Tropheryma whipplei and Giardia intestinalis Co-Infection: Metagenomic Analysis During Infection and the Recovery Follow-Up
Abstract
1. Introduction
2. Case Description
3. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CNS | Central Nervous System |
| CRP | C-Reactive Protein |
| CT | Computed Tomography |
| CSF | Cerebrospinal Fluid |
| HLA | Histocompatibility Leukocyte Antigen |
| Ig | Immunoglobulin |
| IFN-γ | Interferon-γ |
| IL | Interleukin |
| NGS | Next-Generation Sequencing |
| PAS | Periodic Acid-Schiff |
| PCR | Polymerase Chain Reaction |
| PET | Positron Emission Tomography |
| Th | T-cell type |
| WD | Whipple’s Diseases |
References
- Fenollar, F.; Puéchal, X.; Raoult, D. Whipple’s disease. N. Engl. J. Med. 2007, 35, 55–66. [Google Scholar] [CrossRef]
- Rosain, J.; Casanova, J.L.; Bustamante, J. Human genetics of Whipple’s disease. Curr. Opin. Rheumatol. 2025, 37. [Google Scholar] [CrossRef]
- Schneider, T.; Moos, V.; Loddenkemper, C.; Marth, T.; Fenollar, F.; Raoult, D. Whipple’s disease: New aspects of pathogenesis and treatment. Lancet Infect. Dis. 2008, 8, 179–190. [Google Scholar] [CrossRef]
- Marth, T.; Kleen, N.; Stallmach, A.; Ring, S.; Aziz, S.; Schmidt, C.; Strober, W.; Zeitz, M.; Schneider, T. Dysregulated peripheral and mucosal Th1/Th2 response in Whipple’s disease. Gastroenterology. 2002, 123, 1468–1477. [Google Scholar] [CrossRef]
- Moos, V.; Kunkel, D.; Marth, T.; Feurle, G.E.; LaScola, B.; Ignatius, R.; Zeitz, M.; Schneider, T. Reduced peripheral and mucosal Tropheryma whipplei-specific Th1 response in patients with Whipple’s disease. J. Immunol. 2006, 177, 2015–2022. [Google Scholar] [CrossRef]
- Martinetti, M.; Biagi, F.; Badulli, C.; Feurle, G.E.; Müller, C.; Moos, V.; Schneider, T.; Marth, T.; Marchese, A.; Trotta, L.; et al. The HLA alleles DRB1*13 and DQB1*06 are associated to Whipple’s disease. Gastroenterology 2009, 136, 2289–2294. [Google Scholar] [CrossRef]
- Dolmans, R.A.; Boel, C.H.; Lacle, M.M.; Kusters, J.G. Clinical Manifestations, treatment, and diagnosis of Tropheryma whipplei infections. Clin. Microbiol. Rev. 2017, 30, 529–555. [Google Scholar] [CrossRef]
- Bassotti, G.; Pelli, M.A.; Ribacchi, R.; Miglietti, M.; Cavalletti, M.L.; Rossodivita, M.E.; Giovenali, P.; Morelli, A. Giardia lamblia infestation reveals underlying Whipple’s disease in a patient with longstanding constipation. Am. J. Gastroenterol. 1991, 86, 371–374. [Google Scholar]
- Fenollar, F.; Lepidi, H.; Gérolami, R.; Drancourt, M.; Raoult, D. Whipple disease associated with giardiasis. J. Infect. Dis. 2003, 188, 828–834. [Google Scholar] [CrossRef]
- Gil Ruiz, J.A.; Gil Simón, P.; Aparicio Duque, R.; Mayor Jerez, J.L. Association between Whipple’s disease and Giardia lamblia infection. Rev. Esp. Enferm. Dig. 2005, 97, 521–526. [Google Scholar] [CrossRef]
- Moro, L.; Pomari, E.; Leonardi, M.; La Marca, G.; Pajola, B.; Mazzi, C.; Piubelli, C.; Beltrame, A. Tropheryma whipplei, Helicobacter pylori, and intestinal protozoal co-infections in Italian and immigrant populations: A cross-sectional study. Microorganisms 2022, 10, 769. [Google Scholar] [CrossRef]
- Dreier, J.; Szabados, F.; von Herbay, A.; Kroger, T.; Kleesiek, K. Tropheryma whipplei infection of an acellular porcine heart valve bioprosthesis in a patient who did not have intestinal Whipple’s disease. J. Clin. Microbiol. 2004, 42, 4487–4493. [Google Scholar] [CrossRef]
- Hinrikson, H.P.; Dutly, F.; Nair, S.; Altwegg, M. Detection of three different types of “Tropheryma whippelii” directly from clinical specimens by sequencing, single-strand conformation polymorphism (SSCP) analysis and type-specific PCR of their 16S-23S ribosomal intergenic spacer region. Int. J. Syst. Bacteriol. 1999, 49, 1701–1706. [Google Scholar] [CrossRef]
- Hinrikson, H.P.; Dutly, F.; Altwegg, M. Evaluation of a specific nested PCR targeting domain III of the 23S rRNA gene of “Tropheryma whippelii” and proposal of a classification system for its molecular variants. J. Clin. Microbiol. 2000, 38, 595–599. [Google Scholar] [CrossRef]
- Clarke, M.C.C.; Price, R.N. Delayed diagnosis of Whipple’s disease complicated by Jarisch-Herxheimer reaction to ceftriaxone treatment: A case report and literature review. Trop. Med. Infect. Dis. 2022, 7, 40. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data [Online]. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 20 January 2025).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Lu, J.; Rincon, N.; Wood, D.E.; Breitwieser, F.P.; Pockrandt, C.; Langmead, B.; Salzberg, S.L.; Steinegger, M. Metagenome analysis using the Kraken software suite. Nat. Protoc. 2022, 17, 2815–2839. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:13033997. [Google Scholar]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Giga Sci. 2021, 10, giab008. [Google Scholar] [CrossRef]
- Sultan, S.; El-Mowafy, M.; Elgaml, A.; Ahmed, T.A.E.; Hassan, H.; Mottawea, W. Metabolic Influences of gut microbiota dysbiosis on inflammatory bowel disease. Front. Physiol. 2021, 27, 715506. [Google Scholar] [CrossRef]
- Maciel-Fiuza, M.F.; Muller, G.C.; Campos, D.M.S.; do Socorro Silva Costa, P.; Peruzzo, J.; Bonamigo, R.R.; Veit, T.; Vianna, F.S.L. Role of gut microbiota in infectious and inflammatory diseases. Front. Microbiol. 2023, 27, 1098386. [Google Scholar] [CrossRef]
- Marth, T. Tropheryma whipplei, immunosuppression and Whipple’s Disease: From a low-pathogenic, environmental infectious organism to a rare, multifaceted inflammatory complex. Dig. Dis. 2015, 33, 190–199. [Google Scholar] [CrossRef]
- Moos, V.; Schneider, T. Changing paradigms in Whipple’s disease and infection with Tropheryma whipplei. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 1151–1158. [Google Scholar] [CrossRef]
- Cappellini, A.; Minerba, P.; Maimaris, S.; Biagi, F. Whipple’s disease: A rare disease that can be spotted by many doctors. Eur. J. Intern. Med. 2024, 121, 25–29. [Google Scholar] [CrossRef]
- Iyer, S.S.; Cheng, G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit. Rev. Immunol. 2012, 32, 23–63. [Google Scholar] [CrossRef]
- Ghigo, E.; Barry, A.O.; Pretat, L.; Al Moussawi, K.; Desnues, B.; Capo, C.; Kornfeld, H.; Mege, J.L. IL-16 promotes T. whipplei replication by inhibiting phagosome conversion and modulating macrophage activation. PLoS ONE 2010, 5, e13561. [Google Scholar] [CrossRef]
- Moos, V.; Schmidt, C.; Geelhaar, A.; Kunkel, D.; Allers, K.; Schinnerling, K.; Loddenkemper, C.; Fenollar, F.; Moter, A.; Raoult, D.; et al. Impaired immune functions of monocytes and macrophages in Whipple’s disease. Gastroenterology 2010, 138, 210–220. [Google Scholar] [CrossRef]
- Eck, M.; Kreipe, H.; Harmsen, D.; Müller-Hermelink, H.K. Invasion and destruction of mucosal plasma cells by Tropheryma whippelii. Hum. Pathol. 1997, 28, 1424–1428. [Google Scholar] [CrossRef]
- Huangfu, L.; Li, R.; Huang, Y.; Wang, S. The IL-17 family in diseases: From bench to bedside. Signal Transduct. Target Ther. 2023, 8, 402. [Google Scholar] [CrossRef] [PubMed]
- Faubert, G. Immune Response to Giardia duodenalis. Clin. Microbiol. Rev. 2000, 13, 35–54. [Google Scholar] [CrossRef]
- Gisbertz, I.A.; Bergmans, D.C.; van Marion-Kievit, J.A.; Haak, H.R. Concurrent Whipple’s disease and Giardia lamblia infection in a patient presenting with weight loss. Eur. J. Intern. Med. 2001, 12, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Ruttenberg, D.; Ress, S.R.; Price, S.K.; Girdwood, A.H.; Marks, I.N. Common variable hypogammaglobulinemia. A case report. J. Clin. Gastroenterol. 1990, 12, 336–340. [Google Scholar] [CrossRef]
- Dutly, F.; Altwegg, M. Whipple’s disease and “Tropheryma whippelii”. Clin. Microbiol. Rev. 2001, 14, 561–583. [Google Scholar] [CrossRef]
- Fekete, E.; Allain, T.; Siddiq, A.; Sosnowski, O.; Buret, A.G. Giardia spp. and the gut microbiota: Dangerous liaisons. Front. Microbiol. 2021, 11, 618106. [Google Scholar] [CrossRef] [PubMed]
- Ankarklev, J.; Hestvik, E.; Lebbad, M.; Lindh, J.; Kaddu-Mulindwa, D.H.; Andersson, J.O.; Tylleskär, T.; Tumwine, J.K.; Svärd, S.G. Common coinfections of Giardia intestinalis and Helicobacter pylori in non-symptomatic Ugandan children. PLoS Negl. Trop. Dis. 2012, 6, e1780. [Google Scholar] [CrossRef]
- Chen, C.C.; Liou, J.M.; Lee, Y.C.; Hong, T.C.; El-Omar, E.M.; Wu, M.S. The interplay between Helicobacter pylori and gastrointestinal microbiota. Gut Microbes 2021, 13, 1909459. [Google Scholar] [CrossRef]
- Church, J.A.; Fitzgerald, F.; Walker, A.S.; Gibb, D.M.; Prendergast, A.J. The expanding role of co-trimoxazole in developing countries. Lancet Infect. Dis. 2015, 15, 327–339. [Google Scholar] [CrossRef]
- Boumaza, A.; Ben Azzouz, E.; Arrindell, J.; Lepidi, H.; Mezouar, S.; Desnues, B. Whipple’s disease and Tropheryma whipplei infections: From bench to bedside. Lancet Infect. Dis. 2022, 22, e280–e291. [Google Scholar] [CrossRef]
- Delgado, S.; Suárez, A.; Mayo, B. Identification, typing and characterization of Propionibacterium strains from healthy mucosa of the human stomach. Int. J. Food Microbiol. 2011, 149, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Kumpitsch, C.; Koskinen, K.; Schöpf, V.; Moissl-Eichinger, C. The microbiome of the upper respiratory tract in health and disease. BMC Biol. 2019, 17, 87. [Google Scholar] [CrossRef] [PubMed]
- Mayslich, C.; Grange, P.A.; Dupin, N. Cutibacterium acnes as an opportunistic pathogen: An update of its virulence-associated factors. Microorganisms 2021, 9, 303. [Google Scholar] [CrossRef] [PubMed]
- Limon, J.J.; Skalski, J.H.; Underhill, D.M. Commensal fungi in health and disease. Cell Host Microbe. 2017, 22, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability, and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef]
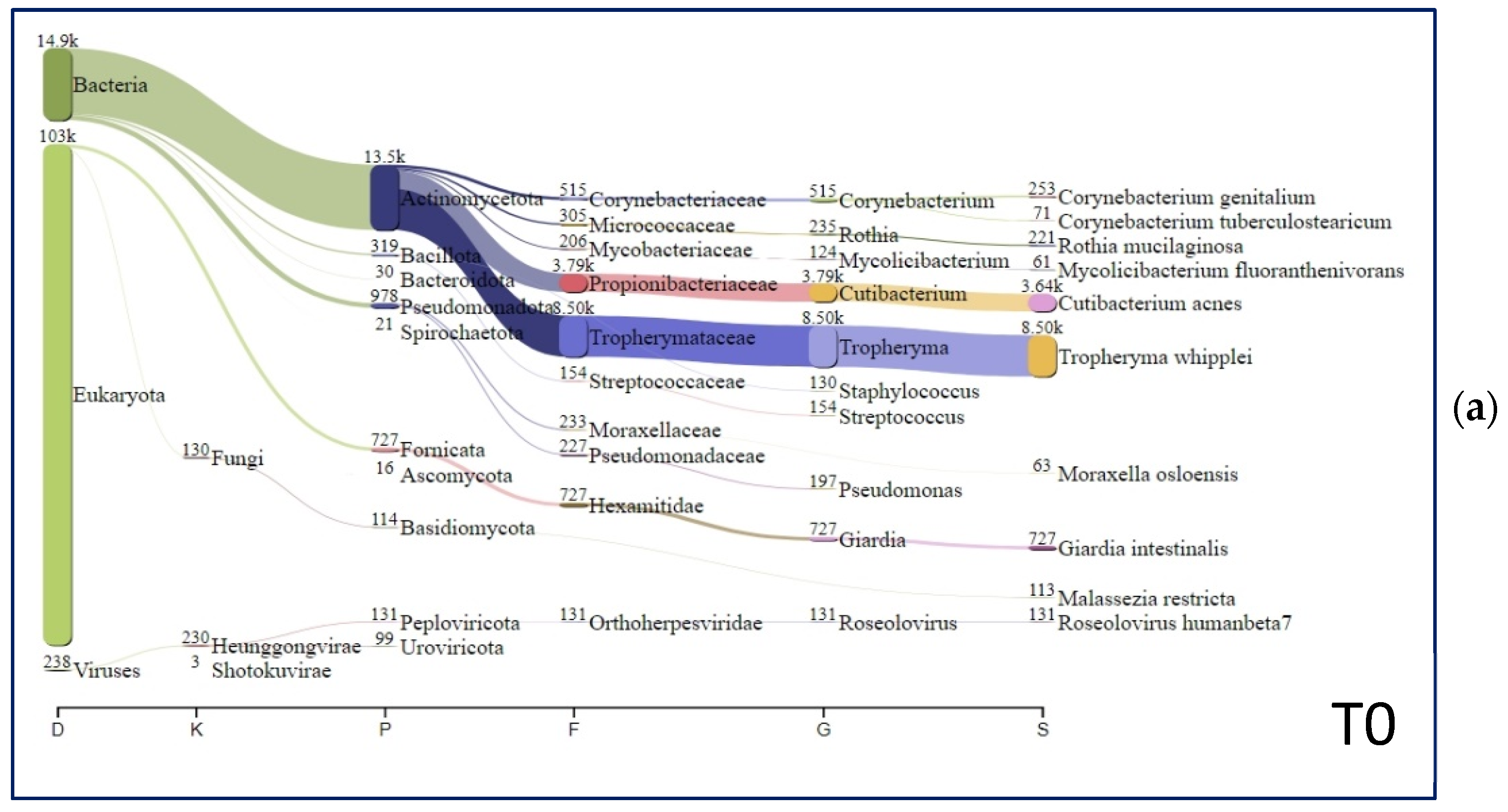
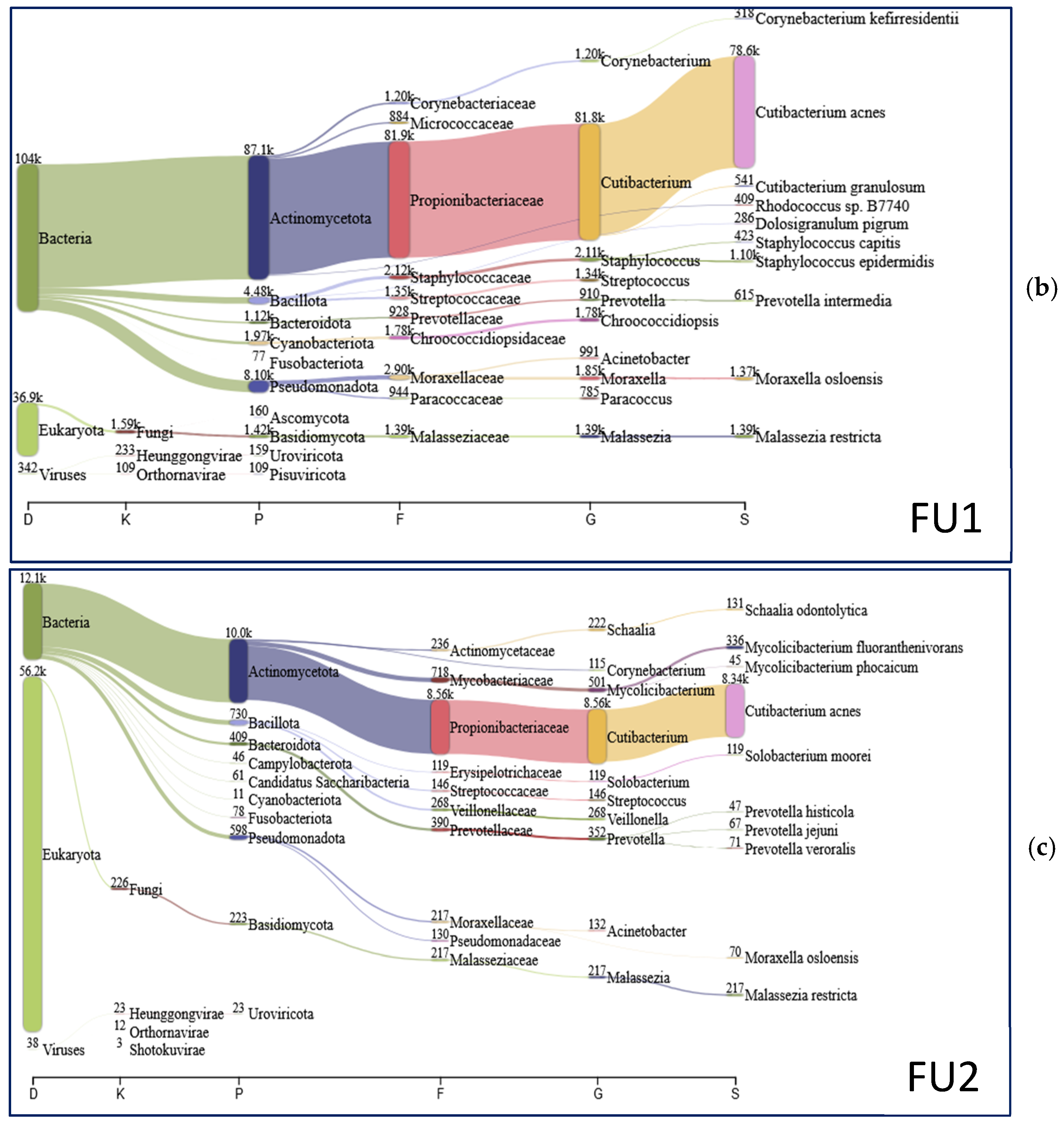
| Laboratory Examinations (Reference Range) | T0 | FU1 | FU2 |
|---|---|---|---|
| Blood cell count, ×1012/L (4.3–5.9) | 2.58 | 4.5 | 4.9 |
| Hemoglobin, g/dL (13–16) | 6.6 | 13.6 | 13.9 |
| Platelet, ×109/L (150–450) | 171 | 201 | 183 |
| White blood cells, ×109/L (4.0–10) | 5.0 | 6.3 | 6.9 |
| Erythrocyte sedimentation rate, mm/h (0–20) | 28 | 13 | 12 |
| INR, (0.91–1.08) | 0.92 | 0.96 | 0.93 |
| Iron, mcg/dL (60–160) | 9.0 | 70 | 85 |
| Sodium, mmol/L (130–150) | 135 | 136 | 135 |
| Potassium, nmol/L (3.5–5.0) | 3.6 | 3.7 | 3.9 |
| C-Reactive protein, mg/L (<5) | 27 | 6 | 2 |
| Ferritin, mcg/L (20–270) | 98 | 101 | 107 |
| Transferrin saturation, % (15–50) | 3.2 | 18 | 20 |
| Alpha-globulins, g/dL (0.11–0.35) | 0.56 | 0.22 | 0.28 |
| Gamma-globulins, g/dL (0.58–1.75) | 0.84 | 0.91 | 0.85 |
| IgM, g/dL (0.5–2.0) | 0.6 | 0.8 | 1.1 |
| IgG, g/dL (5.3–16.5) | 9.0 | 8.9 | 9.2 |
| IgA, g/dL (0.8–5.3) | 3.6 | 2.5 | 3.1 |
| Serum creatinine, mg/dL (0.67–1.2) | 0.98 | 0.86 | 0.91 |
| Albumin, g/dL (>3.5) | 2.7 | 4.6 | 4.7 |
| Bilirubin, mg/dL (0.3–1.2) | 0.3 | 0.6 | 0.8 |
| Glutamic oxaloacetic Transaminase, IU/L (<34) | 33 | 21 | 20 |
| Glutamic pyruvic transaminase, IU/L (<49) | 20 | 11 | 11 |
| Alkaline phosphatase, IU/L (46–116) | 100 | 97 | 85 |
| Lactate dehydrogenase, IU/L (<250) | 160 | 135 | 130 |
| Species | T0 (%) | FU1 (%) | FU2 (%) |
|---|---|---|---|
| Tropheryma whipplei | 51.79 | 0.00 | 0.00 |
| Cutibacterium acnes | 22.18 | 70.50 | 66.84 |
| Giardia intestinalis | 4.43 | 0.00 | 0.03 |
| Corynebacterium genitalium | 1.54 | 0.00 | 0.00 |
| Rothia mucilaginosa | 1.35 | 0.01 | 0.06 |
| Roseolovirus humanbeta7 | 0.80 | 0.07 | 0.00 |
| Malassezia restricta | 0.69 | 1.24 | 1.74 |
| Moraxella osloensis | 0.38 | 1.23 | 0.56 |
| Mycolicibacterium fluoranthenivorans | 0.37 | 0.12 | 2.69 |
| Staphylococcus epidermidis | 0.32 | 0.99 | 0.10 |
| Staphylococcus capitis | 0.09 | 0.38 | 0.05 |
| Cutibacterium granulosum | 0.08 | 0.49 | 0.00 |
| Corynebacterium kroppenstedtii | 0.08 | 0.13 | 0.19 |
| Corynebacterium kefirresidentii | 0.05 | 0.29 | 0.09 |
| Streptococcus thermophilus | 0.05 | 0.10 | 0.03 |
| Schaalia odontolytica | 0.04 | 0.00 | 1.05 |
| Paracoccus yeei | 0.04 | 0.19 | 0.01 |
| Kocuria rhizophila | 0.04 | 0.09 | 0.03 |
| Acinetobacter johnsonii | 0.02 | 0.10 | 0.03 |
| Finegoldia magna | 0.01 | 0.09 | 0,00 |
| Streptococcus sanguinis | 0.01 | 0.12 | 0.02 |
| Dolosigranulum pigrum | 0.00 | 0.26 | 0.00 |
| Streptococcus mitis | 0.00 | 0.15 | 0.02 |
| Prevotella intermedia | 0.00 | 0.55 | 0.02 |
| Acidovorax temperans | 0.00 | 0.18 | 0.00 |
| Lactobacillus iners | 0.00 | 0.17 | 0.00 |
| Haemophilus parainfluenzae | 0.00 | 0.16 | 0.26 |
| Rhodococcus sp. B7740 | 0.00 | 0.37 | 0.00 |
| Solobacterium moorei | 0.00 | 0.00 | 0.95 |
| Corynebacterium accolens | 0.00 | 0.17 | 0.02 |
| Lactobacillus crispatus | 0.00 | 0.13 | 0.00 |
| Staphylococcus pasteuri | 0.00 | 0.11 | 0.00 |
| Others | 15.64 | 21.64 | 25.20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anselmo, A.; Rizzo, F.; Gervasi, E.; Corrent, L.; Ciammaruconi, A.; Fillo, S.; Fortunato, A.; Marella, A.M.; Costantini, S.; Baldassari, L.; et al. Tropheryma whipplei and Giardia intestinalis Co-Infection: Metagenomic Analysis During Infection and the Recovery Follow-Up. Infect. Dis. Rep. 2025, 17, 62. https://doi.org/10.3390/idr17030062
Anselmo A, Rizzo F, Gervasi E, Corrent L, Ciammaruconi A, Fillo S, Fortunato A, Marella AM, Costantini S, Baldassari L, et al. Tropheryma whipplei and Giardia intestinalis Co-Infection: Metagenomic Analysis During Infection and the Recovery Follow-Up. Infectious Disease Reports. 2025; 17(3):62. https://doi.org/10.3390/idr17030062
Chicago/Turabian StyleAnselmo, Anna, Fabiana Rizzo, Elena Gervasi, Luca Corrent, Andrea Ciammaruconi, Silvia Fillo, Antonella Fortunato, Anna Maria Marella, Silvia Costantini, Luca Baldassari, and et al. 2025. "Tropheryma whipplei and Giardia intestinalis Co-Infection: Metagenomic Analysis During Infection and the Recovery Follow-Up" Infectious Disease Reports 17, no. 3: 62. https://doi.org/10.3390/idr17030062
APA StyleAnselmo, A., Rizzo, F., Gervasi, E., Corrent, L., Ciammaruconi, A., Fillo, S., Fortunato, A., Marella, A. M., Costantini, S., Baldassari, L., Lista, F., & Ciervo, A. (2025). Tropheryma whipplei and Giardia intestinalis Co-Infection: Metagenomic Analysis During Infection and the Recovery Follow-Up. Infectious Disease Reports, 17(3), 62. https://doi.org/10.3390/idr17030062








