From Dysbiosis to Prediction: AI-Powered Microbiome Insights into IBD and CRC
Abstract
1. Introduction
- (1)
- Supervised learning models (e.g., Random Forest (RF), Support Vector Machines (SVMs), and Extreme Gradient Boosting (xGBoost)).
- (2)
- Unsupervised learning models (e.g., Principal Component Analysis (PCA) and k-means clustering).
- (3)
- Deep learning models (e.g., Artificial Neural Networks (ANNs) and Convolutional Neural Networks (CNNs)).
- (4)
- Graph-based models (e.g., Graph Neural Networks (GNNs) and Graph Convolutional Networks (GCNs)).
- (5)
- Explainable AI (xAI) (e.g., SHapley Additive exPlanations (SHAP) and Local Interpretable Model-Agnostic Explanations (LIMEs)) [26].
2. From Dysbiosis to Carcinogenesis: A Pathophysiological Perspective
2.1. Etiology and Epidemiologic Relationship
2.2. Pathophysiology vs. Healthy Control (HC)
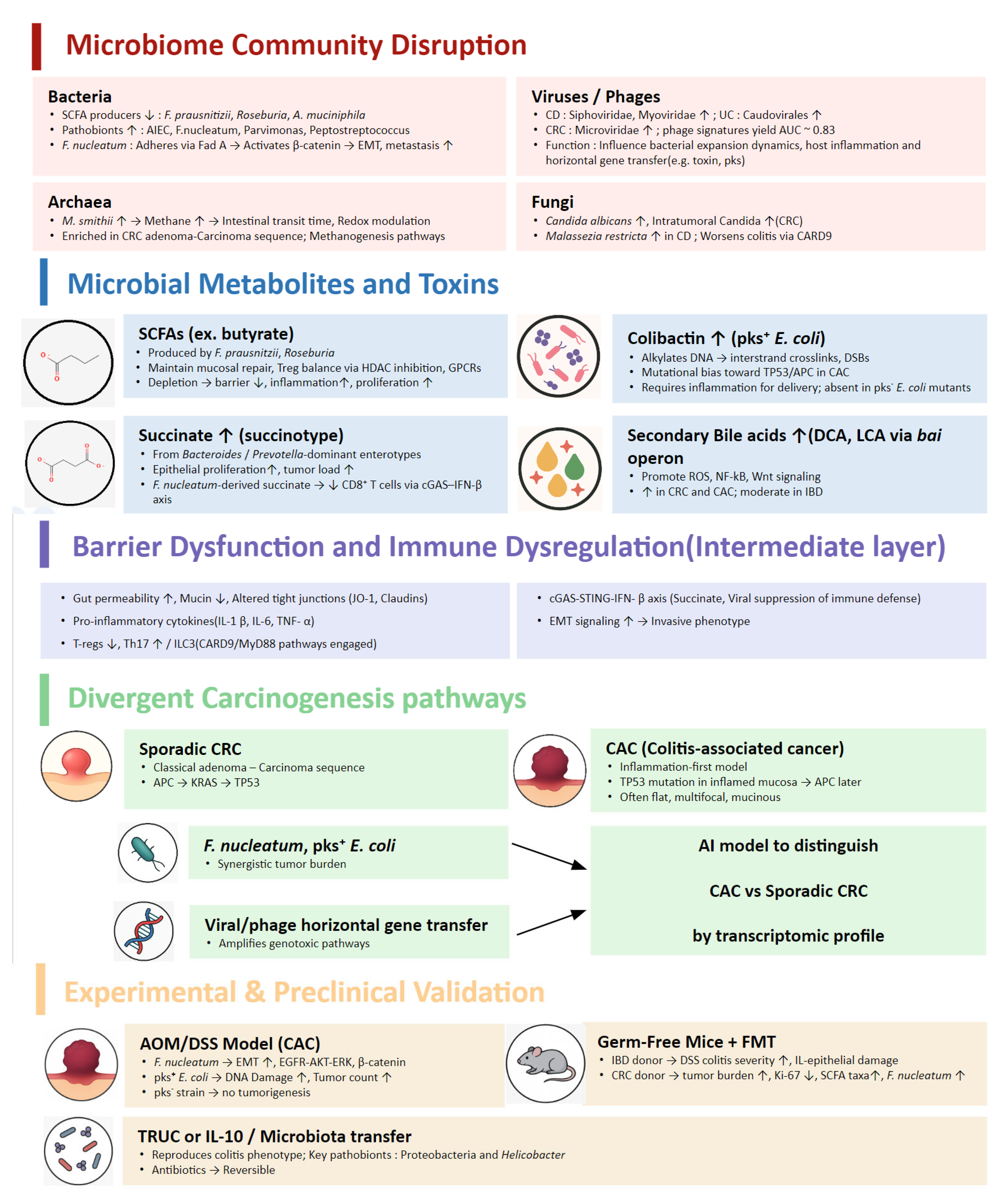
2.2.1. Microbiome Community Disruption
Bacteria
Archaea
Virus/Phages
Fungi
2.2.2. Microbial Metabolites and Toxins
Short-Chain Fatty Acids (SCFAs)
Bile Acids and the Bai Operon
Colibactin and Genotoxic E. coli
Succinate and the “Succinotype”
2.2.3. Carcinogenesis Pathways
Bacterial Oncogenesis
Viral and Phage Contributions
Host–Microbiome Interaction and Immune-Metabolic Signaling
2.3. Experimental Validation
2.3.1. AOM/DSS Model
2.3.2. Germ-Free Mice + FMT
2.3.3. Colitis-Microbiome Transfers
2.3.4. Colibactin-Deficient Strains
2.4. Regional and Demographic Variability in Microbiome Research
3. Translating Microbiome Signals into Clinical Action: Diagnosis, Treatment, and Prognosis
3.1. Microbiome-Based Diagnosis and Classification
3.1.1. CRC
Detection Across the Disease Spectrum
Classification of CRC Subtypes
Methodological Advances
3.1.2. IBD
IBD vs. HC
Differentiating UC and CD
Pediatric IBD (PIBD)
3.1.3. IBD-Associated CRC (CAC)
Diagnostic Challenges in the IBD Context
3.2. Prediction of Treatment Response
3.2.1. CRC
3.2.2. IBD
3.3. Prognosis from Risk Stratification to Surveillance
3.3.1. Prognosis in CRC
3.3.2. Prediction of Flare, Relapse, and Progression in IBD
3.3.3. Early Detection and Shared Modeling
4. Beyond Feces: Expanding the Microbiome Landscape
4.1. Oral Microbiome
4.1.1. Oral–Gut Axis: Microbial Translocation and Systemic Inflammation
4.1.2. Dysbiosis of Oral Microbiota in IBD and CRC
4.1.3. Diagnostic Potential of Salivary Microbiome
4.1.4. AI-Driven Approaches
4.2. Mucosal-Associated Microbiota (MAM)
4.3. Small Intestine (SI) Microbiome
5. Considerations When Applying AI to Microbiome-Based Prediction
5.1. Limitations of Previous Machine Learning Approaches
5.2. Considerations for Robust Generalization
5.3. Strategies for Data Optimization and Preprocessing
5.4. Explainability as a Translational Consideration
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- The Integrative HMP (iHMP) Research Network Consortium; Proctor, L.M.; Creasy, H.H.; Fettweis, J.M.; Lloyd-Price, J.; Ma-hurkar, A.; Zhou, W.; Buck, G.A.; Snyder, M.P.; Strauss, J.F.; et al. The Integrative Human Microbiome Project. Nature 2019, 569, 641–648. [Google Scholar] [CrossRef]
- Raskov, H.; Burcharth, J.; Pommergaard, H.-C.; Rosenberg, J. Irritable Bowel Syndrome, the Microbiota and the Gut-Brain Axis. Gut Microbes 2016, 7, 365–383. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.L.; Schnabl, B. The Gut–Liver Axis and Gut Microbiota in Health and Liver Disease. Nat. Rev. Microbiol. 2023, 21, 719–733. [Google Scholar] [CrossRef] [PubMed]
- Haneishi, Y.; Furuya, Y.; Hasegawa, M.; Picarelli, A.; Rossi, M.; Miyamoto, J. Inflammatory Bowel Diseases and Gut Microbiota. Int. J. Mol. Sci. 2023, 24, 3817. [Google Scholar] [CrossRef] [PubMed]
- Rebersek, M. Gut Microbiome and Its Role in Colorectal Cancer. BMC Cancer 2021, 21, 1325. [Google Scholar] [CrossRef]
- Axelrad, J.E.; Lichtiger, S.; Yajnik, V. Inflammatory Bowel Disease and Cancer: The Role of Inflammation, Immunosuppression, and Cancer Treatment. World J. Gastroenterol. 2016, 22, 4794. [Google Scholar] [CrossRef]
- Sato, Y.; Tsujinaka, S.; Miura, T.; Kitamura, Y.; Suzuki, H.; Shibata, C. Inflammatory Bowel Disease and Colorectal Cancer: Epidemiology, Etiology, Surveillance, and Management. Cancers 2023, 15, 4154. [Google Scholar] [CrossRef]
- Quaglio, A.E.V.; Grillo, T.G.; Oliveira, E.C.S.D.; Stasi, L.C.D.; Sassaki, L.Y. Gut Microbiota, Inflammatory Bowel Disease and Colorectal Cancer. World J. Gastroenterol. 2022, 28, 4053–4060. [Google Scholar] [CrossRef]
- Cai, J.; Sun, L.; Gonzalez, F.J. Gut Microbiota-Derived Bile Acids in Intestinal Immunity, Inflammation, and Tumorigenesis. Cell Host Microbe 2022, 30, 289–300. [Google Scholar] [CrossRef]
- Wang, R.; Li, Z.; Liu, S.; Zhang, D. Global, Regional and National Burden of Inflammatory Bowel Disease in 204 Countries and Territories from 1990 to 2019: A Systematic Analysis Based on the Global Burden of Disease Study 2019. BMJ Open 2023, 13, e065186. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Wagle, N.S.; Nogueira, L.; Devasia, T.P.; Mariotto, A.B.; Yabroff, K.R.; Islami, F.; Jemal, A.; Alteri, R.; Ganz, P.A.; Siegel, R.L. Cancer Treatment and Survivorship Statistics, 2025. CA. Cancer J. Clin. 2025, 75, 308–340. [Google Scholar] [CrossRef] [PubMed]
- Kafel, A.J.; Muzalyova, A.; Schnoy, E. Malignancy and Inflammatory Bowel Disease (IBD): Incidence and Prevalence of Malignancy in Correlation to IBD Therapy and Disease Activity—A Retrospective Cohort Analysis over 5 Years. Biomedicines 2025, 13, 1395. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Wang, F.; Ju, Y.; He, Q.; Sun, T.; Deng, W.; Ding, R.; Zhang, C.; Xu, Q.; Qi, C.; et al. Application and Development of Noninvasive Biomarkers for Colorectal Cancer Screening: A Systematic Review. Int. J. Surg. 2023, 109, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Massaro, C.A.; Meade, S.; Lemarié, F.L.; Kaur, G.; Bressler, B.; Rosenfeld, G.; Leung, Y.; Williams, A.-J.; Lunken, G. Gut Microbiome Predictors of Advanced Therapy Response in Crohn’s Disease: Protocol for the OPTIMIST Prospective, Longitudinal, Observational Pilot Study in Canada. BMJ Open 2025, 15, e094280. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, Y.; Cheng, L.; Cao, X.; Liu, C. Gut Microbiota in Colorectal Cancer: A Review of Its Influence on Tumor Immune Surveillance and Therapeutic Response. Front. Oncol. 2025, 15, 1557959. [Google Scholar] [CrossRef]
- Galasso, L.; Termite, F.; Mignini, I.; Esposto, G.; Borriello, R.; Vitale, F.; Nicoletti, A.; Paratore, M.; Ainora, M.E.; Gasbarrini, A.; et al. Unraveling the Role of Fusobacterium Nucleatum in Colorectal Cancer: Molecular Mechanisms and Pathogenic Insights. Cancers 2025, 17, 368. [Google Scholar] [CrossRef]
- Yu, M.R.; Kim, H.J.; Park, H.R. Fusobacterium Nucleatum Accelerates the Progression of Colitis-Associated Colorectal Cancer by Promoting EMT. Cancers 2020, 12, 2728. [Google Scholar] [CrossRef]
- Rosendahl Huber, A.; Pleguezuelos-Manzano, C.; Puschhof, J.; Ubels, J.; Boot, C.; Saftien, A.; Verheul, M.; Trabut, L.T.; Groenen, N.; Van Roosmalen, M.; et al. Improved Detection of Colibactin-Induced Mutations by Genotoxic E. Coli in Organoids and Colorectal Cancer. Cancer Cell 2024, 42, 487–496. [Google Scholar] [CrossRef]
- Arthur, J.C.; Perez-Chanona, E.; Mühlbauer, M.; Tomkovich, S.; Uronis, J.M.; Fan, T.-J.; Campbell, B.J.; Abujamel, T.; Dogan, B.; Rogers, A.B.; et al. Intestinal Inflammation Targets Cancer-Inducing Activity of the Microbiota. Science 2012, 338, 120–123. [Google Scholar] [CrossRef]
- Wang, X.; Fang, Y.; Liang, W.; Wong, C.C.; Qin, H.; Gao, Y.; Liang, M.; Song, L.; Zhang, Y.; Fan, M.; et al. Fusobacterium Nucleatum Facilitates Anti-PD-1 Therapy in Microsatellite Stable Colorectal Cancer. Cancer Cell 2024, 42, 1729–1746. [Google Scholar] [CrossRef]
- Zhang, H.; Tian, Y.; Xu, C.; Chen, M.; Xiang, Z.; Gu, L.; Xue, H.; Xu, Q. Crosstalk between Gut Microbiotas and Fatty Acid Metabolism in Colorectal Cancer. Cell Death Discov. 2025, 11, 78. [Google Scholar] [CrossRef]
- Kim, K.S.; Noh, J.; Kim, B.-S.; Koh, H.; Lee, D.-W. Refining Microbiome Diversity Analysis by Concatenating and Integrating Dual 16S rRNA Amplicon Reads. npj Biofilms Microbiomes 2025, 11, 57. [Google Scholar] [CrossRef] [PubMed]
- Wirbel, J.; Pyl, P.T.; Kartal, E.; Zych, K.; Kashani, A.; Milanese, A.; Fleck, J.S.; Voigt, A.Y.; Palleja, A.; Ponnudurai, R.; et al. Meta-Analysis of Fecal Metagenomes Reveals Global Microbial Signatures That Are Specific for Colorectal Cancer. Nat. Med. 2019, 25, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Preto, A.J.; Chanana, S.; Ence, D.; Healy, M.D.; Domingo-Fernández, D.; West, K.A. Multi-Omics Data Integration Identifies Novel Biomarkers and Patient Subgroups in Inflammatory Bowel Disease. J. Crohns Colitis 2025, 19, jjae197. [Google Scholar] [CrossRef] [PubMed]
- Novielli, P.; Romano, D.; Magarelli, M.; Bitonto, P.D.; Diacono, D.; Chiatante, A.; Lopalco, G.; Sabella, D.; Venerito, V.; Filannino, P.; et al. Explainable Artificial Intelligence for Microbiome Data Analysis in Colorectal Cancer Biomarker Identification. Front. Microbiol. 2024, 15, 1348974. [Google Scholar] [CrossRef]
- Hernández Medina, R.; Kutuzova, S.; Nielsen, K.N.; Johansen, J.; Hansen, L.H.; Nielsen, M.; Rasmussen, S. Machine Learning and Deep Learning Applications in Microbiome Research. ISME Commun. 2022, 2, 98. [Google Scholar] [CrossRef]
- Aysel, H.I.; Cai, X.; Prugel-Bennett, A. Explainable Artificial Intelligence: Advancements and Limitations. Appl. Sci. 2025, 15, 7261. [Google Scholar] [CrossRef]
- Salih, A.M.; Raisi-Estabragh, Z.; Galazzo, I.B.; Radeva, P.; Petersen, S.E.; Lekadir, K.; Menegaz, G. A Perspective on Explainable Artificial Intelligence Methods: SHAP and LIME. Adv. Intell. Syst. 2025, 7, 2400304. [Google Scholar] [CrossRef]
- Eaden, J.A. The Risk of Colorectal Cancer in Ulcerative Colitis: A Meta-Analysis. Gut 2001, 48, 526–535. [Google Scholar] [CrossRef]
- Jess, T.; Simonsen, J.; Jørgensen, K.T.; Pedersen, B.V.; Nielsen, N.M.; Frisch, M. Decreasing Risk of Colorectal Cancer in Patients With Inflammatory Bowel Disease Over 30 Years. Gastroenterology 2012, 143, 375–381. [Google Scholar] [CrossRef]
- Baker, A.-M.; Cross, W.; Curtius, K.; Al Bakir, I.; Choi, C.-H.R.; Davis, H.L.; Temko, D.; Biswas, S.; Martinez, P.; Williams, M.J.; et al. Evolutionary History of Human Colitis-Associated Colorectal Cancer. Gut 2019, 68, 985–995. [Google Scholar] [CrossRef]
- Terzić, J.; Grivennikov, S.; Karin, E.; Karin, M. Inflammation and Colon Cancer. Gastroenterology 2010, 138, 2101–2114. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.-J.; Blugeon, S.; Bridonneau, C.; Furet, J.-P.; Corthier, G.; et al. Faecalibacterium prausnitzii Is an Anti-Inflammatory Commensal Bacterium Identified by Gut Microbiota Analysis of Crohn Disease Patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef] [PubMed]
- Martín, R.; Chain, F.; Miquel, S.; Lu, J.; Gratadoux, J.-J.; Sokol, H.; Verdu, E.F.; Bercik, P.; Bermúdez-Humarán, L.G.; Langella, P. The Commensal Bacterium Faecalibacterium prausnitzii Is Protective in DNBS-Induced Chronic Moderate and Severe Colitis Models. Inflamm. Bowel Dis. 2014, 20, 417–430. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Dong, X.; Yang, K.; Chevarin, C.; Zhang, J.; Lin, Y.; Zuo, T.; Chu, L.C.; Sun, Y.; Zhang, F.; et al. Association of Adherent-Invasive Escherichia Coli with Severe Gut Mucosal Dysbiosis in Hong Kong Chinese Population with Crohn’s Disease. Gut Microbes 2021, 13, 1994833. [Google Scholar] [CrossRef]
- Darfeuille-Michaud, A.; Boudeau, J.; Bulois, P.; Neut, C.; Glasser, A.-L.; Barnich, N.; Bringer, M.-A.; Swidsinski, A.; Beaugerie, L.; Colombel, J.-F. High Prevalence of Adherent-Invasive Escherichia Coli Associated with Ileal Mucosa in Crohn’s Disease. Gastroenterology 2004, 127, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium Nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/β-Catenin Signaling via Its FadA Adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L.; et al. Fusobacterium Nucleatum Potentiates Intestinal Tumorigenesis and Modulates the Tumor-Immune Microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium Nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017, 170, 548–563. [Google Scholar] [CrossRef]
- Biondi, A.; Basile, F.; Vacante, M. Familial Adenomatous Polyposis and Changes in the Gut Microbiota: New Insights into Colorectal Cancer Carcinogenesis. World J. Gastrointest. Oncol. 2021, 13, 495–508. [Google Scholar] [CrossRef]
- Li, T.; Coker, O.O.; Sun, Y.; Li, S.; Liu, C.; Lin, Y.; Wong, S.H.; Miao, Y.; Sung, J.J.Y.; Yu, J. Multi-Cohort Analysis Reveals Altered Archaea in Colorectal Cancer Fecal Samples Across Populations. Gastroenterology 2025, 168, 525–538. [Google Scholar] [CrossRef]
- Norman, J.M.; Handley, S.A.; Baldridge, M.T.; Droit, L.; Liu, C.Y.; Keller, B.C.; Kambal, A.; Monaco, C.L.; Zhao, G.; Fleshner, P.; et al. Disease-Specific Alterations in the Enteric Virome in Inflammatory Bowel Disease. Cell 2015, 160, 447–460. [Google Scholar] [CrossRef]
- Chen, F.; Li, S.; Guo, R.; Song, F.; Zhang, Y.; Wang, X.; Huo, X.; Lv, Q.; Ullah, H.; Wang, G.; et al. Meta-Analysis of Fecal Viromes Demonstrates High Diagnostic Potential of the Gut Viral Signatures for Colorectal Cancer and Adenoma Risk Assessment. J. Adv. Res. 2023, 49, 103–114. [Google Scholar] [CrossRef]
- Iliev, I.D.; Leonardi, I. Fungal Dysbiosis: Immunity and Interactions at Mucosal Barriers. Nat. Rev. Immunol. 2017, 17, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Yuan, K.; Xu, H.; Li, S.; Coker, O.O.; Liu, W.; Wang, L.; Zhang, X.; Yu, J. Intraneoplastic Fungal Dysbiosis Is Associated with Colorectal Cancer Progression and Host Gene Mutation. eBioMedicine 2025, 113, 105608. [Google Scholar] [CrossRef] [PubMed]
- Limon, J.J.; Tang, J.; Li, D.; Wolf, A.J.; Michelsen, K.S.; Funari, V.; Gargus, M.; Nguyen, C.; Sharma, P.; Maymi, V.I.; et al. Malassezia Is Associated with Crohn’s Disease and Exacerbates Colitis in Mouse Models. Cell Host Microbe 2019, 25, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Mok, K.; Suratanon, N.; Roytrakul, S.; Charoenlappanit, S.; Patumcharoenpol, P.; Chatchatee, P.; Vongsangnak, W.; Nakphaichit, M. ITS2 Sequencing and Targeted Meta-Proteomics of Infant Gut Mycobiome Reveal the Functional Role of Rhodotorula Sp. during Atopic Dermatitis Manifestation. J. Fungi 2021, 7, 748. [Google Scholar] [CrossRef]
- El Mouzan, M.; Al Quorain, A.; Assiri, A.; Almasoud, A.; Alsaleem, B.; Aladsani, A.; Al Sarkhy, A. Gut Fungal Profile in New Onset Treatment-Naïve Ulcerative Colitis in Saudi Children. Saudi J. Gastroenterol. 2025, 31, 28–33. [Google Scholar] [CrossRef]
- Parada Venegas, D.; De La Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef]
- Anthamatten, L.; Von Bieberstein, P.R.; Menzi, C.; Zünd, J.N.; Lacroix, C.; de Wouters, T.; Leventhal, G.E. Stratification of Human Gut Microbiomes by Succinotype Is Associated with Inflammatory Bowel Disease Status. Microbiome 2024, 12, 186. [Google Scholar] [CrossRef] [PubMed]
- Funabashi, M.; Grove, T.L.; Wang, M.; Varma, Y.; McFadden, M.E.; Brown, L.C.; Guo, C.; Higginbottom, S.; Almo, S.C.; Fischbach, M.A. A Metabolic Pathway for Bile Acid Dehydroxylation by the Gut Microbiome. Nature 2020, 582, 566–570. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Xie, G.; Jia, W. Bile Acid–Microbiota Crosstalk in Gastrointestinal Inflammation and Carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 111–128. [Google Scholar] [CrossRef]
- Pleguezuelos-Manzano, C.; Puschhof, J.; Rosendahl Huber, A.; Van Hoeck, A.; Wood, H.M.; Nomburg, J.; Gurjao, C.; Manders, F.; Dalmasso, G.; Stege, P.B.; et al. Mutational Signature in Colorectal Cancer Caused by Genotoxic Pks+ E. Coli. Nature 2020, 580, 269–273. [Google Scholar] [CrossRef]
- Lv, C.; Abdullah, M.; Su, C.-L.; Chen, W.; Zhou, N.; Cheng, Z.; Chen, Y.; Li, M.; Simpson, K.W.; Elsaadi, A.; et al. Correction: Genomic Characterization of Escherichia Coli with a Polyketide Synthase (Pks) Island Isolated from Ulcerative Colitis Patients. BMC Genom. 2025, 26, 91. [Google Scholar] [CrossRef]
- Tian, S.; Paudel, D.; Hao, F.; Neupane, R.; Castro, R.; Patterson, A.D.; Tiwari, A.K.; Prabhu, K.S.; Singh, V. Refined Fiber Inulin Promotes Inflammation-associated Colon Tumorigenesis by Modulating Microbial Succinate Production. Cancer Rep. 2023, 6, e1863. [Google Scholar] [CrossRef]
- Jiang, S.-S.; Xie, Y.-L.; Xiao, X.-Y.; Kang, Z.-R.; Lin, X.-L.; Zhang, L.; Li, C.-S.; Qian, Y.; Xu, P.-P.; Leng, X.-X.; et al. Fusobacterium Nucleatum-Derived Succinic Acid Induces Tumor Resistance to Immunotherapy in Colorectal Cancer. Cell Host Microbe 2023, 31, 781–797. [Google Scholar] [CrossRef]
- Li, J.; Ji, Y.; Chen, N.; Dai, L.; Deng, H. Colitis-Associated Carcinogenesis: Crosstalk between Tumors, Immune Cells and Gut Microbiota. Cell Biosci. 2023, 13, 194. [Google Scholar] [CrossRef]
- Silveira, C.B.; Rohwer, F.L. Piggyback-the-Winner in Host-Associated Microbial Communities. npj Biofilms Microbiomes 2016, 2, 16010. [Google Scholar] [CrossRef]
- Hannigan, G.D.; Duhaime, M.B.; Ruffin, M.T.; Koumpouras, C.C.; Schloss, P.D. Diagnostic Potential and Interactive Dynamics of the Colorectal Cancer Virome. mBio 2018, 9, e02248-18. [Google Scholar] [CrossRef]
- Zeller, G.; Tap, J.; Voigt, A.Y.; Sunagawa, S.; Kultima, J.R.; Costea, P.I.; Amiot, A.; Böhm, J.; Brunetti, F.; Habermann, N.; et al. Potential of Fecal Microbiota for Early-stage Detection of Colorectal Cancer. Mol. Syst. Biol. 2014, 10, 766. [Google Scholar] [CrossRef]
- Qin, Y.; Wang, Q.; Lin, Q.; Liu, F.; Pan, X.; Wei, C.; Chen, J.; Huang, T.; Fang, M.; Yang, W.; et al. Multi-Omics Analysis Reveals Associations between Gut Microbiota and Host Transcriptome in Colon Cancer Patients. mSystems 2025, 10, e00805-24. [Google Scholar] [CrossRef] [PubMed]
- Nagao-Kitamoto, H.; Shreiner, A.B.; Gillilland, M.G.; Kitamoto, S.; Ishii, C.; Hirayama, A.; Kuffa, P.; El-Zaatari, M.; Grasberger, H.; Seekatz, A.M.; et al. Functional Characterization of Inflammatory Bowel Disease–Associated Gut Dysbiosis in Gnotobiotic Mice. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 468–481. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.H.; Zhao, L.; Zhang, X.; Nakatsu, G.; Han, J.; Xu, W.; Xiao, X.; Kwong, T.N.Y.; Tsoi, H.; Wu, W.K.K.; et al. Gavage of Fecal Samples From Patients With Colorectal Cancer Promotes Intestinal Carcinogenesis in Germ-Free and Conventional Mice. Gastroenterology 2017, 153, 1621–1633. [Google Scholar] [CrossRef] [PubMed]
- Garrett, W.S.; Lord, G.M.; Punit, S.; Lugo-Villarino, G.; Mazmanian, S.K.; Ito, S.; Glickman, J.N.; Glimcher, L.H. Communicable Ulcerative Colitis Induced by T-Bet Deficiency in the Innate Immune System. Cell 2007, 131, 33–45. [Google Scholar] [CrossRef]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human Gut Microbiome Viewed across Age and Geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of Diet in Shaping Gut Microbiota Revealed by a Comparative Study in Children from Europe and Rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef]
- El Mouzan, M.; Savidge, T.C.; Al Sarkhy, A.; Badu, S.; Alsaleem, B.; Al Mofarreh, M.; Almasood, A.; Assiri, A. Gut Virome Profile in New Onset Treatment Naïve Saudi Children with Ulcerative Colitis. Saudi J. Gastroenterol. 2025, 31, 212–218. [Google Scholar] [CrossRef]
- Kubinski, R.; Djamen-Kepaou, J.-Y.; Zhanabaev, T.; Hernandez-Garcia, A.; Bauer, S.; Hildebrand, F.; Korcsmaros, T.; Karam, S.; Jantchou, P.; Kafi, K.; et al. Benchmark of Data Processing Methods and Machine Learning Models for Gut Microbiome-Based Diagnosis of Inflammatory Bowel Disease. Front. Genet. 2022, 13, 784397. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, J.; Ji, D.; Liu, Y.; Lu, S.; Lin, Z.; Chen, T.; Ao, L. Microbiome Analysis Reveals Universal Diagnostic Biomarkers for Colorectal Cancer across Populations and Technologies. Front. Microbiol. 2022, 13, 1005201. [Google Scholar] [CrossRef]
- Wu, Y.; Jiao, N.; Zhu, R.; Zhang, Y.; Wu, D.; Wang, A.-J.; Fang, S.; Tao, L.; Li, Y.; Cheng, S.; et al. Identification of Microbial Markers across Populations in Early Detection of Colorectal Cancer. Nat. Commun. 2021, 12, 3063. [Google Scholar] [CrossRef]
- Zhou, D.; Chen, Y.; Wang, Z.; Zhu, S.; Zhang, L.; Song, J.; Bai, T.; Hou, X. Integrating Clinical and Cross-Cohort Metagenomic Features: A Stable and Non-Invasive Colorectal Cancer and Adenoma Diagnostic Model. Front. Mol. Biosci. 2024, 10, 1298679. [Google Scholar] [CrossRef]
- Casimiro-Soriguer, C.S.; Loucera, C.; Peña-Chilet, M.; Dopazo, J. Towards a Metagenomics Machine Learning Interpretable Model for Understanding the Transition from Adenoma to Colorectal Cancer. Sci. Rep. 2022, 12, 450. [Google Scholar] [CrossRef]
- Khannous-Lleiffe, O.; Willis, J.R.; Saus, E.; Moreno, V.; Castellví-Bel, S.; Gabaldón, T.; on behalf of the CRIPREV Consortium. Microbiome Profiling from Fecal Immunochemical Test Reveals Microbial Signatures with Potential for Colorectal Cancer Screening. Cancers 2022, 15, 120. [Google Scholar] [CrossRef]
- Zhou, Z.; Ma, Y.; Zhang, D.; Ji, R.; Wang, Y.; Zhao, J.; Ma, C.; Zhu, H.; Shen, H.; Jiang, X.; et al. Microbiome and Fragmentation Pattern of Blood Cell-Free DNA and Fecal Metagenome Enhance Colorectal Cancer Micro-Dysbiosis and Diagnosis Analysis: A Proof-of-Concept Study. mSystems 2025, 10, e00276-25. [Google Scholar] [CrossRef]
- Guodong, W.; Yinhang, W.; Xinyue, W.; Hong, S.; Jian, C.; Zhanbo, Q.; Shuwen, H. Fecal Occult Blood Affects Intestinal Microbial Community Structure in Colorectal Cancer. BMC Microbiol. 2025, 25, 34. [Google Scholar] [CrossRef]
- Xiang, J.; Chai, N.; Li, L.; Hao, X.; Linghu, E. Alterations of Gut Microbiome in Patients with Colorectal Advanced Adenoma by Metagenomic Analyses. Turk. J. Gastroenterol. 2024, 35, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Zhuang, J.; Pan, Y.; Wu, W.; Ding, K. Different Characteristics in Gut Microbiome between Advanced Adenoma Patients and Colorectal Cancer Patients by Metagenomic Analysis. Microbiol. Spectr. 2022, 10, e01593-22. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.-H.; Sun, G. Improve the Colorectal Cancer Diagnosis Using Gut Microbiome Data. Front. Mol. Biosci. 2022, 9, 921945. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.-Q.; Zhao, W.-F.; Lu, Q.-W.; Zha, F.-R.; Lv, L.-B.; Ye, G.-L.; Gao, H.-L. Fecal Microbial Biomarkers Combined with Multi-Target Stool DNA Test Improve Diagnostic Accuracy for Colorectal Cancer. World J. Gastrointest. Oncol. 2023, 15, 1424–1435. [Google Scholar] [CrossRef]
- Kolisnik, T.; Sulit, A.K.; Schmeier, S.; Frizelle, F.; Purcell, R.; Smith, A.; Silander, O. Identifying Important Microbial and Genomic Biomarkers for Differentiating Right- versus Left-Sided Colorectal Cancer Using Random Forest Models. BMC Cancer 2023, 23, 647. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Shi, J.; Wang, H.; Du, H.; Yang, J.; Wei, K.; Zhou, Z.; Li, M.; Huang, S.; Zhan, L.; et al. The Characteristics of Tissue Microbiota in Different Anatomical Locations and Different Tissue Types of the Colorectum in Patients with Colorectal Cancer. mSystems 2025, 10, e00198-25. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Zhibo, Z.; Jing, Z.; Zhanbo, Q.; Shugao, H.; Weili, J.; Jiang, L.; Shuwen, H. Prediction Model of Poorly Differentiated Colorectal Cancer (CRC) Based on Gut Bacteria. BMC Microbiol. 2022, 22, 312. [Google Scholar] [CrossRef] [PubMed]
- Qingbo, L.; Jing, Z.; Zhanbo, Q.; Jian, C.; Yifei, S.; Yinhang, W.; Shuwen, H. Identification of Enterotype and Its Predictive Value for Patients with Colorectal Cancer. Gut Pathog. 2024, 16, 12. [Google Scholar] [CrossRef]
- Bars-Cortina, D.; Ramon, E.; Rius-Sansalvador, B.; Guinó, E.; Garcia-Serrano, A.; Mach, N.; Khannous-Lleiffe, O.; Saus, E.; Gabaldón, T.; Ibáñez-Sanz, G.; et al. Comparison between 16S rRNA and Shotgun Sequencing in Colorectal Cancer, Advanced Colorectal Lesions, and Healthy Human Gut Microbiota. BMC Genom. 2024, 25, 730. [Google Scholar] [CrossRef]
- Liu, G.; Li, T.; Zhu, X.; Zhang, X.; Wang, J. An Independent Evaluation in a CRC Patient Cohort of Microbiome 16S rRNA Sequence Analysis Methods: OTU Clustering, DADA2, and Deblur. Front. Microbiol. 2023, 14, 1178744. [Google Scholar] [CrossRef]
- Xu, C.; Zhou, M.; Xie, Z.; Li, M.; Zhu, X.; Zhu, H. LightCUD: A Program for Diagnosing IBD Based on Human Gut Microbiome Data. BioData Min. 2021, 14, 2. [Google Scholar] [CrossRef]
- Kim, H.; Na, J.E.; Kim, S.; Kim, T.-O.; Park, S.-K.; Lee, C.-W.; Kim, K.O.; Seo, G.-S.; Kim, M.S.; Cha, J.M.; et al. A Machine Learning-Based Diagnostic Model for Crohn’s Disease and Ulcerative Colitis Utilizing Fecal Microbiome Analysis. Microorganisms 2023, 12, 36. [Google Scholar] [CrossRef]
- Barberio, B.; Facchin, S.; Patuzzi, I.; Ford, A.C.; Massimi, D.; Valle, G.; Sattin, E.; Simionati, B.; Bertazzo, E.; Zingone, F.; et al. A Specific Microbiota Signature Is Associated to Various Degrees of Ulcerative Colitis as Assessed by a Machine Learning Approach. Gut Microbes 2022, 14, 2028366. [Google Scholar] [CrossRef]
- Gacesa, R.; Vich Vila, A.; Collij, V.; Mujagic, Z.; Kurilshikov, A.; Voskuil, M.D.; Festen, E.A.M.; Wijmenga, C.; Jonkers, D.M.A.E.; Dijkstra, G.; et al. A Combination of Fecal Calprotectin and Human Beta-Defensin 2 Facilitates Diagnosis and Monitoring of Inflammatory Bowel Disease. Gut Microbes 2021, 13, 1943288. [Google Scholar] [CrossRef]
- Jangi, S.; Hsia, K.; Zhao, N.; Kumamoto, C.A.; Friedman, S.; Singh, S.; Michaud, D.S. Dynamics of the Gut Mycobiome in Patients With Ulcerative Colitis. Clin. Gastroenterol. Hepatol. 2024, 22, 821–830. [Google Scholar] [CrossRef]
- Song, K.; Zhou, Y.-H. Leveraging Scheme for Cross-Study Microbiome Machine Learning Prediction and Feature Evaluations. Bioengineering 2023, 10, 231. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.-Y.; Park, J.-L.; Yeo, M.-K.; Kang, S.-B.; Kim, J.-M.; Kim, J.S.; Kim, S.-Y. Diagnosis of Crohn’s Disease and Ulcerative Colitis Using the Microbiome. BMC Microbiol. 2023, 23, 336. [Google Scholar] [CrossRef] [PubMed]
- Liñares-Blanco, J.; Fernandez-Lozano, C.; Seoane, J.A.; López-Campos, G. Machine Learning Based Microbiome Signature to Predict Inflammatory Bowel Disease Subtypes. Front. Microbiol. 2022, 13, 872671. [Google Scholar] [CrossRef]
- Sarrabayrouse, G.; Elias, A.; Yáñez, F.; Mayorga, L.; Varela, E.; Bartoli, C.; Casellas, F.; Borruel, N.; Herrera De Guise, C.; Machiels, K.; et al. Fungal and Bacterial Loads: Noninvasive Inflammatory Bowel Disease Biomarkers for the Clinical Setting. mSystems 2021, 6, 10.1128/msystems.01277-20. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xiao, Y.; Xu, X.; Guo, L.; Yu, Y.; Li, N.; Xu, C. Characteristics of Fecal Microbiota and Machine Learning Strategy for Fecal Invasive Biomarkers in Pediatric Inflammatory Bowel Disease. Front. Cell. Infect. Microbiol. 2021, 11, 711884. [Google Scholar] [CrossRef]
- Chen, W.; Wang, D.; Deng, X.; Zhang, H.; Dong, D.; Su, T.; Lu, Q.; Jiang, C.; Ni, Q.; Cui, Y.; et al. Bile Acid Profiling as an Effective Biomarker for Staging in Pediatric Inflammatory Bowel Disease. Gut Microbes 2024, 16, 2323231. [Google Scholar] [CrossRef]
- Vermeer, E.; Jagt, J.Z.; Lap, E.M.; Struys, E.A.; Budding, A.E.; Verhoeven-Duif, N.M.; Bosma, M.; Van Limbergen, J.E.; Koot, B.G.P.; De Jonge, R.; et al. Fecal Gut Microbiota and Amino Acids as Noninvasive Diagnostic Biomarkers of Pediatric Inflammatory Bowel Disease. Gut Microbes 2025, 17, 2517828. [Google Scholar] [CrossRef]
- Masaadeh, A.H.; Eletrebi, M.; Parajuli, B.; De Jager, N.; Bosch, D.E. Human Colitis-Associated Colorectal Carcinoma Progression Is Accompanied by Dysbiosis with Enriched Pathobionts. Gut Microbes 2025, 17, 2479774. [Google Scholar] [CrossRef]
- Liu, K.; Yang, X.; Zeng, M.; Yuan, Y.; Sun, J.; He, P.; Sun, J.; Xie, Q.; Chang, X.; Zhang, S.; et al. The Role of Fecal Fusobacterium Nucleatum and Pks+ Escherichia Coli as Early Diagnostic Markers of Colorectal Cancer. Dis. Markers 2021, 2021, 1–11. [Google Scholar] [CrossRef]
- Li, Z.; Liu, Y.; Guo, P.; Wei, Y. Construction and Validation of a Novel Angiogenesis Pattern to Predict Prognosis and Immunotherapy Efficacy in Colorectal Cancer. Aging 2023, 15, 12413–12450. [Google Scholar] [CrossRef] [PubMed]
- Hillege, L.E.; Trepka, K.R.; Ziemons, J.; Aarnoutse, R.; Guthrie, B.G.H.; De Vos-Geelen, J.; Iersel, L.V.; Van Hellemond, I.E.G.; Baars, A.; Vestjens, J.H.M.J.; et al. Metagenomic Analysis during Capecitabine Therapy Reveals Microbial Chemoprotective Mechanisms and Predicts Drug Toxicity in Colorectal Cancer Patients. Oncology 2024, 12, 2024.10.11.24315249. [Google Scholar] [CrossRef]
- Hillege, L.E.; Trepka, K.R.; Guthrie, B.G.H.; Fu, X.; Aarnoutse, R.; Paymar, M.R.; Olson, C.; Zhang, C.; Ortega, E.; Ramirez, L.; et al. Microbial Vitamin Biosynthesis Links Gut Microbiota Dynamics to Chemotherapy Toxicity. mBio 2025, 16, e00930-25. [Google Scholar] [CrossRef] [PubMed]
- Xiaofeng, N.; Jian, C.; Jingjing, W.; Zhanbo, Q.; Yifei, S.; Jing, Z.; Shuwen, H. Correlation of Gut Microbiota with Leukopenia after Chemotherapy in Patients with Colorectal Cancer. BMC Microbiol. 2023, 23, 349. [Google Scholar] [CrossRef]
- Nichols, B.; Briola, A.; Logan, M.; Havlik, J.; Mascellani, A.; Gkikas, K.; Milling, S.; Ijaz, U.Z.; Quince, C.; Svolos, V.; et al. Gut Metabolome and Microbiota Signatures Predict Response to Treatment with Exclusive Enteral Nutrition in a Prospective Study in Children with Active Crohn’s Disease. Am. J. Clin. Nutr. 2024, 119, 885–895. [Google Scholar] [CrossRef]
- Dang, Y.; Xu, X.; Ma, J.; Zhou, M.; Xu, C.; Huang, X.; Xu, F.; Wang, Z.; Shi, H.; Zhang, S. Gut Microbiome Signatures Predict 5-ASA Efficacy in Ulcerative Colitis. iScience 2025, 28, 112568. [Google Scholar] [CrossRef]
- Zheng, Q.; Zhong, Y.; Lian, H.; Zhuang, J.; Wang, L.; Chen, J.; Wang, H.; Wang, H.; Ye, X.; Huang, Z.; et al. Gut Microbial Signatures Associated With Clinical Remission in Inflammatory Bowel Disease Treated With Biologics: A Comprehensive Multi-Cohort Analysis. United Eur. Gastroenterol. J. 2025, ueg2.70064. [Google Scholar] [CrossRef]
- Sakurai, T.; Nishiyama, H.; Sakai, K.; De Velasco, M.A.; Nagai, T.; Komeda, Y.; Kashida, H.; Okada, A.; Kawai, I.; Nishio, K.; et al. Mucosal Microbiota and Gene Expression Are Associated with Long-Term Remission after Discontinuation of Adalimumab in Ulcerative Colitis. Sci. Rep. 2020, 10, 19186. [Google Scholar] [CrossRef]
- Al Radi, Z.M.A.; Prins, F.M.; Collij, V.; Vich Vila, A.; Festen, E.A.M.; Dijkstra, G.; Weersma, R.K.; Klaassen, M.A.Y.; Gacesa, R. Exploring the Predictive Value of Gut Microbiome Signatures for Therapy Intensification in Patients with Inflammatory Bowel Disease: A 10-Year Follow-up Study. Inflamm. Bowel Dis. 2024, 30, 1642–1653. [Google Scholar] [CrossRef]
- He, R.; Li, P.; Wang, J.; Cui, B.; Zhang, F.; Zhao, F. The Interplay of Gut Microbiota between Donors and Recipients Determines the Efficacy of Fecal Microbiota Transplantation. Gut Microbes 2022, 14, 2100197. [Google Scholar] [CrossRef]
- Smyth, J.; Godet, J.; Choudhary, A.; Das, A.; Gkoutos, G.V.; Acharjee, A. Microbiome-Based Colon Cancer Patient Stratification and Survival Analysis. Cancer Med. 2024, 13, e70434. [Google Scholar] [CrossRef]
- Shiroma, H.; Shiba, S.; Erawijantari, P.P.; Takamaru, H.; Yamada, M.; Sakamoto, T.; Kanemitsu, Y.; Mizutani, S.; Soga, T.; Saito, Y.; et al. Surgical Treatment for Colorectal Cancer Partially Restores Gut Microbiome and Metabolome Traits. mSystems 2022, 7, e00018-22. [Google Scholar] [CrossRef]
- Trivieri, N.; Pracella, R.; Cariglia, M.G.; Panebianco, C.; Parrella, P.; Visioli, A.; Giani, F.; Soriano, A.A.; Barile, C.; Canistro, G.; et al. BRAFV600E Mutation Impinges on Gut Microbial Markers Defining Novel Biomarkers for Serrated Colorectal Cancer Effective Therapies. J. Exp. Clin. Cancer Res. 2020, 39, 285. [Google Scholar] [CrossRef]
- Serrano-Gómez, G.; Mayorga, L.; Oyarzun, I.; Roca, J.; Borruel, N.; Casellas, F.; Varela, E.; Pozuelo, M.; Machiels, K.; Guarner, F.; et al. Dysbiosis and Relapse-Related Microbiome in Inflammatory Bowel Disease: A Shotgun Metagenomic Approach. Comput. Struct. Biotechnol. J. 2021, 19, 6481–6489. [Google Scholar] [CrossRef]
- O’Sullivan, J.; Patel, S.; Leventhal, G.E.; Fitzgerald, R.S.; Laserna-Mendieta, E.J.; Huseyin, C.E.; Konstantinidou, N.; Rutherford, E.; Lavelle, A.; Dabbagh, K.; et al. Host-Microbe Multi-Omics and Succinotype Profiling Have Prognostic Value for Future Relapse in Patients with Inflammatory Bowel Disease. Gut Microbes 2025, 17, 2450207. [Google Scholar] [CrossRef]
- Jacobs, J.P.; Sauk, J.S.; Ahdoot, A.I.; Liang, F.; Katzka, W.; Ryu, H.J.; Khandadash, A.; Lagishetty, V.; Labus, J.S.; Naliboff, B.D.; et al. Microbial and Metabolite Signatures of Stress Reactivity in Ulcerative Colitis Patients in Clinical Remission Predict Clinical Flare Risk. Inflamm. Bowel Dis. 2024, 30, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, J.P.; Goudarzi, M.; Lagishetty, V.; Li, D.; Mak, T.; Tong, M.; Ruegger, P.; Haritunians, T.; Landers, C.; Fleshner, P.; et al. Crohn’s Disease in Endoscopic Remission, Obesity, and Cases of High Genetic Risk Demonstrate Overlapping Shifts in the Colonic Mucosal-Luminal Interface Microbiome. Genome Med. 2022, 14, 91. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Meng, J.; Lin, S.; Peng, Z.; Zhang, R.; Shen, X.; Zheng, W.; Zheng, Q.; Wu, L.; Wang, X.; et al. Integrating Gut Microbiome and Metabolomics with Magnetic Resonance Enterography to Advance Bowel Damage Prediction in Crohn’s Disease. J. Inflamm. Res. 2025, 18, 7631–7649. [Google Scholar] [CrossRef] [PubMed]
- Machiels, K.; Pozuelo Del Río, M.; Martinez-De La Torre, A.; Xie, Z.; Pascal Andreu, V.; Sabino, J.; Santiago, A.; Campos, D.; Wolthuis, A.; D’Hoore, A.; et al. Early Postoperative Endoscopic Recurrence in Crohn’s Disease Is Characterised by Distinct Microbiota Recolonisation. J. Crohns Colitis 2020, 14, 1535–1546. [Google Scholar] [CrossRef]
- Syama, K.; Jothi, J.A.A.; Khanna, N. Automatic Disease Prediction from Human Gut Metagenomic Data Using Boosting GraphSAGE. BMC Bioinform. 2023, 24, 126. [Google Scholar] [CrossRef]
- Zhou, Y.-L.; Deng, J.-W.; Liu, Z.-H.; Ma, X.-Y.; Zhu, C.-Q.; Xie, Y.-H.; Zhou, C.-B.; Fang, J.-Y. Derivation and Validation of Lifestyle-Based and Microbiota-Based Models for Colorectal Adenoma Risk Evaluation and Self-Prediction. BMJ Open Gastroenterol. 2025, 12, e001597. [Google Scholar] [CrossRef]
- Huang, A.; Torres, A.; Patel, R.; Saxena, A.; Patel, M. Fusobacterium Nucleatum as a Marker for Epithelial to Mesenchymal Transition in Colorectal Cancer. FASEB J. 2022, 36, fasebj.2022.36.S1.L7993. [Google Scholar] [CrossRef]
- Pignatelli, P.; Nuccio, F.; Piattelli, A.; Curia, M.C. The Role of Fusobacterium Nucleatum in Oral and Colorectal Carcinogenesis. Microorganisms 2023, 11, 2358. [Google Scholar] [CrossRef]
- Zhang, S.; Li, C.; Liu, J.; Geng, F.; Shi, X.; Li, Q.; Lu, Z.; Pan, Y. Fusobacterium Nucleatum Promotes Epithelial-mesenchymal Transiton through Regulation of the lncRNA MIR4435-2HG/miR-296-5p/Akt2/SNAI1 Signaling Pathway. FEBS J. 2020, 287, 4032–4047. [Google Scholar] [CrossRef]
- Shimomura, Y.; Sugi, Y.; Kume, A.; Tanaka, W.; Yoshihara, T.; Matsuura, T.; Komiya, Y.; Ogata, Y.; Suda, W.; Hattori, M.; et al. Strain-Level Detection of Fusobacterium Nucleatum in Colorectal Cancer Specimens by Targeting the CRISPR–Cas Region. Microbiol. Spectr. 2023, 11, e05123-22. [Google Scholar] [CrossRef]
- Yang, C.; Zhao, Y.; Chen, J.; Chen, N. P1350 The Role of Oral Porphyromonas Gingivalis and Its Outer Membrane Vesicles in Inflammatory Bowel Disease. J. Crohns Colitis 2025, 19 (Suppl. S1), i2431. [Google Scholar] [CrossRef]
- Abdelbary, M.M.H.; Hatting, M.; Bott, A.; Dahlhausen, A.; Keller, D.; Trautwein, C.; Conrads, G. The Oral-Gut Axis: Salivary and Fecal Microbiome Dysbiosis in Patients with Inflammatory Bowel Disease. Front. Cell. Infect. Microbiol. 2022, 12, 1010853. [Google Scholar] [CrossRef]
- Zhou, T.; Xu, W.; Wang, Q.; Jiang, C.; Li, H.; Chao, Y.; Sun, Y.; Lan, A. The Effect of the “Oral-Gut” Axis on Periodontitis in Inflammatory Bowel Disease: A Review of Microbe and Immune Mechanism Associations. Front. Cell. Infect. Microbiol. 2023, 13, 1132420. [Google Scholar] [CrossRef]
- Xiang, B.; Runzhi, C.; Min, Z.; Yuanqi, Z.; Xiang, P.; Wei, W.; Qi, Z.; Zhiyin, G.; Jun, H.; Tao, D.; et al. P1214 The Crosstalk between Oral and Intestinal Microbiota in Inflammatory Bowel Disease Patients. J. Crohns Colitis 2024, 18 (Suppl. S1), i2145. [Google Scholar] [CrossRef]
- Wands, D.; Whelan, R.; Hansen, R.; Rimmer, P.; Iqbal, T.; Ho, G.T. P1328 Reduced Salivary Alpha-Diversity in Inflammatory Bowel Disease: Systematic Review and Meta-Analysis of 1000 IBD Patients—A Role for Oral Microbiome as a Marker of Downstream Dysbiosis in IBD. J. Crohns Colitis 2025, 19 (Suppl. S1), i2390–i2392. [Google Scholar] [CrossRef]
- Camañes-Gonzalvo, S.; Montiel-Company, J.M.; Lobo-de-Mena, M.; Safont-Aguilera, M.J.; Fernández-Diaz, A.; López-Roldán, A.; Paredes-Gallardo, V.; Bellot-Arcís, C. Relationship between Oral Microbiota and Colorectal Cancer: A Systematic Review. J. Periodontal Res. 2024, 59, 1071–1082. [Google Scholar] [CrossRef]
- Ji, S.; Kook, J.-K.; Park, S.-N.; Lim, Y.K.; Choi, G.H.; Jung, J.-S. Characteristics of the Salivary Microbiota in Periodontal Diseases and Potential Roles of Individual Bacterial Species To Predict the Severity of Periodontal Disease. Microbiol. Spectr. 2023, 11, e04327-22. [Google Scholar] [CrossRef]
- Akase, T.; Inubushi, J.; Hayashi-Okada, Y.; Shimizu, Y. Association of Fusobacterium Nucleatum in Human Saliva with Periodontal Status and Composition of the Salivary Microbiome Including Periodontopathogens. Microbiol. Spectr. 2024, 12, e00855-24. [Google Scholar] [CrossRef]
- Chen, Q.; Huang, X.; Zhang, H.; Jiang, X.; Zeng, X.; Li, W.; Su, H.; Chen, Y.; Lin, F.; Li, M.; et al. Characterization of Tongue Coating Microbiome from Patients with Colorectal Cancer. J. Oral Microbiol. 2024, 16, 2344278. [Google Scholar] [CrossRef]
- Rezasoltani, S.; Aghdaei, H.A.; Jasemi, S.; Gazouli, M.; Dovrolis, N.; Sadeghi, A.; Schlüter, H.; Zali, M.R.; Sechi, L.A.; Feizabadi, M.M. Oral Microbiota as Novel Biomarkers for Colorectal Cancer Screening. Cancers 2022, 15, 192. [Google Scholar] [CrossRef]
- Kang, S.-B.; Kim, H.; Kim, S.; Kim, J.; Park, S.-K.; Lee, C.-W.; Kim, K.O.; Seo, G.-S.; Kim, M.S.; Cha, J.M.; et al. Potential Oral Microbial Markers for Differential Diagnosis of Crohn’s Disease and Ulcerative Colitis Using Machine Learning Models. Microorganisms 2023, 11, 1665. [Google Scholar] [CrossRef]
- Rezasoltani, S.; Azizmohammad Looha, M.; Asadzadeh Aghdaei, H.; Jasemi, S.; Sechi, L.A.; Gazouli, M.; Sadeghi, A.; Torkashvand, S.; Baniali, R.; Schlüter, H.; et al. 16S rRNA Sequencing Analysis of the Oral and Fecal Microbiota in Colorectal Cancer Positives versus Colorectal Cancer Negatives in Iranian Population. Gut Pathog. 2024, 16, 9. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Wang, Z.; Tang, J.; Cao, D.-X.; Qian, Y.; Xie, Y.-H.; Chen, H.-Y.; Chen, Y.-X.; Chen, Z.-F.; et al. A Clinical Nomogram Incorporating Salivary Desulfovibrio Desulfuricans Level and Oral Hygiene Index for Predicting Colorectal Cancer. Ann. Transl. Med. 2021, 9, 754. [Google Scholar] [CrossRef]
- Čipčić Paljetak, H.; Barešić, A.; Panek, M.; Perić, M.; Matijašić, M.; Lojkić, I.; Barišić, A.; Vranešić Bender, D.; Ljubas Kelečić, D.; Brinar, M.; et al. Gut Microbiota in Mucosa and Feces of Newly Diagnosed, Treatment-Naïve Adult Inflammatory Bowel Disease and Irritable Bowel Syndrome Patients. Gut Microbes 2022, 14, 2083419. [Google Scholar] [CrossRef]
- Juge, N. Relationship between Mucosa-Associated Gut Microbiota and Human Diseases. Biochem. Soc. Trans. 2022, 50, 1225–1236. [Google Scholar] [CrossRef]
- Lepage, P.; Seksik, P.; Sutren, M.; De La Cochetière, M.-F.; Jian, R.; Marteau, P.; Doré, J. Biodiversity of the Mucosa-Associated Microbiota Is Stable Along the Distal Digestive Tract in Healthy Individuals and Patients With Ibd. Inflamm. Bowel Dis. 2005, 11, 473–480. [Google Scholar] [CrossRef]
- Effendi, R.M.R.A.; Anshory, M.; Kalim, H.; Dwiyana, R.F.; Suwarsa, O.; Pardo, L.M.; Nijsten, T.E.C.; Thio, H.B. Akkermansia Muciniphila and Faecalibacterium Prausnitzii in Immune-Related Diseases. Microorganisms 2022, 10, 2382. [Google Scholar] [CrossRef]
- Goldiș, A.; Dragomir, R.; Mercioni, M.A.; Goldiș, C.; Sirca, D.; Enătescu, I.; Olariu, L.; Belei, O. Personalized Microbiome Modulation to Improve Clinical Outcomes in Pediatric Inflammatory Bowel Disease: A Multi-Omics and Interventional Approach. Microorganisms 2025, 13, 1047. [Google Scholar] [CrossRef]
- Zhu, B.; Bai, Y.; Yeo, Y.Y.; Lu, X.; Rovira-Clavé, X.; Chen, H.; Yeung, J.; Gerber, G.K.; Angelo, M.; Shalek, A.K.; et al. A Spatial Multi-Modal Dissection of Host-Microbiome Interactions within the Colitis Tissue Microenvironment. Immunology 2024, 2024.03.04.583400. [Google Scholar] [CrossRef]
- Hu, S.; Bourgonje, A.R.; Gacesa, R.; Jansen, B.H.; Björk, J.R.; Bangma, A.; Hidding, I.J.; Van Dullemen, H.M.; Visschedijk, M.C.; Faber, K.N.; et al. Mucosal Host-Microbe Interactions Associate with Clinical Phenotypes in Inflammatory Bowel Disease. Nat. Commun. 2024, 15, 1470. [Google Scholar] [CrossRef]
- Santangelo, B.E.; Bada, M.; Hunter, L.E.; Lozupone, C. Hypothesizing Mechanistic Links between Microbes and Disease Using Knowledge Graphs. Sci. Rep. 2025, 15, 6905. [Google Scholar] [CrossRef]
- Kuang, J.; Zheng, X.; Jia, W. Investigating Regional-Specific Gut Microbial Distribution: An Uncharted Territory in Disease Therapeutics. Protein Cell 2024, 16, 623–640. [Google Scholar] [CrossRef]
- Kastl, A.J.; Terry, N.A.; Wu, G.D.; Albenberg, L.G. The Structure and Function of the Human Small Intestinal Microbiota: Current Understanding and Future Directions. Cell. Mol. Gastroenterol. Hepatol. 2020, 9, 33–45. [Google Scholar] [CrossRef]
- Nagayama, M.; Yano, T.; Atarashi, K.; Tanoue, T.; Sekiya, M.; Kobayashi, Y.; Sakamoto, H.; Miura, K.; Sunada, K.; Kawaguchi, T.; et al. TH1 Cell-Inducing Escherichia Coli Strain Identified from the Small Intestinal Mucosa of Patients with Crohn’s Disease. Gut Microbes 2020, 12, 1788898. [Google Scholar] [CrossRef]
- Ma, X.; Lu, X.; Zhang, W.; Yang, L.; Wang, D.; Xu, J.; Jia, Y.; Wang, X.; Xie, H.; Li, S.; et al. Gut Microbiota in the Early Stage of Crohn’s Disease Has Unique Characteristics. Gut Pathog. 2022, 14, 46. [Google Scholar] [CrossRef]
- Briggs, K.; Tomar, V.; Ollberding, N.; Haberman, Y.; Bourgonje, A.R.; Hu, S.; Chaaban, L.; Sunuwar, L.; Weersma, R.K.; Denson, L.A.; et al. Crohn’s Disease–Associated Pathogenic Mutation in the Manganese Transporter ZIP8 Shifts the Ileal and Rectal Mucosal Microbiota Implicating Aberrant Bile Acid Metabolism. Inflamm. Bowel Dis. 2024, 30, 1379–1388. [Google Scholar] [CrossRef]
- Hale, M.F. Capsule Endoscopy: Current Practice and Future Directions. World J. Gastroenterol. 2014, 20, 7752. [Google Scholar] [CrossRef]
- Matsuzawa, H.; Munakata, S.; Kawai, M.; Sugimoto, K.; Kamiyama, H.; Takahashi, M.; Kojima, Y.; Sakamoto, K. Analysis of Ileostomy Stool Samples Reveals Dysbiosis in Patients with High-Output Stomas. Biosci. Microbiota Food Health 2021, 40, 135–143. [Google Scholar] [CrossRef]
- Wirbel, J.; Zych, K.; Essex, M.; Karcher, N.; Kartal, E.; Salazar, G.; Bork, P.; Sunagawa, S.; Zeller, G. Microbiome Meta-Analysis and Cross-Disease Comparison Enabled by the SIAMCAT Machine Learning Toolbox. Genome Biol. 2021, 22, 93. [Google Scholar] [CrossRef]
- Lee, Y.; Cappellato, M.; Di Camillo, B. Machine Learning–Based Feature Selection to Search Stable Microbial Biomarkers: Application to Inflammatory Bowel Disease. GigaScience 2022, 12, giad083. [Google Scholar] [CrossRef]
- Unal, M.; Bostanci, E.; Ozkul, C.; Acici, K.; Asuroglu, T.; Guzel, M.S. Crohn’s Disease Prediction Using Sequence Based Machine Learning Analysis of Human Microbiome. Diagnostics 2023, 13, 2835. [Google Scholar] [CrossRef]
- Lee, S.; Lee, I. Comprehensive Assessment of Machine Learning Methods for Diagnosing Gastrointestinal Diseases through Whole Metagenome Sequencing Data. Gut Microbes 2024, 16, 2375679. [Google Scholar] [CrossRef]
- Rynazal, R.; Fujisawa, K.; Shiroma, H.; Salim, F.; Mizutani, S.; Shiba, S.; Yachida, S.; Yamada, T. Leveraging Explainable AI for Gut Microbiome-Based Colorectal Cancer Classification. Genome Biol. 2023, 24, 21. [Google Scholar] [CrossRef]
- DiMucci, D.; Kon, M.; Segrè, D. BowSaw: Inferring Higher-Order Trait Interactions Associated With Complex Biological Phenotypes. Front. Mol. Biosci. 2021, 8, 663532. [Google Scholar] [CrossRef]
- Muller, E.; Shiryan, I.; Borenstein, E. Multi-Omic Integration of Microbiome Data for Identifying Disease-Associated Modules. Nat. Commun. 2024, 15, 2621. [Google Scholar] [CrossRef]
- Pope, Q.; Varma, R.; Tataru, C.; David, M.M.; Fern, X. Learning a Deep Language Model for Microbiomes: The Power of Large Scale Unlabeled Microbiome Data. PLOS Comput. Biol. 2025, 21, e1011353. [Google Scholar] [CrossRef] [PubMed]
- Imangaliyev, S.; Schlötterer, J.; Meyer, F.; Seifert, C. Diagnosis of Inflammatory Bowel Disease and Colorectal Cancer through Multi-View Stacked Generalization Applied on Gut Microbiome Data. Diagnostics 2022, 12, 2514. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Liu, Y.; Liu, Y.; Wang, J. Comprehensive Data Optimization and Risk Prediction Framework: Machine Learning Methods for Inflammatory Bowel Disease Prediction Based on the Human Gut Microbiome Data. Front. Microbiol. 2024, 15, 1483084. [Google Scholar] [CrossRef] [PubMed]
- Mulenga, M.; Rajamanikam, A.; Kumar, S.; Muhammad, S.B.; Bhassu, S.; Samudid, C.; Sabri, A.Q.M.; Seera, M.; Eke, C.I. Revolutionizing Colorectal Cancer Detection: A Breakthrough in Microbiome Data Analysis. PLoS ONE 2025, 20, e0316493. [Google Scholar] [CrossRef]
- Pusa, T.; Rousu, J. Stable Biomarker Discovery in Multi-Omics Data via Canonical Correlation Analysis. PLoS ONE 2024, 19, e0309921. [Google Scholar] [CrossRef]
- Onwuka, S.; Bravo-Merodio, L.; Gkoutos, G.V.; Acharjee, A. Explainable AI-Prioritized Plasma and Fecal Metabolites in Inflammatory Bowel Disease and Their Dietary Associations. iScience 2024, 27, 110298. [Google Scholar] [CrossRef]

| Reference | Objective | DL Model (Highest Performance) | Input | Output | Publication Date |
|---|---|---|---|---|---|
| [44] | Diagnosis of CRC using gut viral signatures | RF | Stool virome (405 CRC-associated vOTUs) | AUC 0.830(cross-cohort, CRC vs. HC) | 2 October 2022 |
| [70] | Diagnosis of CRC using microbial and functional profiles | RF | Stool microbiota (genus-level), KEGG functional profiles | Genus-level AUC 0.84(CRC vs. HC), 0.73(CRC vs. CA) 3-genus signature AUC 0.87(CRC vs. HC). 0.67(CRC vs. CA) | 3 November 2022 |
| [71] | Diagnosis of colorectal adenoma and CRC | RF | Stool microbiota (ASV-level), age, sex, and BMI | AUC 0.78 (adenoma vs. control), AUC 0.84 (adenoma vs. CRC, external validation) | 24 May 2021 |
| [72] | CRC and adenoma diagnosis integrating microbiome and clinical data | RF | Large-scale metagenomic data + clinical variables | AUC 0.939 (CRC), 0.925 (adenoma) | 22 January 2024 |
| [73] | Interpretable ML model for CRC and adenoma classification using functional profiles | Explainable Boosting Machine (EBM) | Stool microbiota(WGS), KEGG, eggNOG functional profile | eggNOG profile 100% hit ratio for all the performed tests | 10 January 2022 |
| [74] | Risk stratification of FIT-positive individuals using microbiome signatures | Neural Network | 16S rRNA(V3–V4) 8 selected taxa, age, sex, fecal hemoglobin concentration (FIT) | Sensitivity: 98.98% (CRC), 97.98% (CR lesions) | 25 December 2022 |
| [75] | Diagnosis of adenoma and CRC using blood cfDNA microbiome | RF | Blood cfDNA | AUC 0.8849 (adenoma), AUC 0.9824 (CRC) | 29 April 2025 |
| [76] | To evaluate the impact of FOBT on gut microbiota composition and improve CRC prediction models | SVM | Gut microbiota(16S rRNA, genus-level), FOBT status | Accuracy without FOBT: 89.71% → with FOBT: 92% | 20 January 2025 |
| [77] | Diagnosis of advanced adenoma using gut microbiota | RF | Shotgun metagenomic data (species-level abundance) | AUC 0.799 (adenoma vs. control) | 10 October 2024 |
| [78] | Discriminating Advanced Adenoma from CRC using metagenomic microbiome and SNP data | RF (SNP model) | Microbial SNPs from fecal metagenomic data | Accuracy 92.31% | 1 December 2022 |
| [79] | Improve the Colorectal Cancer Diagnosis Using Gut Microbiome | RF, BART | 16S rRNA/Shotgun metagenomics | AUC 0.867 (RF), 0.882 (Bart) | 12 August 2022 |
| [80] | Improve CRC diagnostic accuracy by combining gut microbiota, MT-sDNA, and tumor markers | RF | 16S rRNA(genus-level), MT-sDNA, CEA | Accuracy 97.1% Sensitivity 98.1%, Specificity 92.3% | 15 August 2023 |
| [81] | Classification of right- vs. left-sided colorectal cancer using tumor-associated microbial features | RF | Tumor-derived microbial RNA-seq expression data | AUC 0.9, 0.76, and 0.89 for the human genomic, microbial, and combined feature sets, respectively | 11 July 2023 |
| [82] | Tissue microbiome analysis by site and type in CRC patients | RF | Tissue-derived metagenomic data(WGS) | Site- and tissue-specific microbial signatures | 27 May 2025 |
| [83] | Classification of poorly vs. moderately differentiated colorectal cancer using gut microbiota | RF | Fecal 16S rRNA data (V1–V4) | Accuracy 100% | 20 December 2022 |
| [84] | Classification of healthy, adenoma, and CRC based on enterotype-specific gut microbiota | RF | Fecal 16S rRNA sequencing data (genus-level), stratified by enterotype | AUC 0.78, S_E model, CRC vs. non-CRC | 27 February 2024 |
| [85] | CRC and adenoma diagnosis integrating microbiome and clinical data | RF | Large-scale metagenomic data + clinical variables | AUC 0.939 (CRC), 0.925 (adenoma) | 22 January 2024 |
| Reference | Objective | DL Model (Highest Performance) | Input | Output | Publication Date |
|---|---|---|---|---|---|
| [87] | Development of LightCUD for IBD and UC/CD diagnosis | LightGBM | WGS (strain-level), 16S rRNA (genus-level) | AUC 0.984 (IBD vs. HC-WGS), AUC 0.989 (CD vs. UC-WGS) | 19 January 2021 |
| [88] | Diagnosis of IBD and subtype differentiation (CD vs. UC) | sPLS-DA | 16S rRNA (phylotype-level, V3–V4) | AUC 0.992 (IBD vs. HC), AUC 0.988 (CD vs. UC) | 24 December 2023 |
| [89] | Identify the microbiota signature by UC disease activity | sPLS-DA | 16S rRNA (V3–V4) | Perfect class prediction (Active UC/Inactive UC/HC) | 28 December 2021 |
| [90] | Non-invasive diagnosis and monitoring of IBD using fecal biomarkers and microbiome | Logistic Regression | Fecal HBD2, FCal, 16S rRNA (Genus-level) | AUC 0.93 (IBD vs. IBS) | 7 June 2021 |
| [91] | Classify disease activity in UC based on gut fungal (mycobiome) signatures | RF | ITS2 | AUC ~0.80 (Active vs. Remission UC) | 19 April 2024 |
| [92] | Incorporating external samples to improve model robustness and generalizability. | RF, among others | 16S rRNA (Genus level) | AUC increased by up to 0.075 | 8 February 2023 |
| [93] | Diagnosis of UC and CD patients | Regularized Logistic Regression | Fecal WGS (species-level) | AUC 0.873 (train), 0.778 (test), 0.633 (validation) | 11 November 2023 |
| [94] | Diagnosis of UC and CD patients | RF | 16s rRNA (genus-level OTU) | AUC 0.76 (HC vs. CD), 0.74 (HC vs. UC) | 17 May 2022 |
| [95]. | Including absolute microbial load and clinical markers | RF | Fungal 18S rDNA copies, bacterial 16S rDNA copies | AUC 0.86 (UC vs. CD) | 27 April 2021 |
| [96] | Diagnosis of PIBD using fecal microbiota | RF | Stool microbiota (11 OTUs) | AUC 0.88 (HC vs. PIBD), 0.84 (IBS vs. PIBD) | 7 December 2021 |
| [97] | Classifying PIBD activity using bile acid | RF | Serum BAs | AUC 0.84 | 21 February 2024 |
| [98] | Noninvasive diagnosis of Pediatric IBD using fecal AAs and microbiota | Logistic Regression | Fecal microbiota, AAs | AUC 0.94 (Discovery), 0.84 (Validation) | 4 June 2025 |
| Domain. | Oral Microbiome | Mucosal-Associated Microbiota (MAM) | Small Intestine Microbiome |
|---|---|---|---|
| Diagnostic Utility | AUC ≈ 0.90–0.97; Fully non-invasive screening | Outperforms fecal profiles in IBD/CRC stratification | Early-stage evidence; Limited clinical use |
| Key Bacterial Taxa | F. nucleatum, S. anginosus (CRC); P. intermedia, Veillonella (IBD) | Beneficial - F. prausnitzii, A. muciniphila Pathogenic - AIEC | E. coli 35A1, R. gnavus (CD) |
| Sampling Methods | Saliva, dental plaque, tongue coating - repeatable, non-invasive | Endoscopic biopsy - high precision, invasive | Endoscopy, capsule/string tests, stoma effluent - technically demanding |
| Challenges | High inter-individual variability; Periodontal confounders | Invasiveness, spatial heterogeneity, small sample size | Low biomass, contamination risk, scarce longitudinal data |
| AI/ML Integration | Mature ML / xAI models for CRC & IBD early prediction | Active ML / graph-AI mapping host-microbe networks | Limited by data sparsity; Models under development |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.; Gim, D.; Kim, S.; Park, S.; Eom, T.P.; Seol, J.; Yeo, J.; Jo, C.; Seo, G.; Ku, H.; et al. From Dysbiosis to Prediction: AI-Powered Microbiome Insights into IBD and CRC. Gastroenterol. Insights 2025, 16, 34. https://doi.org/10.3390/gastroent16030034
Kim M, Gim D, Kim S, Park S, Eom TP, Seol J, Yeo J, Jo C, Seo G, Ku H, et al. From Dysbiosis to Prediction: AI-Powered Microbiome Insights into IBD and CRC. Gastroenterology Insights. 2025; 16(3):34. https://doi.org/10.3390/gastroent16030034
Chicago/Turabian StyleKim, Minkwan, Donghyeon Gim, Sunghan Kim, Sungsu Park, Tehyun Phillip Eom, Jaehoon Seol, Junyeong Yeo, Changmin Jo, Gunha Seo, Hyungjune Ku, and et al. 2025. "From Dysbiosis to Prediction: AI-Powered Microbiome Insights into IBD and CRC" Gastroenterology Insights 16, no. 3: 34. https://doi.org/10.3390/gastroent16030034
APA StyleKim, M., Gim, D., Kim, S., Park, S., Eom, T. P., Seol, J., Yeo, J., Jo, C., Seo, G., Ku, H., & Kim, J. H. (2025). From Dysbiosis to Prediction: AI-Powered Microbiome Insights into IBD and CRC. Gastroenterology Insights, 16(3), 34. https://doi.org/10.3390/gastroent16030034






