Liquid Biopsy in Pancreatic Ductal Adenocarcinoma: Clinical Utility, Trials, and Future Directions
Abstract
1. Introduction
2. Methods
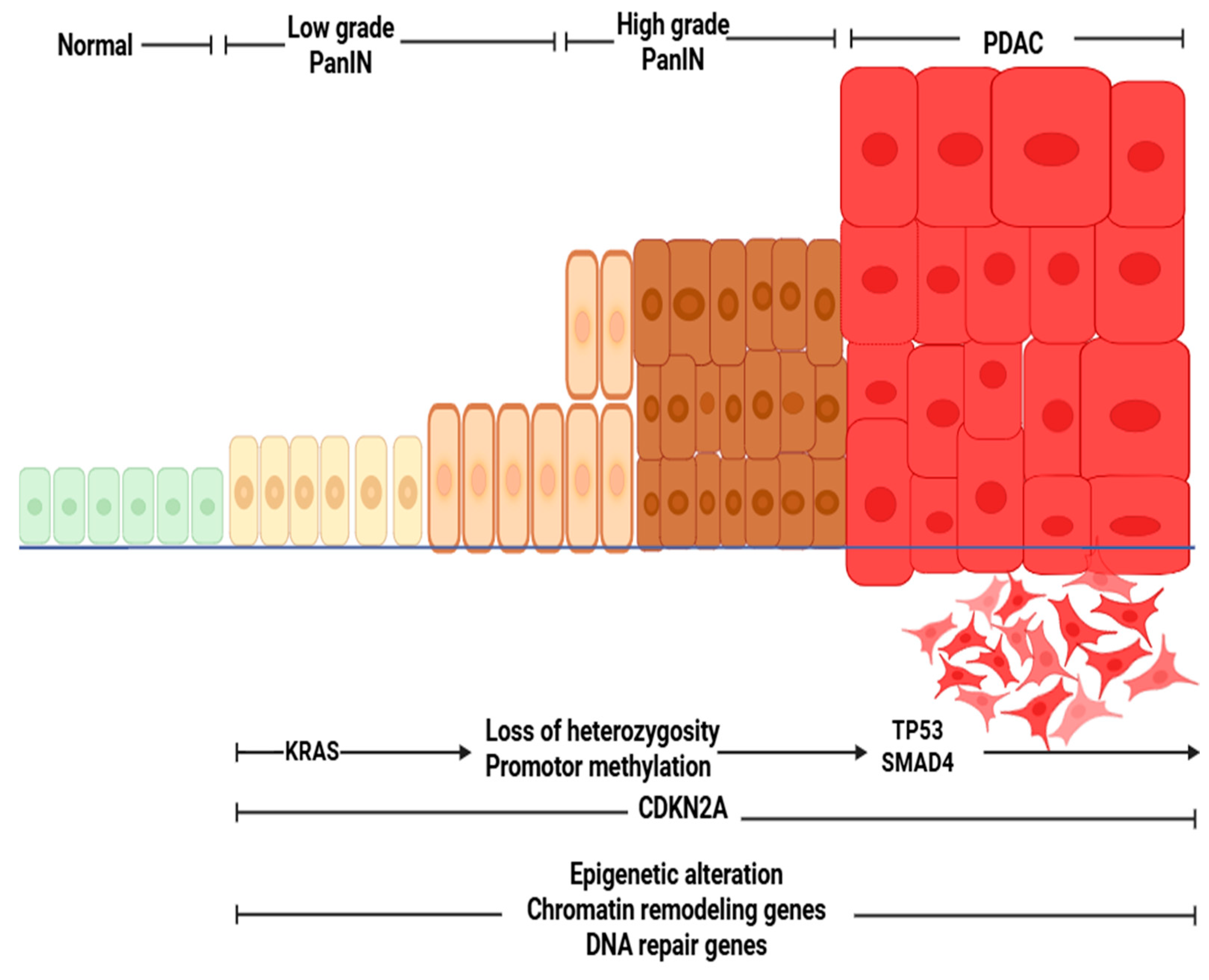
3. Pathophysiology of Pancreatic Ductal Adenocarcinoma (PDAC)
3.1. Genetic Landscape of PDAC
3.2. Tumor Microenvironment
3.3. Mechanisms of Tumor Dissemination
4. Overview of Liquid Biopsy in Oncology
4.1. Biological Basis of Liquid Biopsy
4.2. Comparison with Tissue Biopsy
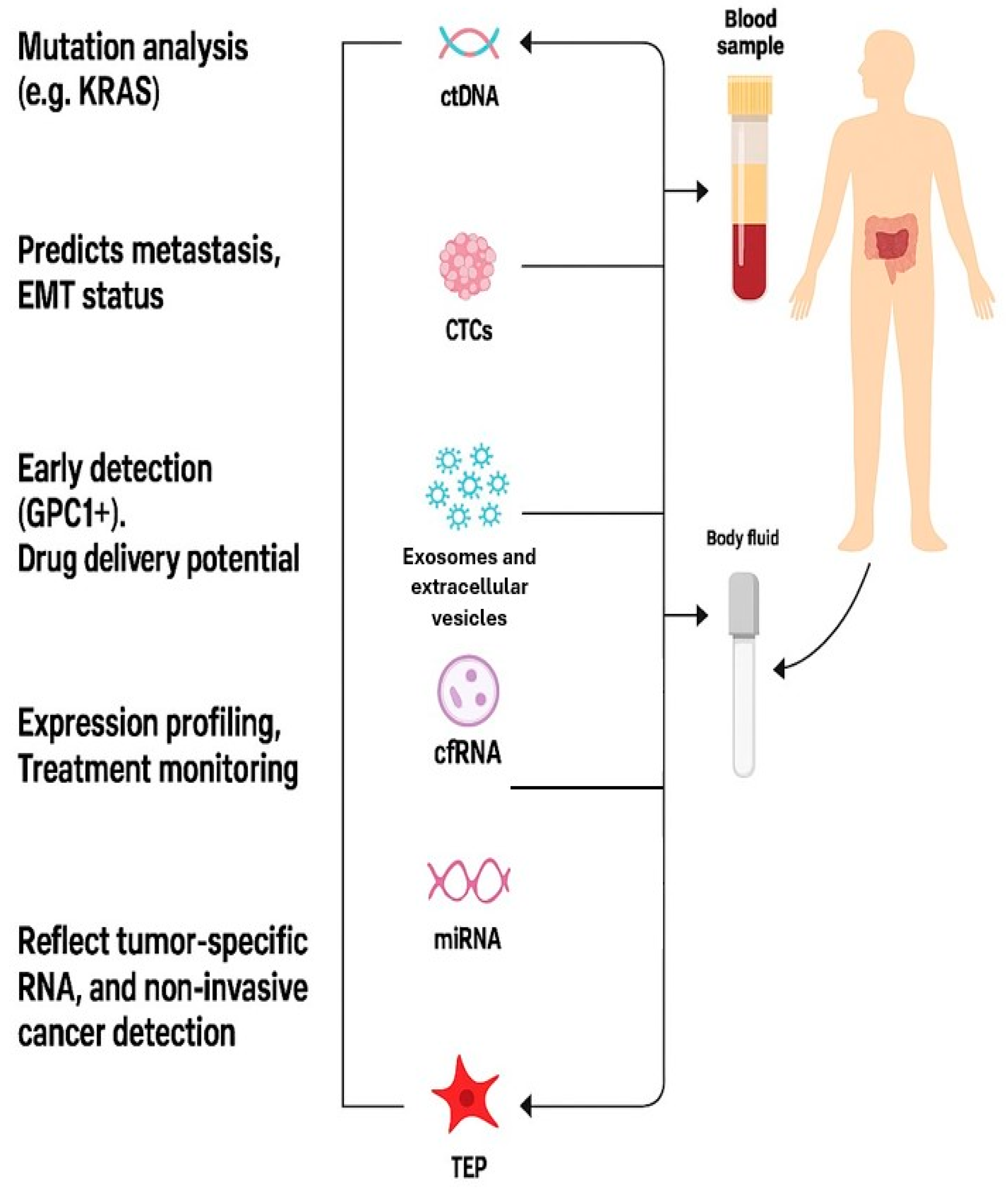
5. Molecular and Cellular Components of Liquid Biopsy in PDAC
5.1. Circulating Tumor DNA (ctDNA)
5.1.1. Detection Techniques
5.1.2. Clinical Applications
5.2. Circulating Tumor Cells (CTCs)
5.2.1. Isolation Techniques
Label-Dependent (Immunoaffinity-Based) Techniques:
Label-Independent (Physical Property-Based) Techniques:
5.2.2. Clinical Relevance in PDAC
5.3. Exosomes and Extracellular Vesicles
5.3.1. Isolation Techniques
5.3.2. Diagnostic Potential
5.4. Tumor-Educated Platelets (TEPs)
5.4.1. Detection and Isolation Techniques
Platelet Isolation
RNA Sequencing of TEPs
Bioinformatics and Machine Learning
Multiplexed Platforms
5.4.2. Diagnostic Potential
- -Early Detection and Diagnosis
- -Prognostic Value
- -Monitoring Therapeutic Response
- -Potential for Personalized Medicine
5.5. Cell-Free RNAs (cfRNAs) and miRNAs
6. Techniques and Analytical Platforms in Liquid Biopsy
6.1. Sample Types and Collection
6.2. Biomarker-Specific Isolation Techniques
6.3. Molecular Detection Techniques
6.4. Bioinformatics and Data Interpretation
6.5. Biological Limitations
7. Clinical Trials and Ongoing Research
8. Future Directions in Liquid Biopsy for Pancreatic Cancer
8.1. Multianalyte and Multi-Omics Liquid Biopsy Platforms
8.2. Artificial Intelligence and Predictive Modeling
8.3. Minimal Residual Disease (MRD) and Personalized Surveillance
8.4. Integration into Precision Oncology and Adaptive Trials
8.5. Knowledge Gaps and Limitations
8.6. Future Directions
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Garrido-Laguna, I.; Hidalgo, M. Pancreatic cancer: From state-of-the-art treatments to promising novel therapies. Nat. Rev. Clin. Oncol. 2015, 12, 319–334. [Google Scholar] [CrossRef] [PubMed]
- Siegel Rebecca, L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA A Cancer J. Clin. 2018, 68, 7–30. [Google Scholar]
- McGuigan, A.; Kelly, P.; Turkington, R.C.; Jones, C.; Coleman, H.G.; McCain, R.S. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 2018, 24, 4846–4861. [Google Scholar] [CrossRef] [PubMed]
- Sarantis, P.; Koustas, E.; Papadimitropoulou, A.; Papavassiliou, A.G.; Karamouzis, M.V. Pancreatic ductal adenocarcinoma: Treatment hurdles, tumor microenvironment and immunotherapy. World J. Gastrointest. Oncol. 2020, 12, 173–181. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Knox, J.J.; Jang, G.H.; Grant, R.C.; Zhang, A.; Ma, L.; Elimova, E.; Jang, R.; Moore, M.; Biagi, J.; Tehfe, M.; et al. Whole genome and transcriptome profiling in advanced pancreatic cancer patients on the COMPASS trial. Nat. Commun. 2025, 16, 5919. [Google Scholar] [CrossRef]
- Bilreiro, C.; Andrade, L.; Santiago, I.; Marques, R.M.; Matos, C. Imaging of pancreatic ductal adenocarcinoma—An update for all stages of patient management. Eur. J. Radiol. Open 2024, 12, 100553. [Google Scholar] [CrossRef]
- Dhar, J.; Samanta, J.; Nabi, Z.; Aggarwal, M.; Conti Bellocchi, M.C.; Facciorusso, A.; Frulloni, L.; Crinò, S.F. Endoscopic Ultrasound-Guided Pancreatic Tissue Sampling: Lesion Assessment, Needles, and Techniques. Medicina 2024, 60, 2021. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Grossberg, A.J.; Chu, L.C.; Deig, C.R.; Fishman, E.K.; Hwang, W.L.; Maitra, A.; Marks, D.L.; Mehta, A.; Nabavizadeh, N.; Simeone, D.M.; et al. Multidisciplinary standards of care and recent progress in pancreatic ductal adenocarcinoma. CA A Cancer J. Clin. 2020, 70, 375–403. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ren, B.; Liu, X.; Suriawinata, A.A. Pancreatic ductal adenocarcinoma and its precursor lesions: Histopathology, cytopathology, and molecular pathology. Am. J. Pathol. 2019, 189, 9–21. [Google Scholar] [CrossRef]
- Yousef, M.M.; Hurd, M.W.; Yousef, A.M.; A Snyder, R.; Knafl, M.; Fanaeian, M.M.; Chacko, R.; Peterson, J.; Smaglo, B.G.; Wolff, R.A.; et al. Frequency and oncologic outcomes of KRAS mutations in circulating tumor DNA of patients with pancreatic ductal adenocarcinoma. J. Clin. Oncol. 2024, 42, 636. [Google Scholar] [CrossRef]
- Waters, A.M.; Der, C.J. KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb. Perspect. Med. 2018, 8, a031435. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Iwatate, Y.; Hoshino, I.; Ishige, F.; Itami, M.; Chiba, S.; Arimitsu, H.; Yanagibashi, H.; Nagase, H.; Yokota, H.; Takayama, W. Prognostic significance of p16 protein in pancreatic ductal adenocarcinoma. Mol. Clin. Oncol. 2020, 13, 83–91. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mello, S.S.; Flowers, B.M.; Mazur, P.K.; Lee, J.J.; Müller, F.; Denny, S.K.; Ferreira, S.; Hanson, K.; Kim, S.K.; Greenleaf, W.J.; et al. Multifaceted role for p53 in pancreatic cancer suppression. Proc. Natl. Acad. Sci. USA 2023, 120, e2211937120. [Google Scholar] [CrossRef]
- Stefanoudakis, D.; Frountzas, M.; Schizas, D.; Michalopoulos, N.V.; Drakaki, A.; Toutouzas, K.G. Significance of TP53, CDKN2A, SMAD4 and KRAS in Pancreatic Cancer. Curr. Issues Mol. Biol. 2024, 46, 2827–2844. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of cancer: New dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Pishvaian, M.J.; Bender, R.J.; Halverson, D.; Rahib, L.; Hendifar, A.E.; Mikhail, S.; Chung, V.; Picozzi, V.J.; Sohal, D.; Blais, E.M.; et al. Molecular Profiling of Patients with Pancreatic Cancer: Initial Results from the Know Your Tumor Initiative. Clin. Cancer Res. 2018, 24, 5018–5027. [Google Scholar] [CrossRef] [PubMed]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.-M.; Gingras, M.-C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.C.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef]
- Furukawa, T.; Kuboki, Y.; Tanji, E.; Yoshida, S.; Hatori, T.; Yamamoto, M.; Shibata, N.; Shimizu, K.; Kamatani, N.; Shiratori, K. Whole-exome sequencing uncovers frequent GNAS mutations in intraductal papillary mucinous neoplasms of the pancreas. Sci. Rep. 2011, 531, 161. [Google Scholar] [CrossRef]
- National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology: Pancreatic Adenocarcinoma; Version 2.2025; National Comprehensive Cancer Network: Plymouth Meeting, PA, USA, 2025; Available online: https://www.nccn.org/professionals/physician_gls/pdf/pancreatic.pdf (accessed on 8 October 2025).
- Neesse, A.; Algül, H.; Tuveson, D.A.; Gress, T.M. Stromal biology and therapy in pancreatic cancer. Gut 2015, 64, 1379–1389. [Google Scholar] [CrossRef] [PubMed]
- Sherman, M.H.; Beatty, G.L. Tumor microenvironment in pancreatic cancer pathogenesis and therapeutic resistance. Annu. Rev. Pathol. Mech. Dis. 2023, 18, 123–148. [Google Scholar] [CrossRef] [PubMed]
- Beatty, G.L.; Winograd, R.; Evans, R.A.; Long, K.B.; Luque, S.L.; Lee, J.W.; Clendenin, C.; Gladney, W.L.; Knoblock, D.M.; Guirnalda, P.D.; et al. Exclusion of T cells from pancreatic carcinomas in mice is regulated by Ly6Clow F4/80+ extratumoral macrophages. Cancer Immunol. Res. 2015, 3, 233–239. [Google Scholar] [CrossRef]
- Ju, Y.; Xu, D.; Liao, M.; Sun, Y.; Bao, W.-D.; Yao, F.; Ma, L. Barriers and opportunities in pancreatic cancer immunotherapy. NPJ Precis. Oncol. 2024, 8, 199. [Google Scholar] [CrossRef] [PubMed]
- Elyada, E.; Bolisetty, M.; Laise, P.; Flynn, W.F.; Courtois, E.T.; Burkhart, R.A.; Teinor, J.A.; Belleau, P.; Biffi, G.; Lucito, M.S.; et al. Cross-species single-cell analysis reveals a conserved myeloid population in pancreatic ductal adenocarcinoma. Cancer Discov. 2019, 9, 1102–1123. [Google Scholar] [CrossRef]
- Öhlund, D.; Handly-Santana, A.; Biffi, G.; Elyada, E.; Almeida, A.S.; Ponz-Sarvise, M.; Corbo, V.; Oni, T.E.; Hearn, S.A.; Lee, E.J.; et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. Cancer Cell. 2017, 31, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Mello, A.M.; Ngodup, T.; Lee, Y.; Donahue, K.L.; Li, J.; Rao, A.; Carpenter, E.S.; Crawford, H.C.; di Magliano, M.P.; Lee, K.E. Hypoxia promotes an inflammatory phenotype of fibroblasts in pancreatic cancer. Oncogenesis 2022, 11, 56. [Google Scholar] [CrossRef]
- Yang, J.; Liu, Y.; Liu, S. The role of epithelial-mesenchymal transition and autophagy in pancreatic ductal adenocarcinoma invasion. Cell Death Dis. 2023, 14, 506. [Google Scholar] [CrossRef]
- Zheng, X.; Carstens, J.; Kim, J.; Scheible, M.; Kaye, J.; Sugimoto, H.; Wu, C.-C.; LeBleu, V.S.; Kalluri, R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 2015, 527, 525–530. [Google Scholar] [CrossRef]
- Kinugasa, H.; Nouso, K.; Miyahara, K.; Morimoto, Y.; Dohi, C.; Tsutsumi, K.; Kato, H.; Matsubara, T.; Okada, H.; Yamamoto, K. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer 2015, 121, 2271–2280. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pantel, K.; Alix-Panabières, C. Liquid biopsy in 2016: Circulating tumour cells and cell-free DNA in gastrointestinal cancer. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 73–74. [Google Scholar] [CrossRef] [PubMed]
- Bartolomucci, A.; Nobrega, M.; Ferrier, T.; Dickinson, K.; Kaorey, N.; Nadeau, A.; Castillo, A.; Burnier, J.V. Circulating tumor DNA to monitor treatment response in solid tumors and advance precision oncology. NPJ Precis. Oncol. 2025, 9, 84. [Google Scholar] [CrossRef]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Jung, E.H.; Shin, H.; Park, C.S.; Park, S.B.; Jung, D.E.; Leem, G.; Kim, S.J.; Jo, J.H.; Chung, M.J.; et al. Phenotypic characteristics of circulating tumor cells and predictive impact for efficacy of chemotherapy in patients with pancreatic cancer: A prospective study. Front. Oncol. 2023, 13, 1206565. [Google Scholar] [CrossRef] [PubMed]
- Mantini, G.; Meijer, L.L.; Glogovitis, I.; In ‘t Veld, S.G.J.G.; Paleckyte, R.; Capula, M.; Le Large, T.Y.S.; Morelli, L.; Pham, T.V.; Piersma, S.R.; et al. Omics Analysis of Educated Platelets in Cancer and Benign Disease of the Pancreas. Cancers 2020, 13, 66. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Heitzer, E.; Ulz, P.; Geigl, J.B. Circulating tumor DNA as a liquid biopsy for cancer. Clin. Chem. 2015, 61, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci. Transl. Med. 2014, 6, 224. [Google Scholar] [CrossRef]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Allenson, K.; Castillo, J.; San Lucas, F.A.; Scelo, G.; Kim, D.U.; Bernard, V.; Davis, G.; Kumar, T.; Katz, M.; Overman, M.J.; et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann. Oncol. 2017, 28, 741–747. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ma, L.; Guo, H.; Zhao, Y.; Liu, Z.; Wang, C.; Bu, J.; Sun, T.; Wei, J. Liquid biopsy in cancer: Current status, challenges and future prospects. Signal Transduct. Target. Ther. 2024, 9, 336. [Google Scholar] [CrossRef] [PubMed]
- Parikh, A.R.; Leshchiner, I.; Elagina, L.; Goyal, L.; Levovitz, C.; Siravegna, G.; Livitz, D.; Rhrissorrakrai, K.; Martin, E.E.; Van Seventer, E.E.; et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 2019, 25, 1415–1421. [Google Scholar] [CrossRef]
- Firpo, M.A.; Boucher, K.M.; Bleicher, J.; Khanderao, G.D.; Rosati, A.; Poruk, K.E.; Kamal, S.; Marzullo, L.; De Marco, M.; Falco, A.; et al. Multianalyte Serum Biomarker Panel for Early Detection of Pancreatic Adenocarcinoma. JCO Clin. Cancer Inform. 2023, 7, e2200160. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hata, T.; Mizuma, M.; Motoi, F.; Ohtsuka, H.; Nakagawa, K.; Morikawa, T.; Unno, M. Prognostic impact of postoperative circulating tumor DNA as a molecular minimal residual disease marker in patients with pancreatic cancer undergoing surgical resection. J. Hepato-Biliary-Pancreat. Sci. 2023, 30, 815–824. [Google Scholar] [CrossRef] [PubMed]
- Tjensvoll, K.; Lapin, M.; Buhl, T.; Oltedal, S.; Steen-Ottosen Berry, K.; Gilje, B.; Søreide, J.A.; Javle, M.; Nordgård, O.; Smaaland, R. Clinical relevance of circulating KRAS mutated DNA in plasma from patients with advanced pancreatic cancer. Mol. Oncol. 2016, 10, 635–643. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zill, O.A.; Greene, C.; Sebisanovic, D.; Siew, L.M.; Leng, J.; Vu, M.; Hendifar, A.E.; Wang, Z.; Atreya, C.E.; Kelley, R.K.; et al. Cell-Free DNA Next-Generation Sequencing in Pancreatobiliary Carcinomas. Cancer Discov. 2015, 5, 1040–1048. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Poruk, K.E.; Blackford, A.L.; Weiss, M.J.; Cameron, J.L.; He, J.; Goggins, M.; Rasheed, Z.A.; Wolfgang, C.L.; Wood, L.D. Circulating Tumor Cells Expressing Markers of Tumor-Initiating Cells Predict Poor Survival and Cancer Recurrence in Patients with Pancreatic Ductal Adenocarcinoma. Clin. Cancer Res. 2017, 23, 2681–2690. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Stoecklein, N.H.; Fluegen, G.; Guglielmi, R.; Neves, R.P.; Hackert, T.; Birgin, E.; Cieslik, S.A.; Sudarsanam, M.; Driemel, C.; van Dalum, G.; et al. Ultra-sensitive CTC-based liquid biopsy for pancreatic cancer enabled by large blood volume analysis. Mol. Cancer 2023, 22, 181. [Google Scholar] [CrossRef]
- Zhao, X.H.; Wang, Z.R.; Chen, C.L.; Di, L.; Bi, Z.F.; Li, Z.H.; Liu, Y.M. Molecular detection of epithelial-mesenchymal transition markers in circulating tumor cells from pancreatic cancer patients: Potential role in clinical practice. World J. Gastroenterol. 2019, 25, 138–150. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mishra, A.; Huang, S.B.; Dubash, T.; Burr, R.; Edd, J.F.; Wittner, B.S.; Cunneely, Q.E.; Putaturo, V.R.; Deshpande, A.; Antmen, E.; et al. Tumor cell-based liquid biopsy using high-throughput microfluidic enrichment of entire leukapheresis product. Nat. Commun. 2025, 16, 32. [Google Scholar] [CrossRef]
- Franses, J.W.; Philipp, J.; Missios, P.; Bhan, I.; Liu, A.; Yashaswini, C.; Tai, E.; Zhu, H.; Ligorio, M.; Nicholson, B.; et al. Pancreatic circulating tumor cell profiling identifies LIN28B as a metastasis driver and drug target. Nat. Commun. 2020, 11, 3303. [Google Scholar] [CrossRef]
- Macaraniag, C.; Khan, I.; Barabanova, A.; Valle, V.; Zhou, J.; Giulianotti, P.C.; Borgeat, A.; Votta-Velis, G.; Papautsky, I. Benchmarking microfluidic and immunomagnetic platforms for isolating circulating tumor cells in pancreatic cancer. Lab A Chip 2025, 25, 5292–5301. [Google Scholar] [CrossRef]
- Yasui, K.; Saito, T.; Ueda, S.; Shinohara, K.; Fukami, Y.; Sano, T.; Nakanishi, H. Sequential Changes in Circulating Tumor Cells in the Peripheral Blood of Pancreatic Cancer Patients with Preoperative Chemotherapy Using a New Immunocytology-Based, Light Microscopic CTC Detection Platform. Diagnostics 2025, 15, 752. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dimitrov-Markov, S.; Perales-Patón, J.; Bockorny, B.; Dopazo, A.; Muñoz, M.; Baños, N.; Bonilla, V.; Menendez, C.; Duran, Y.; Huang, L.; et al. Discovery of New Targets to Control Metastasis in Pancreatic Cancer by Single-cell Transcriptomics Analysis of Circulating Tumor Cells. Mol. Cancer Ther. 2020, 19, 1751–1760. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R. The biology and function of extracellular vesicles in immune response and immunity. Immunity 2024, 57, 1752–1768. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dilsiz, N. A comprehensive review on recent advances in exosome isolation and characterization: Toward clinical applications. Transl. Oncol. 2024, 50, 102121. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lobb, R.J.; Becker, M.; Wen, S.W.; Wong, C.S.; Wiegmans, A.P.; Leimgruber, A.; Möller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Moutinho-Ribeiro, P.; Adem, B.; Batista, I.; Silva, M.; Silva, S.; Ruivo, C.F.; Morais, R.; Peixoto, A.; Coelho, R.; Costa-Moreira, P.; et al. Exosomal glypican-1 discriminates pancreatic ductal adenocarcinoma from chronic pancreatitis. Dig. Liver Dis. 2022, 54, 871–877. [Google Scholar] [CrossRef] [PubMed]
- Kowkabany, G.; Bao, Y. Nanoparticle Tracking Analysis: An Effective Tool to Characterize Extracellular Vesicles. Molecules 2024, 29, 4672. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liga, A.; Vliegenthart, A.D.; Oosthuyzen, W.; Dear, J.W.; Kersaudy-Kerhoas, M. Exosome isolation: A microfluidic road-map. Lab A Chip 2015, 15, 2388–2394. [Google Scholar] [CrossRef] [PubMed]
- Abue, M.; Yokoyama, M.; Shibuya, R.; Tamai, K.; Yamaguchi, K.; Sato, I.; Tanaka, N.; Hamada, S.; Shimosegawa, T.; Sugamura, K.; et al. Circulating miR-483-3p and miR-21 is highly expressed in plasma of pancreatic cancer. Int. J. Oncol. 2015, 46, 539–547. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kamerkar, S.; LeBleu, V.S.; Sugimoto, H.; Yang, S.; Ruivo, C.F.; Melo, S.A.; Lee, J.J.; Kalluri, R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017, 546, 498–503. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Best, M.G.; Sol, N.; Kooi, I.; Tannous, J.; Westerman, B.A.; Rustenburg, F.; Schellen, P.; Verschueren, H.; Post, E.; Koster, J.; et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell 2015, 28, 666–676. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sol, N.; Veld, S.G.I.; Vancura, A.; Tjerkstra, M.; Leurs, C.; Rustenburg, F.; Schellen, P.; Verschueren, H.; Post, E.; Zwaan, K.; et al. Tumor-educated platelet RNA for the detection and (pseudo) progression monitoring of glioblastoma. Cell Rep. Med. 2020, 1, 100101. [Google Scholar] [CrossRef]
- Krishnan, A.; Thomas, S. Toward platelet transcriptomics in cancer diagnosis, prognosis and therapy. Br. J. Cancer 2022, 126, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Ju, S.; Chen, C.; Zhang, J.; Xu, L.; Zhang, X.; Li, Z.; Chen, Y.; Zhou, J.; Ji, F.; Wang, L. Detection of circulating tumor cells: Opportunities and challenges. Biomark. Res. 2022, 10, 58. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, G.A.D. MicroRNAs: Circulating biomarkers for the early detection of imperceptible cancers via biosensor and machine-learning advances. Oncogene 2024, 43, 2135–2142. [Google Scholar] [CrossRef]
- Huang, B.; Huang, H.; Zhang, S.; Zhang, D.; Shi, Q.; Liu, J.; Guo, J. Artificial intelligence in pancreatic cancer. Theranostics 2022, 12, 6931–6954. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2019, 20, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Aravanis, A.M.; Lee, M.; Klausner, R.D. Next-Generation Sequencing of Circulating Tumor DNA for Early Cancer Detection. Cell 2017, 168, 571–574. [Google Scholar] [CrossRef] [PubMed]
- Jensen, T.J.; Goodman, A.M.; Kato, S.; Ellison, C.K.; Daniels, G.A.; Kim, L.; Nakashe, P.; McCarthy, E.; Mazloom, A.R.; McLennan, G.; et al. Genome-Wide Sequencing of Cell-Free DNA Identifies Copy-Number Alterations That Can Be Used for Monitoring Response to Immunotherapy in Cancer Patients. Mol. Cancer Ther. 2019, 18, 448–458. [Google Scholar] [CrossRef] [PubMed]
- Guler, G.D.; Ning, Y.; Ku, C.J.; Phillips, T.; McCarthy, E.; Ellison, C.K.; Bergamaschi, A.; Collin, F.; Lloyd, P.; Scott, A.; et al. Detection of early stage pancreatic cancer using 5-hydroxymethylcytosine signatures in circulating cell free DNA. Nat. Commun. 2020, 11, 5270. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cristiano, S.; Leal, A.; Phallen, J.; Fiksel, J.; Adleff, V.; Bruhm, D.C.; Jensen, S.Ø.; Medina, J.E.; Hruban, C.; White, J.R.; et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019, 570, 385–389. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lapin, M.; Edland, K.H.; Tjensvoll, K.; Oltedal, S.; Austdal, M.; Garresori, H.; Rozenholc, Y.; Gilje, B.; Nordgård, O. Comprehensive ctDNA Measurements Improve Prediction of Clinical Outcomes and Enable Dynamic Tracking of Disease Progression in Advanced Pancreatic Cancer. Clin. Cancer Res. 2023, 29, 1267–1278. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
| Analyte | Main Clinical Applications | Typical Sensitivity/Specificity Range | Sample Type | Analytical Platform(s) | Critical Pre-Analytic Considerations |
|---|---|---|---|---|---|
| TEPs [35] | Early detection, treatment response, molecular profiling | Sensitivity: 70–95%/Specificity: 85–95% | Whole blood (platelet-rich plasma) | RNA-seq, qRT-PCR, NanoString, bioinformatics classifiers | Process within 2 h; avoid platelet activation; centrifuge gently |
| cfDNA/ctDNA mutations [38] | Early detection, prognosis, treatment monitoring, MRD detection | Sensitivity: ~72% Specificity: >90% | Plasma (preferred) or serum | ddPCR, BEAMing, NGS (CAPP-Seq, Guardant360) | Use EDTA or Streck tubes; plasma separation within 2–4 h; avoid hemolysis |
| CTCs [65] | Prognosis, treatment monitoring, MRD detection | Sensitivity: 40–70%/Specificity: >95% | Whole blood | CellSearch®, microfluidic chips (CTC-iChip, Parsortix), immunomagnetic enrichment | Collect in CellSave or EDTA tubes; process within 24 h; gentle handling to preserve viability |
| Exosomes/EVs [66] | Early detection, prognosis, therapy resistance monitoring | Sensitivity: 70–90%/Specificity: 80–95% | Plasma, serum, urine | Ultracentrifugation, SEC, microfluidic chips, NTA | Store at −80 °C; avoid freeze–thaw cycles; standardize isolation method |
| cfRNA/miRNA [67] | Diagnosis, prognosis, treatment response | Sensitivity: 60–90%/Specificity: 80–95% | Plasma, serum, saliva | qRT-PCR, NGS, microarrays | Use RNA-stabilizing tubes; rapid plasma separation; store at −80 °C |
| Fragmentomics (cfDNA size) [68] | Early detection, tumor origin classification | Sensitivity: 70–85%/Specificity: 85–95% | Plasma | WGS/WES, cfDNA fragmentation analysis, AI-based models | Specialized software; prompt processing to preserve fragment integrity |
| Protein biomarkers (CA19-9, CEA) | Diagnosis, disease monitoring, risk stratification | Sensitivity: 60–80%/Specificity: 70–90% | Serum, plasma | ELISA, mass spectrometry, multiplex immunoassays | Avoid hemolysis; standardize sample dilution and temperature |
| Trial Name/Identifier | Phase & Design | Objective | Patient Population | Status & Completion |
|---|---|---|---|---|
| AMPLIFY--201(NCT04853017) | Phase I | Evaluate safety & efficacy of KRAS-targeted vaccine ELI--002--2P using ctDNA clearance | Post-surgery PDAC with KRAS mutation | Completed (January 2023) |
| AMPLIFY--7P(NCT05726864) | Phase I/II | Assess ELI--002--2P in delaying recurrence via ctDNA monitoring | Post-surgery RAS-mutated PDAC | Recruiting (April 2023–November 2026) |
| CASPER(NCT05634931) | Observational cohort | Prognostic assessment of ctDNA in surgical resectability & treatment response | Resectable PDAC | Recruiting (December 2022–May 2026) |
| DYNAMIC--Pancreas (NCT03899636) | Phase II | Post-op ctDNA informing adjuvant chemo decisions | Resectable PDAC | Completed (November 2023) |
| PROJECTION (NCT04246203) | Observational | Prognostic role of pre-op ctDNA on disease-free survival (DFS) | Resectable PDAC | Completed (March 2025) |
| ctDNA Diagnostic (pre-op) (NCT03524677) | Observational | Diagnostic role of ctDNA (KRAS, CDKN2A, SMAD4, TP53) pre-op | Non-metastatic PDAC | Completed (January 2020) |
| ctDNA for recurrence surveillance (NCT02934984) | Observational | ctDNA for recurrence surveillance | Resectable PDAC | Completed (January 2021) |
| Exosomal RNA Biomarker (NCT04636788) | Prospective cohort | Exosomal small RNA diagnostic biomarker | PDAC & pancreatic lesions | Completed (November 2022) |
| PRIMUS--002(NCT04176952) | Phase II non-randomized | Prognostic ctDNA in neoadjuvant treatment regimens | Resectable/borderline PDAC | Completed August 2021 |
| ctDNA-based MRD(NCT05479708) | Observational | ctDNA as a biomarker for early detection of MRD and predicting relapse in resected PDAC | after PDAC resection | Recruiting (August 2024–November 2025) |
| PLATON(NCT04484636) | Observational multicohort | Targetable mutations via tissue + liquid biopsy | GI cancer including PDAC (n ≈ 400) | Completed (December 2024) |
| Multi-biomarker Screening (NCT03334708) | Observational | Multi-biomarker (ctDNA, exosome, CTC) screening & treatment response | GI cancers including PDAC (n ≈ 700) | Recruiting (October 2017–October 2025) |
| PANCAID (NCT06283576) | Multiple-cohort diagnostic | AI-driven liquid biopsy for early detection | Early PDAC, high-risk populations | Recruiting (May 2024–December 2027) |
| CIRCPAC (NCT05788744) | Observational case–control study | Tumor DNA and Circular DNA Analysis in localized PDAC to Optimize the Pre- and Post-operative Treatment | Localized PDAC (stage 1–3) | Recruiting (January 2023–January 2030) |
| LIPAC (NCT05400681) | Observational | PLF+ and KRAS ctDNA for curative surgery and to study the prognostic impact in PPAC patients | PDAC pancreatic resection specimen age > 18 years | Completed (December 2024) |
| The Role of MicroRNA in the Diagnosis, Prognosis and Response to Treatment in Pancreatic Cancer (NCT04406831) | Observational | MicroRNA in the diagnosis, prognosis and response to treatment in PDAC | New unresectable PDAC | Recruiting (April 2015-April 2027) |
| DNA Promoter Hypermethylation as a Blood Based Maker for Pancreatic Cancer (NCT02079363) | Observational | Cell-free DNA Promoter Hypermethylation in Plasma From patients PDAC. Diagnostic, prognostic | Patients with chronic pancreatitis and detecting patients with particularly high risk of developing pancreatic cancer. | Completed (January 2018) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bendari, A.; Vele, O.; Baskovich, B.; Bendari, A.; Sebika, M.; Gomez Marti, J.L.; Krishnamurthy, K.; Asiry, S. Liquid Biopsy in Pancreatic Ductal Adenocarcinoma: Clinical Utility, Trials, and Future Directions. Gastroenterol. Insights 2025, 16, 39. https://doi.org/10.3390/gastroent16040039
Bendari A, Vele O, Baskovich B, Bendari A, Sebika M, Gomez Marti JL, Krishnamurthy K, Asiry S. Liquid Biopsy in Pancreatic Ductal Adenocarcinoma: Clinical Utility, Trials, and Future Directions. Gastroenterology Insights. 2025; 16(4):39. https://doi.org/10.3390/gastroent16040039
Chicago/Turabian StyleBendari, Ahmed, Oana Vele, Brett Baskovich, Alaa Bendari, Mona Sebika, Juan Luis Gomez Marti, Kritika Krishnamurthy, and Saeed Asiry. 2025. "Liquid Biopsy in Pancreatic Ductal Adenocarcinoma: Clinical Utility, Trials, and Future Directions" Gastroenterology Insights 16, no. 4: 39. https://doi.org/10.3390/gastroent16040039
APA StyleBendari, A., Vele, O., Baskovich, B., Bendari, A., Sebika, M., Gomez Marti, J. L., Krishnamurthy, K., & Asiry, S. (2025). Liquid Biopsy in Pancreatic Ductal Adenocarcinoma: Clinical Utility, Trials, and Future Directions. Gastroenterology Insights, 16(4), 39. https://doi.org/10.3390/gastroent16040039






