria and SARS-CoV-2 Diagnosis
2.3. Data Analysis
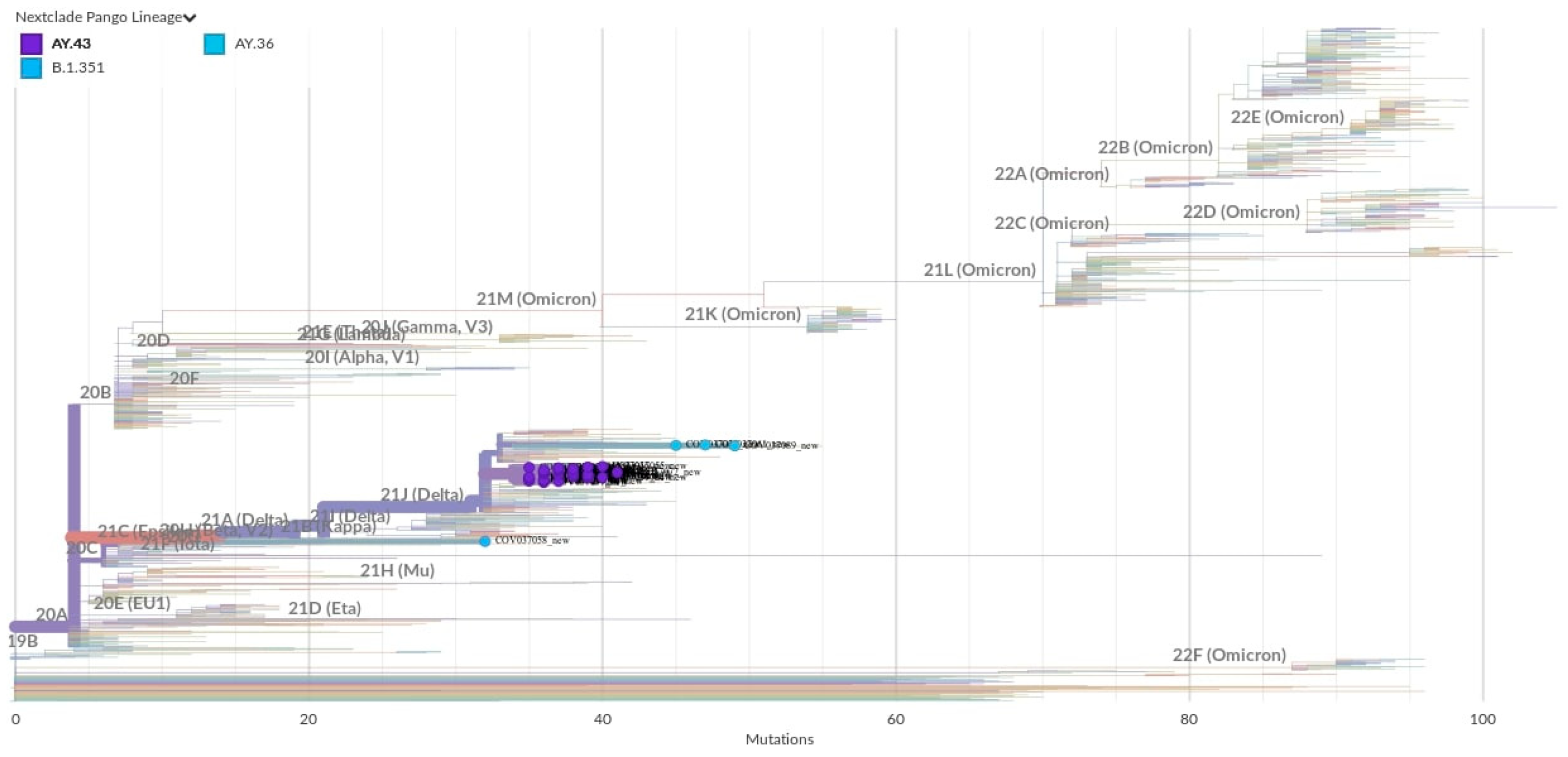
2.4. Whole-Genome Sequencing and Phylogenetic Analysis
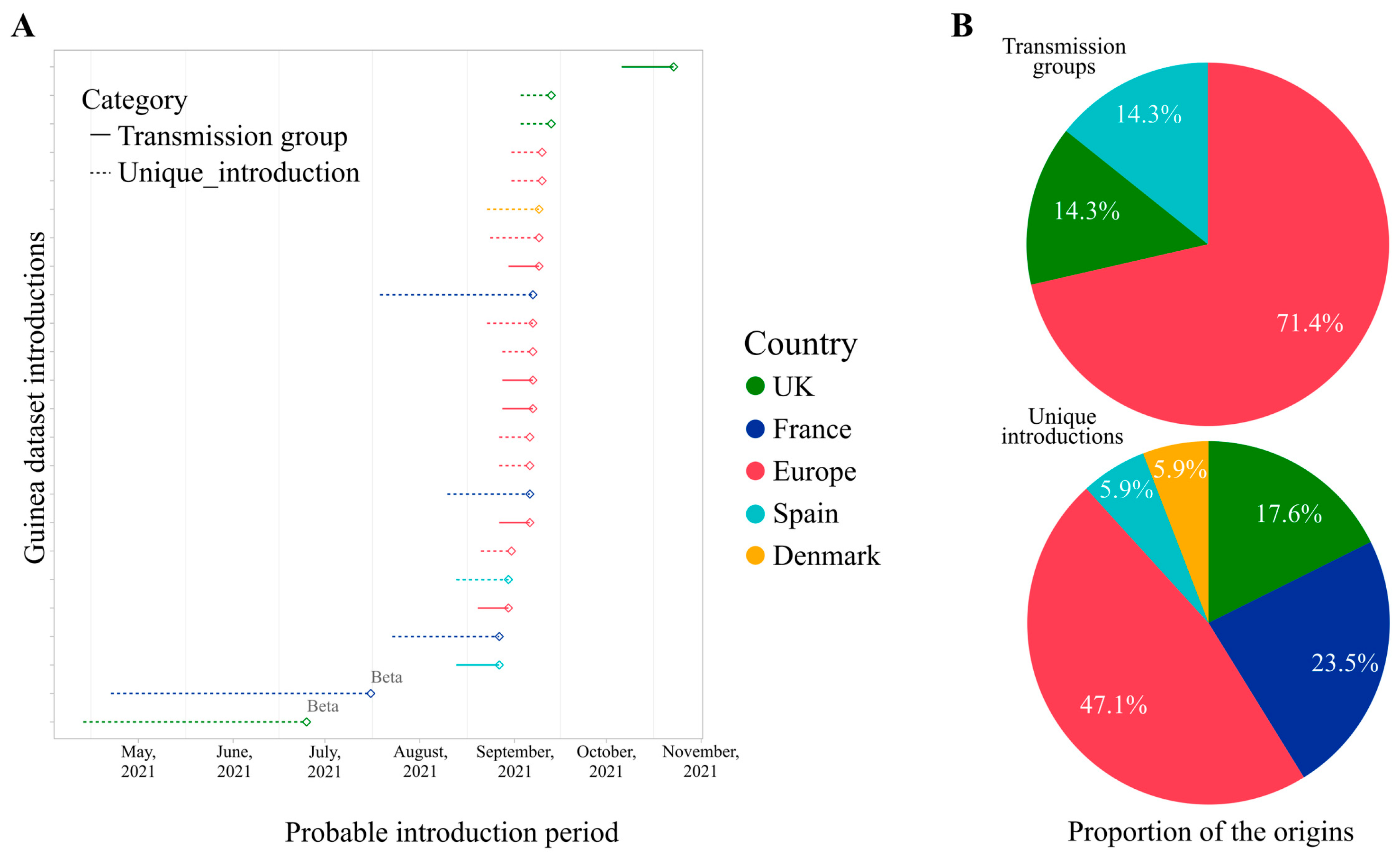
2.5. Genome-Based Epidemiological Analysis
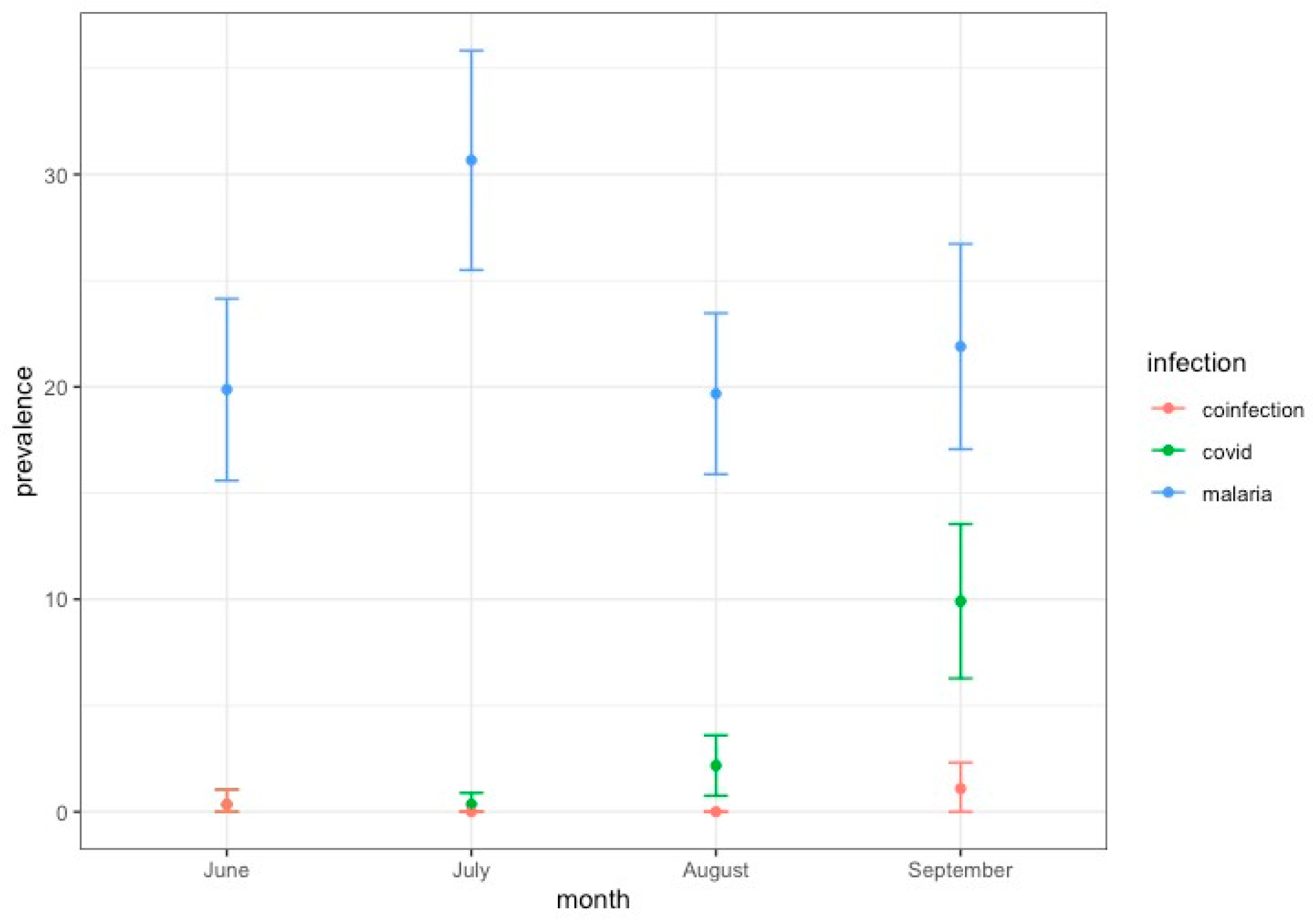
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. WHO COVID-19 Dashboard. Available online: https://covid19.who.int/ (accessed on 28 July 2023).
- Kannan, S.; Ali, P.S.S.; Sheeza, A. Omicron (B.1.1.529)—variant of concern—molecular profile and epidemiology: A mini review. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 8019–8022. [Google Scholar] [CrossRef] [PubMed]
- Tegally, H.; Wilkinson, E.; Lessells, R.J.; Giandhari, J.; Pillay, S.; Msomi, N.; Mlisana, K.; Bhiman, J.N.; von Gottberg, A.; Walaza, S.; et al. Sixteen novel lineages of SARS-CoV-2 in South Africa. Nat. Med. 2021, 27, 440–446. [Google Scholar] [CrossRef]
- Tegally, H.; San, J.E.; Cotten, M.; Moir, M.; Tegomoh, B.; Mboowa, G.; Martin, D.P.; Baxter, C.; Lambisia, A.W.; Diallo, A.; et al. The evolving SARS-CoV-2 epidemic in Africa: Insights from rapidly expanding genomic surveillance. Science 2022, 378, 42. [Google Scholar] [CrossRef]
- Hosch, S.; Mpina, M.; Nyakurungu, E.; Borico, N.S.; Obama, T.M.A.; Ovona, M.C.; Wagner, P.; Rubin, S.E.; Vickos, U.; Milang, D.V.N.; et al. Genomic Surveillance Enables the Identification of Co-infections with Multiple SARS-CoV-2 Lineages in Equatorial Guinea. Front. Public Health 2022, 9, 818401. [Google Scholar] [CrossRef]
- Ozer, E.A.; Simons, L.M.; Adewumi, O.M.; Fowotade, A.A.; Omoruyi, E.C.; Adeniji, J.A.; Olayinka, O.A.; Dean, T.J.; Zayas, J.; Bhimalli, P.P.; et al. Multiple expansions of globally uncommon SARS-CoV-2 lineages in Nigeria. Nat. Commun. 2022, 13, 688. [Google Scholar] [CrossRef]
- MINSABS. Ministerio de Sanidad y Bienestar Social-Guinea Equatorial. Available online: https://guineasalud.org/estadisticas/ (accessed on 23 April 2023).
- Shu, Y.; McCauley, J. GISAID: Global initiative on sharing all influenza data—From vision to reality. Eurosurveillance 2017, 22, 30494. [Google Scholar] [CrossRef]
- World Health Organization. Scaling up Genomic Sequencing in Africa. Available online: https://www.afro.who.int/news/scaling-genomic-sequencing-africa (accessed on 25 June 2023).
- Uwishema, O.; Badri, R.; Onyeaka, H.; Okereke, M.; Akhtar, S.; Mhanna, M.; Zafar, B.; Zahabioun, A.; Said, K.A.; Tovani-Palone, M.R. Fighting Tuberculosis in Africa: The Current Situation Amidst the COVID-19 Pandemic. Disaster Med. Public Health Prep. 2022, 16, 2302–2304. [Google Scholar] [CrossRef]
- Shi, B.; Zheng, J.; Xia, S.; Lin, S.; Wang, X.; Liu, Y.; Zhou, X.-N.; Liu, J. Accessing the syndemic of COVID-19 and malaria intervention in Africa. Infect. Dis. Poverty 2021, 10, 14–25. [Google Scholar] [CrossRef]
- Uwishema, O.; Sapkota, S.; Wellington, J.; Onyeaka, C.V.P.; Onyeaka, H. Leishmaniasis control in the light of the COVID-19 pandemic in Africa. Ann. Med. Surg. 2022, 80, 104263. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. World Malaria Report 2022; WHO: Geneva, Switzerland, 2022. [Google Scholar]
- World Health Organization. The Potential Impact of Health Service Disruptions on the Burden of Malaria: A Modelling Analysis for Countries in Sub-Saharan Africa. 2020. Available online: https://www.who.int/publications/i/item/9789240004641 (accessed on 28 July 2023).
- López-Farfán, D.; Yerbanga, R.S.; Parres-Mercader, M.; Torres-Puente, M.; Gómez-Navarro, I.; Sanou, D.M.S.; Yao, A.F.; Ouédraogo, J.B.; Comas, I.; Irigoyen, N.; et al. Prevalence of SARS-CoV-2 and co-infection with malaria during the first wave of the pandemic (the Burkina Faso case). Front. Public Health 2022, 10, 1048404. [Google Scholar] [CrossRef]
- Amoo, O.S.; Aina, O.O.; Okwuraiwe, A.P.; Onwuamah, C.K.; Shaibu, J.O.; Ige, F.; Owaneze, K.; Agboola, H.O.; Kareithi, D.N.; Onuigbo, T.I.; et al. COVID-19 Spread Patterns Is Unrelated to Malaria Co-Infections in Lagos, Nigeria. Adv. Infect. Dis. 2020, 10, 200–215. [Google Scholar] [CrossRef]
- Sebastião, C.S.; Gaston, C.; Paixão, J.P.; Sacomboio, E.N.M.; Neto, Z.; de Vasconcelos, J.N.; Morais, J. Coinfection between SARS-CoV-2 and vector-borne diseases in Luanda, Angola. J. Med. Virol. 2021, 94, 366–371. [Google Scholar] [CrossRef]
- Ingabire, P.M.; Nantale, R.; Sserwanja, Q.; Nakireka, S.; Musaba, M.W.; Muyinda, A.; Tumuhaise, C.; Namulema, E.; Bongomin, F.; Napyo, A.; et al. Factors associated with prolonged hospitalization of patients with corona virus disease (COVID-19) in Uganda: A retrospective cohort study. Trop. Med. Health 2022, 50, 100. [Google Scholar] [CrossRef]
- Muhammad, Y.; Aminu, Y.K.; Ahmad, A.E.; Iliya, S.; Muhd, N.; Yahaya, M.; Mustapha, A.S.; Tahiru, A.; Abdulkadir, S.S.; Ibrahim, J.S.; et al. An elevated 8-isoprostaglandin F2 alpha (8-iso-PGF2α) in COVID-19 subjects co-infected with malaria. Pan. Afr. Med. J. 2020, 37, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Onosakponome, E.O.; Wogu, M.N.; López-Arellano, M.E. The Role of Sex in Malaria-COVID19 Coinfection and Some Associated Factors in Rivers State, Nigeria. J. Parasitol. Res. 2020, 2020, 8829848. [Google Scholar] [CrossRef]
- Matangila, J.R.; Nyembu, R.K.; Telo, G.M.; Ngoy, C.D.; Sakobo, T.M.; Massolo, J.M.; Muyembe, B.M.; Mvwala, R.K.; Ilunga, C.K.; Limbole, E.B.; et al. Clinical characteristics of COVID-19 patients hospitalized at Clinique Ngaliema, a public hospital in Kinshasa, in the Democratic Republic of Congo: A retrospective cohort study. PLoS ONE 2020, 15, e0244272. [Google Scholar] [CrossRef]
- Achan, J.; Serwanga, A.; Wanzira, H.; Kyagulanyi, T.; Nuwa, A.; Magumba, G.; Kusasira, S.; Sewanyana, I.; Tetteh, K.; Drakeley, C.; et al. Current malaria infection, previous malaria exposure, and clinical profiles and outcomes of COVID-19 in a setting of high malaria transmission: An exploratory cohort study in Uganda. Lancet Microbe 2022, 3, e62–e71. [Google Scholar] [CrossRef]
- Bakamutumaho, B.; Cummings, M.J.; Owor, N.; Kayiwa, J.; Namulondo, J.; Byaruhanga, T.; Muwanga, M.; Nsereko, C.; Rwamutwe, E.; Mutonyi, R.; et al. Severe COVID-19 in uganda across two epidemic phases: A prospective cohort study. Am. J. Trop. Med. Hyg. 2021, 105, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Ben Morton, B.; Barnes, K.G.; Anscombe, C.; Jere, K.; Matambo, P.; Mandolo, J.; Kamng’ona, R.; Brown, C.; Nyirenda, J.; Phiri, T.; et al. Distinct clinical and immunological profiles of patients with evidence of SARS-CoV-2 infection in sub-Saharan Africa. Nat. Commun. 2021, 12, 3554. [Google Scholar] [CrossRef]
- Hussein, R.; Guedes, M.; Ibraheim, N.; Ali, M.M.; El-Tahir, A.; Allam, N.; Abuakar, H.; Pecoits-Filho, R.; Kotanko, P. Impact of COVID-19 and malaria coinfection on clinical outcomes: A retrospective cohort study. Clin. Microbiol. Infect. 2022, 28, 1152.e1–1152.e6. [Google Scholar] [CrossRef] [PubMed]
- Ditombi, B.C.M.; Ngondza, B.P.; Boulingui, C.M.; Nguema, O.A.M.; Ngomo, J.M.N.; M’bondoukwé, N.P.; Moutongo, R.; Mawili-Mboumba, D.P.; Akotet, M.K.B. Malaria and COVID-19 prevalence in a population of febrile children and adolescents living in Libreville. S. Afr. J. Infect. Dis. 2022, 37, 5. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, N.N.; Gajbhiye, R.K.; Bahirat, S.; Lokhande, P.D.; Mathe, A.; Rathi, S.; Warty, N.; Mahajan, K.N.; Srivastava, V.; Kuppusamy, P.; et al. Co-infection of malaria and early clearance of SARS-CoV-2 in healthcare workers. J. Med. Virol. 2021, 93, 2431–2438. [Google Scholar] [CrossRef]
- Berzosa, P.; De Lucio, A.; Romay-Barja, M.; Herrador, Z.; González, V.; García, L.; Fernández-Martínez, A.; Santana-Morales, M.; Ncogo, P.; Valladares, B.; et al. Comparison of three diagnostic methods (microscopy, RDT, and PCR) for the detection of malaria parasites in representative samples from Equatorial Guinea. Malar. J. 2018, 17, 333. [Google Scholar] [CrossRef] [PubMed]
- Datosmacro. Available online: https://datosmacro.expansion.com/demografia/poblacion/guinea-ecuatorial (accessed on 13 December 2023).
- Grubaugh, N.D.; Gangavarapu, K.; Quick, J.; Matteson, N.L.; De Jesus, J.G.; Main, B.J.; Tan, A.L.; Paul, L.M.; Brackney, D.E.; Grewal, S.; et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019, 20, 8. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- O’Toole, Á.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.; et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021, 7, veab064. [Google Scholar] [CrossRef]
- Aksamentov, I.; Roemer, C.; Hodcroft, E.B.; Neher, R.A. Nextclade: Clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw. 2021, 6, 3773. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. NextStrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef]
- Sagulenko, P.; Puller, V.; Neher, R.A. TreeTime: Maximum-likelihood phylodynamic analysis. Virus Evol. 2018, 4, vex042. [Google Scholar] [CrossRef]
- Martínez-Martínez, F.J.; Massinga, A.J.; De Jesus, Á.; Ernesto, R.M.; Cano-Jiménez, P.; Chiner-Oms, Á.; Gómez-Navarro, I.; Guillot-Fernández, M.; Guinovart, C.; Sitoe, A.; et al. Tracking SARS-CoV-2 introductions in Mozambique using pandemic-scale phylogenies: A retrospective observational study. Lancet Glob. Health 2023, 11, e933–e941. [Google Scholar] [CrossRef]
- Turakhia, Y.; Thornlow, B.; Hinrichs, A.S.; De Maio, N.; Gozashti, L.; Lanfear, R.; Haussler, D.; Corbett-Detig, R. Ultrafast Sample placement on Existing tRees (UShER) enables real-time phylogenetics for the SARS-CoV-2 pandemic. Nat. Genet. 2021, 53, 809–816. [Google Scholar] [CrossRef]
- Lanfear, R. Global Phylogenies of SARS-CoV-2 Sequences. Available online: https://github.com/roblanf/sarscov2phylo (accessed on 16 August 2023).
- Thornlow, B. matUtils. Available online: https://usher-wiki.readthedocs.io/en/latest/matUtils.html (accessed on 28 July 2023).
- Sanderson, T. Taxonium, a web-based tool for exploring large phylogenetic trees. eLife 2022, 11, e82392. [Google Scholar] [CrossRef]
- Kusi, K.A.; Frimpong, A.; Partey, F.D.; Lamptey, H.; Amoah, L.E.; Ofori, M.F. High infectious disease burden as a basis for the observed high frequency of asymptomatic SARS-CoV-2 infections in sub-Saharan Africa. AAS Open Res. 2021, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Hussein, M.I.H.; Albashir, A.A.D.; Elawad, O.A.M.A.; Homeida, A. Malaria and COVID-19: Unmasking their ties. Malar. J. 2020, 19, 457. [Google Scholar] [CrossRef] [PubMed]
- Habibzadeh, F. Malaria and the incidence of COVID-19 in Africa: An ecological study. BMC Infect. Dis. 2023, 23, 66. [Google Scholar] [CrossRef] [PubMed]
- Osei, S.A.; Biney, R.P.; Anning, A.S.; Nortey, L.N.; Ghartey-Kwansah, G. Low incidence of COVID-19 case severity and mortality in Africa; Could malaria co-infection provide the missing link? BMC Infect. Dis. 2022, 22, 78. [Google Scholar] [CrossRef] [PubMed]
- Iesa, M.; Osman, M.; Hassan, M.; Dirar, A.; Abuzeid, N.; Mancuso, J.; Pandey, R.; Mohammed, A.; Borad, M.; Babiker, H.; et al. SARS-CoV-2 and Plasmodium falciparum common immunodominant regions may explain low COVID-19 incidence in the malaria-endemic belt. New Microbes New Infect. 2020, 38, 100817. [Google Scholar] [CrossRef]
- Raham, T.F. Influence of malaria endemicity and tuberculosis prevalence on COVID-19 mortality. Public Health 2021, 194, 2021. [Google Scholar] [CrossRef]
- Tapela, K.; Prah, D.A.; Tetteh, B.; Nuokpem, F.; Dosoo, D.; Coker, A.; Kumi-Ansah, F.; Amoako, E.; Assah, K.O.; Kilba, C.; et al. Cellular immune response to SARS-CoV-2 and clinical presentation in individuals exposed to endemic malaria. Cell Rep. 2024, 43, 114533. [Google Scholar] [CrossRef] [PubMed]
- Johns Hopkins University. Johns Hopkins Coronavirus Resource Center. Available online: https://coronavirus.jhu.edu/region/equatorial-guinea (accessed on 10 July 2023).
- Wilkinson, E.; Giovanetti, M.; Tegally, H.; San, J.E.; Lessells, R.; Cuadros, D.; Martin, D.P.; Rasmussen, D.A.; Zekri, A.-R.N.; Sangare, A.K.; et al. A year of genomic surveillance reveals how the SARS-CoV-2 pandemic unfolded in Africa. Science 2021, 374, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://cov-lineages.org/lineage_list.html (accessed on 1 June 2023).
- Mlcochova, P.; Kemp, S.A.; Dhar, M.S.; Papa, G.; Meng, B.; Ferreira, I.A.T.M.; Datir, R.; Collier, D.A.; Albecka, A.; Singh, S.; et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021, 599, 114–119. [Google Scholar] [CrossRef] [PubMed]
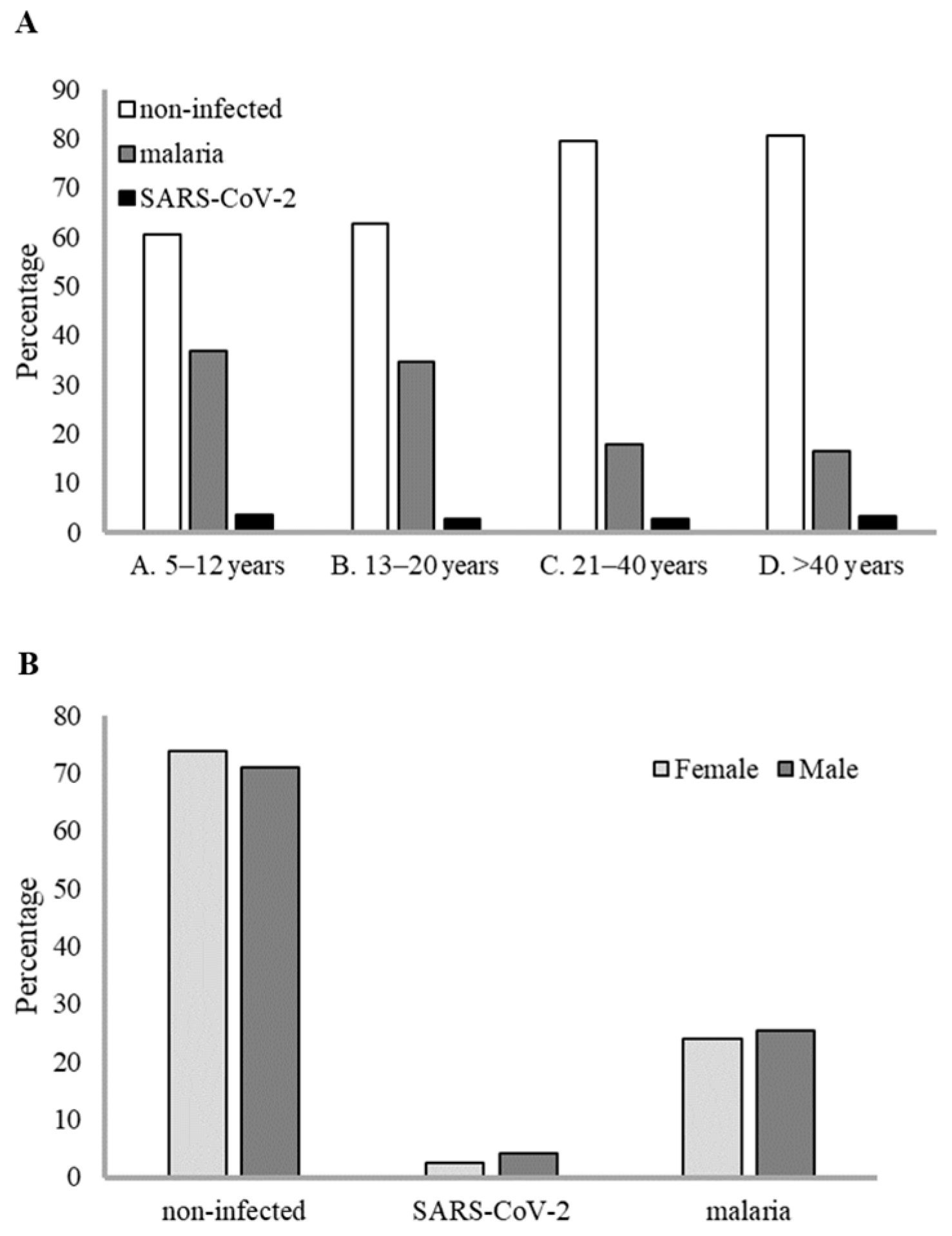
| Age Group (Years) | Gender a | Non-Infected | SARS-CoV-2 | Malaria | SARS-CoV-2 and Malaria | Subtotal | Total |
|---|---|---|---|---|---|---|---|
| A. 5–12 | F | 89 (62.67%) | 4 (2.81%) | 49 (34.51%) | 0 | 142 (54.41%) | |
| M | 69 (57.98%) | 5 (4.20%) | 47 (39.49%) | 2 (1.68%) | 119 (45.59%) | 261 (16.77%) | |
| B. 13–20 | F | 149 (61.08%) | 8 (3.33%) | 84 (35.00%) | 1 (0.42%) | 240 (74.30%) | |
| M | 54 (65.06%) | 1 (1.22%) | 28 (33.73%) | 0 | 83 (25.69%) | 323 (20.76%) | |
| C. 21–40 | F | 444 (79.14%) | 11 (1.96%) | 108 (19.25%) | 2 (0.36%) | 561 (78.02%) | |
| M | 128 (81.01%) | 9 (5.69%) | 21(13.29%) | 0 | 158 (21.97%) | 719 (46.21%) | |
| D. >40 | F | 152 (81.72%) | 5 (2.69%) | 30 (16.13%) | 1 (0.54%) | 186 (73.52%) | |
| M | 52 (77.61%) | 3 (4.48%) | 12 (17.91%) | 0 | 67 (26.48%) | 253 (16.26%) | |
| Subtotal | F | 834 (73.87%) | 28 (2.48%) | 271 (24.00%) | 4 (0.35%) | 1.129 (72.56%) | |
| M | 303 (70.96%) | 18 (4.21%) | 108 (25.29%) | 2 (0.47%) | 427 (27.44%) | ||
| Total | 1.137 (73.07%) | 46 (2.96%) | 379 (24.36%) | 6 (0.39%) | 1.556 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Farfán, D.; Ncogo, P.; Oki, C.; Riloha, M.; Ondo, V.; Cano-Jiménez, P.; Martínez-Martínez, F.J.; Molina-de la Fuente, I.; Comas, I.; Irigoyen, N.; et al. Circulation of SARS-CoV-2 and Co-Infection with Plasmodium falciparum in Equatorial Guinea. Infect. Dis. Rep. 2025, 17, 111. https://doi.org/10.3390/idr17050111
López-Farfán D, Ncogo P, Oki C, Riloha M, Ondo V, Cano-Jiménez P, Martínez-Martínez FJ, Molina-de la Fuente I, Comas I, Irigoyen N, et al. Circulation of SARS-CoV-2 and Co-Infection with Plasmodium falciparum in Equatorial Guinea. Infectious Disease Reports. 2025; 17(5):111. https://doi.org/10.3390/idr17050111
Chicago/Turabian StyleLópez-Farfán, Diana, Policarpo Ncogo, Consuelo Oki, Matilde Riloha, Valero Ondo, Pablo Cano-Jiménez, Francisco José Martínez-Martínez, Irene Molina-de la Fuente, Iñaki Comas, Nerea Irigoyen, and et al. 2025. "Circulation of SARS-CoV-2 and Co-Infection with Plasmodium falciparum in Equatorial Guinea" Infectious Disease Reports 17, no. 5: 111. https://doi.org/10.3390/idr17050111
APA StyleLópez-Farfán, D., Ncogo, P., Oki, C., Riloha, M., Ondo, V., Cano-Jiménez, P., Martínez-Martínez, F. J., Molina-de la Fuente, I., Comas, I., Irigoyen, N., Berzosa, P., Benito Llanes, A., & Gómez-Díaz, E. (2025). Circulation of SARS-CoV-2 and Co-Infection with Plasmodium falciparum in Equatorial Guinea. Infectious Disease Reports, 17(5), 111. https://doi.org/10.3390/idr17050111






