Dynamics in Redox-Active Molecules Following Ischemic Preconditioning in the Brain
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Induction of Cerebral Ischemia-Reperfusion and Ischemic Preconditioning
2.3. Tissue Homogenisation, Isolation of Mitochondria, and Protein Content Determination
2.4. Western Blot and Immunodetection
2.5. Two-Dimensional Electrophoresis (2-DE)
2.6. Protein Identification by Mass Spectrometry
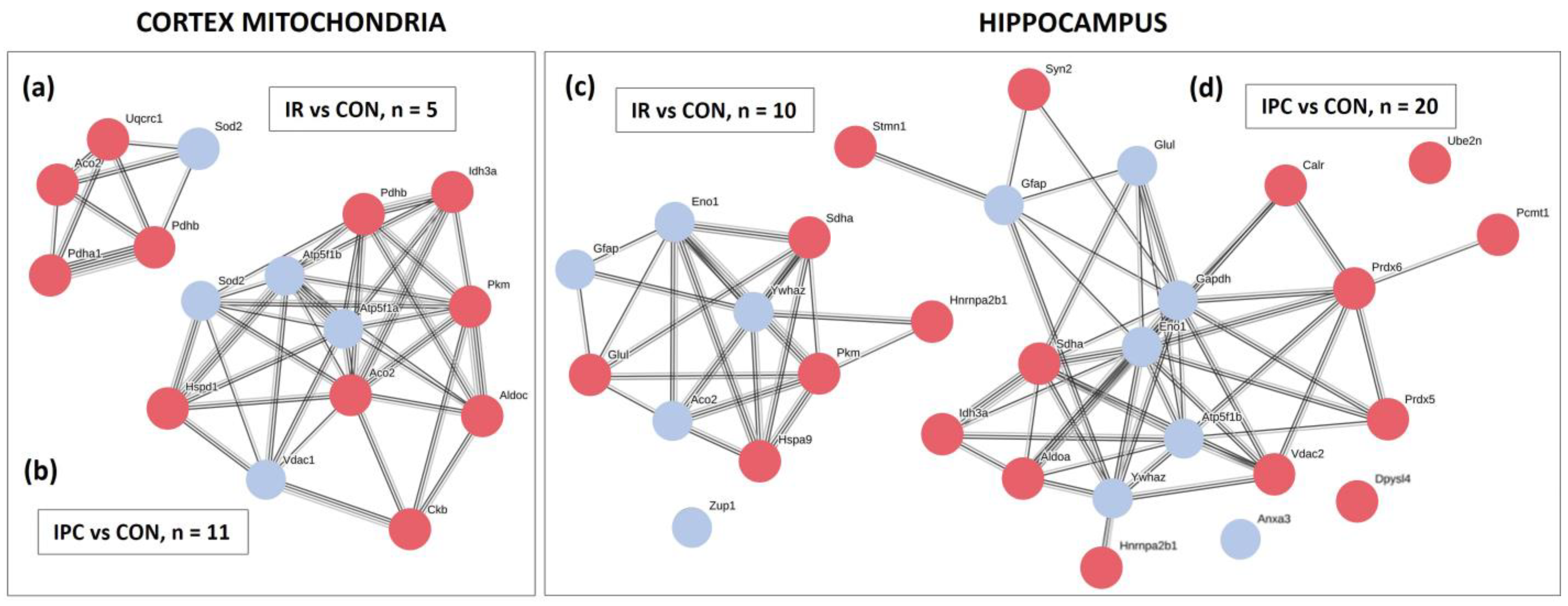
2.7. Quantification, Protein–Protein Interaction Network
2.8. Total Glutathione and the Assay of Enzyme Activities
2.8.1. Glutathione Content, Glutathione Reductase, and Glutathione Peroxidase Activity
2.8.2. The Assay of Superoxide Dismutase and ATP Synthase Activity
2.9. Fluorescent Immunohistochemistry
2.10. Data Analysis
3. Results
3.1. Identification of Altered Proteins in the Cortex and Hippocampus
3.2. Western Blot Analysis and Immunodetection of Modified Proteins
3.3. Fluorescent Immunohistochemical Analysis of Peroxiredoxins
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Spot No. | Protein ID | Protein Name | Gene | Mw (kDa) | pI | IR/CON Fold Change/p-Value | IPC/CON Fold Change/p-Value |
|---|---|---|---|---|---|---|---|
| 1 | QCR1 | Cytochrome b-c1 complex subunit 1, mitochondrial | Uqcrc1 | 53.50 | 5.57 | 0.34 0.011 * | ns |
| 2 | ATPB | ATP synthase subunit beta, mitochondrial | Atp5b | 56.32 | 5.19 | ns | 1.54 0.033 * |
| 3 | ODPB | Pyruvate dehydrogenase E1 component subunit beta | Pdhb | 39.30 | 6.20 | 0.49 0.0045 ** | 0.31 3 × 10−4 *** |
| 4 | IDH3A | Isocitrate dehydrogenase 3 (NAD+) alpha, mitochondrial | Idh3a | 40.04 | 6.47 | ns | 0.53 0.053 |
| 5 | KCRB | Creatine kinase, brain isoform CRA_a | Ckb | 36.15 | 6.27 | ns | 0.35 0.043 * |
| 6 | CH60 | 60kDa heat shock protein, mitochondrial | Hspd1 | 61.09 | 5.91 | ns | 0.36 6 × 10−4 *** |
| 7 | TPIS | Triosephosphate isomerase | Tpi1 | 27.34 | 6.89 | ns | ns |
| 8 | ODPA | Pyruvate dehydrogenase E1 subunit alpha, mitochondrial | Pdha1 | 43.88 | 6.67 | 0.57 0.012 * | ns |
| 9 | DLDH | Dihydrolipoyl dehydrogenase, mitochondrial | Dld | 54.57 | 7.96 | ns | ns |
| 10 | ALDOC | Fructose-bisphosphate aldolase C | Aldoc | 43.88 | 8.49 | ns | 0.39 0.016 * |
| 11 | KPYM | Pyruvate kinase PKM | Pkm | 58.29 | 6.63 | ns | 0.48 0.050 |
| 12 | ACON | Aconitate hydratase, mitochondrial | Aco2 | 86.12 | 7.87 | 0.35 0.0014 ** | 0.27 1 × 10−5 *** |
| 13 | SODM | Superoxide dismutase [Mn], mitochondrial | Sod2 | 24.89 | 8.96 | 1.53 0.018 * | 2.26 0.0086 ** |
| 14 | ATPA | ATP synthase subunit alpha, mitochondrial | Atp5a1 | 59.83 | 9.22 | ns | 1.64 0.043 * |
| 15 | VDAC1 | Voltage-dependent anion-selective channel protein 1 | Vdac1 | 30.85 | 8.62 | ns | 2.85 0.018 * |
| Spot No. | Protein ID | Protein Name | Gene | Mw (kDa) | pI | IR/CON Fold Change/p-Value | IPC/CON Fold Change/p-Value |
|---|---|---|---|---|---|---|---|
| 1 | CALR | Calreticulin | Calr | 48.14 | 4.33 | ns | 0.33 0.0037 ** |
| 2 | GFAP | Glial fibrillary acidic protein beta | Gfap | 38.52 | 5.35 | 1.77 0.047 * | 2.04 0.021 * |
| 3 | STMN1 | Stathmin | Stmn1 | 17.28 | 5.76 | ns | 0.38 0.023 * |
| 4 | UBE2N | Ubiquitin-conjugating enzyme E2 | Ube2n | 17.17 | 6.13 | ns | 0.44 0.068 |
| 5 | 1433Z | 14-3-3 protein zeta/delta | Ywhaz | 27.92 | 4.73 | 1.85 0.028 * | 2.28 0.032 * |
| 6 | PRDX6 | Peroxiredoxin 6 | Prdx6 | 24.86 | 5.64 | ns | 0.36 0.0076 ** |
| 7 | ANXA3 | Annexin 3 | Anxa3 | 36.57 | 5.96 | ns | 1.53 0.0067 ** |
| 8 | IDH3A | Isocitrate dehydrogenase 3 (NAD+) alpha, mitochondrial | Idh3a | 40.04 | 6.47 | ns | 0.38 0.036 * |
| 9 | ATPB | ATP synthase subunit beta, mitochondrial | Atp5b | 56.32 | 5.15 | ns | 2.07 0.033 * |
| 10 | SDHA | Succinate dehydrogenase [Q] flavoprotein subunit | Sdha | 72.60 | 6.75 | 0.62 0.029 * | 0.49 9 × 10−4 *** |
| 11 | GRP75 | 75 kDa glucose-regulated protein | Hspa9 | 74.10 | 5.97 | 0.51 0.0042 ** | ns |
| 12 | ALDR | Aldo-keto reductase family 1 member B1 | Akr1b1 | 36.23 | 6.26 | ns | ns |
| 13 | IVD | Isovaleryl-CoA dehydrogenase, mitochondrial | Ivd | 46.86 | 8.03 | ns | ns |
| 14 | ENOA | Alpha enolase | Eno1 | 47.44 | 6.16 | 3.08 0.031 * | 2.23 0.033 * |
| 15 | HS71A | Heat shock protein70 kDa protein 1A | Hspa1a | 70.43 | 5.61 | Spot missing in the control group | |
| 16 | PRDX5 | Peroxiredoxin 5 | Prdx5 | 22.51 | 8.94 | ns | 0.54 0.0063 ** |
| 17 | G3P | Glyceraldehyde-3 phosphate dehydrogenase | Gapdh | 36.09 | 8.14 | ns | 1.86 0.043 * |
| 18 | ALDOA | Fructose bisphosphate aldolase A | Aldoa | 39.78 | 8.31 | ns | 0.64 0.049 * |
| 19 | GLNA | Glutamine synthetase | Glul | 42.98 | 6.64 | 1.51 0.0018 ** | 1.56 0.0014 ** |
| 20 | DHE3 | Glutamate dehydrogenase, mitochondrial | Glud1 | 61.72 | 8.05 | ns | ns |
| 21 | ZUP1 | Zinc finger-containing ubiquitin peptidase 1 | Zup1 | 66.99 | 6.08 | 1.71 0.050 * | ns |
| 22 | DPYL4 | Dihydropyrimidinase-related protein 4 | Dpysl4 | 61.62 | 6.3 | ns | 0.46 0.012 * |
| 23 | KPYM | Pyruvate kinase | Pkm | 58.29 | 6.63 | 0.60 0.022 * | ns |
| 24 | SYN2 | Synapsin 2 | Syn2 | 63.70 | 8.73 | ns | 0.38 0.015 * |
| 25 | ACON | Aconitate hydratase | Aco2 | 86.12 | 7.87 | 1.68 0.019 * | ns |
| 26 | PIMT | Protein-D-aspartate-O-methyltransferase | Pcmt1 | 24.68 | 7.14 | ns | 0.35 0.0099 ** |
| 27 | KAD1 | Adenylate kinase isoenzyme 1 | Ak1 | 21.68 | 7.66 | ns | ns |
| 28 | VDAC2 | Voltage-dependent anion-selective channel protein 2 | Vdac2 | 32.35 | 7.44 | ns | 0.47 0.024 * |
| 29 | ROA2 | Heterogenous nuclear ribonucleoproteins A2/B1 | Hnmpa2b1 | 37.51 | 8.94 | 0.56 0.029 * | 0.26 0.026 * |
References
- Castro, J.P.; Wardelmann, K.; Grune, T.; Kleinridders, A. Mitochondrial Chaperones in the Brain: Safeguarding Brain Health and Metabolism? Front. Endocrinol. 2018, 9, 196. [Google Scholar] [CrossRef]
- Barros, L.F.; Ruminot, I.; Sandoval, P.Y.; San Martín, A. Enlightening brain energy metabolism. Neurobiol. Dis. 2023, 184, 106211. [Google Scholar] [CrossRef]
- Bonvento, G.; Bolaños, J.P. Astrocyte-neuron metabolic cooperation shapes brain activity. Cell Metab. 2021, 33, 1546–1564. [Google Scholar] [CrossRef]
- Berridge, M.J.; Bootman, M.D.; Roderick, H.L. Calcium signaling: Dynamics, homeostasis, and remodeling. Nat. Rev. Mol. Cell. Biol. 2003, 4, 517–529. [Google Scholar] [CrossRef]
- Wegrzyn, J.; Potla, R.; Chwae, Y.J.; Sepuri, N.B.; Zhang, Q.; Koeck, T.; Derecka, M.; Szczepanek, K.; Szelag, M.; Gornicka, A.; et al. Function of mitochondrial Stat3 in cellular respiration. Science 2009, 323, 793–797. [Google Scholar] [CrossRef]
- Shulga, N.; Pastorino, J.G. GRIM-19-mediated translocation of STAT3 to mitochondria is necessary for TNF-induced necroptosis. J. Cell Sci. 2012, 125, 2995–3003. [Google Scholar] [CrossRef]
- Simcox, E.M.; Reeve, A.K. An introduction to mitochondria, their structure and functions. In Mitochondrial Dysfunction in Neurodegenerative Disorders, 2nd ed.; Reeve, A.K., Simcox, E.M., Duchen, M.R., Turnbull, D.M., Eds.; Springer Nature: Cham, Switzerland, 2016; pp. 3–30. [Google Scholar] [CrossRef]
- Bélanger, M.; Allaman, I.; Magistretti, P.J. Brain energy metabolism: Focus on astrocyte-neuron metabolic cooperation. Cell Metab. 2011, 14, 724–738. [Google Scholar] [CrossRef]
- Hidalgo, C.; Arias-Cavieres, A. Calcium, reactive oxygen species, and synaptic plasticity. Physiology 2016, 31, 201–215. [Google Scholar] [CrossRef]
- Angelova, P.R.; Abramov, A.Y. Role of mitochondrial ROS in the brain: From physiology to neurodegeneration. FEBS Lett. 2018, 592, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Cha, M.; Lee, B.H. Neuroprotective Effect of Antioxidants in the Brain. Int. J. Mol. Sci. 2020, 21, 7152. [Google Scholar] [CrossRef]
- Jelinek, M.; Jurajda, M.; Duris, K. Oxidative Stress in the Brain: Basic Concepts and Treatment Strategies in Stroke. Antioxidants 2021, 10, 1886. [Google Scholar] [CrossRef]
- Levings, D.C.; Pathak, S.S.; Yang, Y.M.; Slattery, M. Limited expression of Nrf2 in neurons across the central nervous system. Redox Biol. 2023, 65, 102830. [Google Scholar] [CrossRef]
- Yang, T.; Sun, Y.; Li, Q.; Li, S.; Shi, Y.; Leak, R.K.; Chen, J.; Zhang, F. Ischemic preconditioning provides long-lasting neuroprotection against ischemic stroke: The role of Nrf2. Exp. Neurol. 2020, 325, 113142. [Google Scholar] [CrossRef]
- Baxter, P.S.; Hardingham, G.E. Adaptive regulation of the brain’s antioxidant defenses by neurons and astrocytes. Free Radic. Biol. Med. 2016, 100, 147–152. [Google Scholar] [CrossRef]
- Chen, Y.; Qin, C.; Huang, J.; Tang, X.; Liu, C.; Huang, K.; Xu, J.; Guo, G.; Tong, A.; Zhou, L. The role of astrocytes in oxidative stress of central nervous system: A mixed blessing. Cell Prolif. 2020, 53, e12781. [Google Scholar] [CrossRef]
- DeSai, C.; Hays Shapshak, A. Cerebral Ischemia. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2024; pp. 2–26. Available online: https://www.ncbi.nlm.nih.gov/books/NBK560510/ (accessed on 8 January 2024).
- Lin, L.; Wang, X.; Yu, Z. Ischemia-reperfusion Injury in the Brain: Mechanisms and Potential Therapeutic Strategies. Biochem. Pharmacol. 2016, 5, 213. [Google Scholar] [CrossRef]
- Li, S.; Hafeez, A.; Noorulla, F.; Geng, X.; Shao, G.; Ren, C.; Lu, G.; Zhao, H.; Ding, Y.; Ji, X. Preconditioning in neuroprotection: From hypoxia to ischemia. Prog. Neurobiol. 2017, 157, 79–91. [Google Scholar] [CrossRef]
- Liu, X.Q.; Sheng, R.; Qin, Z.H. The neuroprotective mechanism of brain ischemic preconditioning. Acta Pharmacol. Sin. 2009, 30, 1071–1080. [Google Scholar] [CrossRef]
- Rodriguez, C.; Agulla, J.; Delgado-Esteban, M. Refocusing the Brain: New Approaches in Neuroprotection Against Ischemic Injury. Neurochem. Res. 2021, 46, 51–63. [Google Scholar] [CrossRef]
- Pulsinelli, W.A.; Brierley, J.B. A new model of bilateral hemispheric ischemia in the unanesthetized rat. Stroke 1979, 10, 267–272. [Google Scholar] [CrossRef]
- Dodd, P.R.; Hardy, J.A.; Oakley, A.E.; Edwardson, J.A.; Perry, E.K.; Delaunoy, J.P.A. rapid method for preparing synaptosomes:comparison with alternative procedures. Brain Res. 1981, 226, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Tatarkova, Z.; Engler, I.; Calkovska, A.; Mokra, D.; Drgova, A.; Hodas, P.; Lehotsky, J.; Dobrota, D.; Kaplan, P. Effect of long-term normobaric hyperoxia on oxidative stress in mitochondria of the guinea pig brain. Neurochem. Res. 2011, 36, 1475–1481. [Google Scholar] [CrossRef]
- Cocco, T.; Sgobbo, P.; Clemente, M.; Lopriore, B.; Grattagliano, I.; Di Paola, M.; Villani, G. Tissue-specific changes of mitochondrial functions in aged rats: Effect of long-term dietary treatment with N-acetylcysteine. Free Radic. Biol. Med. 2005, 38, 796–805. [Google Scholar] [CrossRef] [PubMed]
- Paxinos, G.; Watson, C. The Rat Brain in Stereotaxic Coordinates: Hard Cover Edition, 6th ed.; Academic Press: London, UK, 2006; p. 456. Available online: https://shop.elsevier.com/books/the-rat-brain-in-stereotaxic-coordinates/paxinos/978-0-12-374121-9 (accessed on 7 March 2024).
- Ma, R.; Xie, Q.; Li, Y.; Chen, Z.; Ren, M.; Chen, H.; Li, H.; Li, J.; Wang, J. Animal models of cerebral ischemia: A review. Biomed. Pharmacother. 2020, 131, 110686. [Google Scholar] [CrossRef] [PubMed]
- Onose, G.; Anghelescu, A.; Blendea, D.; Ciobanu, V.; Daia, C.; Firan, F.C.; Oprea, M.; Spinu, A.; Popescu, C.; Ionescu, A.; et al. Cellular and Molecular Targets for Non-Invasive, Non-Pharmacological Therapeutic/Rehabilitative Interventions in Acute Ischemic Stroke. Int. J. Mol. Sci. 2022, 23, 907. [Google Scholar] [CrossRef]
- Roque, C. Cellular and Molecular Effects of Ischemia on Brain Cells. In Reperfusion Injuries-Advances in Understanding, Prevention, Treatment; Jerez, Z.M., Peterson, R., Eds.; IntechOpen: Rijeka, Croatia, 2023. [Google Scholar] [CrossRef]
- Hausenloy, D.J.; Yellon, D.M. The therapeutic potential of ischemic conditioning: An update. Nat. Rev. Cardiol. 2011, 8, 619–629. [Google Scholar] [CrossRef] [PubMed]
- Kardesoglu, E.; Isilak, Z.; Uz, O.; Yiginer, O. Ischemic conditioning: A current concept in reducing reperfusion injury. Chin. Med. J. (Eng.) 2011, 124, 480. [Google Scholar]
- Wen, M.; Jin, Y.; Zhang, H.; Sun, X.; Kuai, Y.; Tan, W. Proteomic Analysis of Rat Cerebral Cortex in the Subacute to Long-Term Phases of Focal Cerebral IR Injury. J. Proteome Res. 2019, 18, 3099–3118. [Google Scholar] [CrossRef]
- Ding, Z.M.; Wu, B.; Zhang, W.Q.; Lu, X.J.; Lin, Y.C.; Geng, Y.J.; Miao, Y.F. Neuroprotective effects of ischemic preconditioning and postconditioning on global brain ischemia in rats through the same effect on inhibition of apoptosis. Int. J. Mol. Sci. 2012, 13, 6089–6101. [Google Scholar] [CrossRef]
- Cho, Y.S.; Cho, J.H.; Shin, B.N.; Cho, G.S.; Kim, I.H.; Park, J.H.; Ahn, J.H.; Ohk, T.G.; Cho, B.R.; Kim, Y.M.; et al. Ischemic preconditioning maintains the immunoreactivities of glucokinase and glucokinase regulatory protein in neurons of the gerbil hippocampal CA1 region following transient cerebral ischemia. Mol. Med. Rep. 2015, 12, 4939–4946. [Google Scholar] [CrossRef]
- Koch, S.; Gonzalez, N. Preconditioning the human brain: Proving the principle in subarachnoid hemorrhage. Stroke 2013, 44, 1748–1753. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sanada, S.; Komuro, I.; Kitakaze, M. Pathophysiology of myocardial reperfusion injury: Preconditioning, postconditioning, and translational aspects of protective measures. Am. J. Physiol. Heart Circ. Physiol. 2011, 301, H1723–H1741. [Google Scholar] [CrossRef]
- Hausenloy, D.J.; Yellon, D.M. Ischaemic conditioning, and reperfusion injury. Nat. Rev. Cardiol. 2016, 13, 193–209. [Google Scholar] [CrossRef]
- Minamino, T. Cardioprotection from ischemia/reperfusion injury: Basic and translational research. Circ. J. 2012, 76, 1074–1082. [Google Scholar] [CrossRef] [PubMed]
- Niedzielska, E.; Smaga, I.; Gawlik, M.; Moniczewski, A.; Stankowicz, P.; Pera, J.; Filip, M. Oxidative stress in neurodegenerative diseases. Mol. Neurobiol. 2016, 53, 4094–4125. [Google Scholar] [CrossRef] [PubMed]
- Federico, A.; Cardaioli, E.; Da Pozzo, P.; Formichi, P.; Gallus, G.N.; Radi, E. Mitochondria, oxidative stress and neurodegeneration. J. Neurol. Sci. 2012, 322, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Scarian, E.; Viola, C.; Dragoni, F.; Di Gerlando, R.; Rizzo, B.; Diamanti, L.; Gagliardi, S.; Bordoni, M.; Pansarasa, O. New Insights into Oxidative Stress and Inflammatory Response in Neurodegenerative Diseases. Int. J. Mol. Sci. 2024, 25, 2698. [Google Scholar] [CrossRef] [PubMed]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014, 94, 909–950. [Google Scholar] [CrossRef]
- Patergnani, S.; Bouhamida, E.; Leo, S.; Pinton, P.; Rimessi, A. Mitochondrial Oxidative Stress and “Mito-Inflammation”: Actors in the Diseases. Biomedicines 2021, 9, 216. [Google Scholar] [CrossRef]
- Zheng, F.; Zhou, Y.T.; Zeng, Y.F.; Liu, T.; Yang, Z.Y.; Tang, T.; Luo, J.K.; Wang, Y. Proteomics Analysis of Brain Tissue in a Rat Model of Ischemic Stroke in the Acute Phase. Front. Mol. Neurosci. 2020, 13, 27. [Google Scholar] [CrossRef] [PubMed]
- Puisieux, F.; Deplanque, D.; Bulckaen, H.; Ma-boudou, P.; Gelé, P.; Lhermitte, M.; Lebuffe, G.; Bordet, R. Brain ischemic preconditioning is abolished by antioxidant drugs but does not up-regulate superoxide dismutase and glutathione peroxidase. Brain Res. 2004, 1027, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Park, J.H.; Kim, I.H.; Cho, G.S.; Ahn, J.H.; Tae, H.J.; Choi, S.Y.; Cho, J.H.; Kim, D.W.; Kwon, Y.G.; et al. Neuroprotection of ischemic preconditioning is mediated by thioredoxin 2 in the hippocampal CA1 region following a subsequent transient cerebral ischemia. Brain Pathol. 2017, 27, 276–291. [Google Scholar] [CrossRef]
- Reaume, A.G.; Elliott, J.L.; Hoffman, E.K.; Kowall, N.W.; Ferrante, R.J.; Siwek, D.F.; Wilcox, H.M.; Flood, D.G.; Beal, M.F.; Brown, R.H., Jr.; et al. Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat. Genet. 1996, 13, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Surmeli, N.B.; Litterman, N.K.; Miller, A.F.; Groves, J.T. Peroxynitrite mediates active site tyrosine nitration in manganese superoxide dismutase. Evidence of a role for the carbonate radical anion. J. Am. Chem. Soc. 2010, 132, 17174–17185. [Google Scholar] [CrossRef] [PubMed]
- Demicheli, V.; Moreno, D.M.; Radi, R. Human Mn-superoxide dismutase inactivation by peroxynitrite: A paradigm of metal-catalyzed tyrosine nitration in vitro and in vivo. Metallomics 2018, 10, 679–695. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Kukreti, R.; Saso, L.; Kukreti, S. Oxidative Stress: A Key Modulator in Neurodegenerative Diseases. Molecules 2019, 24, 1583. [Google Scholar] [CrossRef] [PubMed]
- Kaludercic, N.; Giorgio, V. The Dual Function of Reactive Oxygen/Nitrogen Species in Bioenergetics and Cell Death: The Role of ATP Synthase. Oxid. Med. Cell Longev. 2016, 2016, 3869610. [Google Scholar] [CrossRef] [PubMed]
- Szeliga, M. Peroxiredoxins in Neurodegenerative Diseases. Antioxidants 2020, 9, 1203. [Google Scholar] [CrossRef]
- Monteiro, G.; Horta, B.B.; Pimenta, D.C.; Augusto, O.; Netto, L.E. Reduction of 1-Cys peroxiredoxins by ascorbate changes the thiol-specific antioxidant paradigm, revealing another function of vitamin C. Proc. Natl. Acad. Sci. USA 2007, 104, 4886–4891. [Google Scholar] [CrossRef]
- Jang, H.H.; Lee, K.O.; Chi, Y.H.; Jung, B.G.; Park, S.K.; Park, J.H.; Lee, J.R.; Lee, S.S.; Moon, J.C.; Yun, J.W.; et al. Two Enzymes in One. Cell 2004, 117, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Jang, H.H. Role of Cytosolic 2-Cys Prx1 and Prx2 in Redox Signaling. Antioxidants 2019, 8, 169. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.G.; Woo, H.A.; Kil, I.S.; Bae, S.H. Peroxiredoxin functions as a peroxidase and a regulator and sensor of local peroxides. J. Biol. Chem. 2012, 287, 4403–4410. [Google Scholar] [CrossRef]
- Goemaere, J.; Knoops, B. Peroxiredoxin distribution in the mouse brain with emphasis on neuronal populations affected in neurodegenerative disorders. J. Comp. Neurol. 2011, 520, 258–280. [Google Scholar] [CrossRef]
- Zhu, H.; Santo, A.; Li, Y. The antioxidant enzyme peroxiredoxin and its protective role in neurological disorders. Exp. Biol. Med. 2012, 237, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Pluta, R.; Ułamek, M.; Jabłoński, M. Alzheimer’s mechanisms in ischemic brain degeneration. Anat. Rec. 2009, 292, 1863–1881. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, T.; Hata, R.; Kondo, T.; Takenaka, S. Proteomic analysis of the hippocampus in naïve and ischemic-preconditioned rat. J. Neurol. Sci. 2015, 358, 158–171. [Google Scholar] [CrossRef]
- Lee, Y.M.; Park, S.H.; Shin, D.I.; Hwang, J.Y.; Park, B.; Park, Y.J.; Lee, T.H.; Chae, H.Z.; Jin, B.K.; Oh, T.H.; et al. Oxidative modification of peroxiredoxin is associated with drug-induced apoptotic signaling in experimental models of Parkinson disease. J. Biol. Chem. 2008, 283, 9986–9998. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.S.; Lee, Y.S.; Kim, D.H.; Oh, G.T.; Jeon, W.K.; Han, J.S. Peroxiredoxin 2 deletion impairs hippocampal-dependent memory via exacerbating transient ischemia-induced oxidative damage. Brain Res. Bull. 2022, 184, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Qu, D.; Rashidian, J.; Mount, M.P.; Aleyasin, H.; Parsanejad, M.; Lira, A.; Haque, E.; Zhang, Y.; Callaghan, S.; Daigle, M.; et al. Role of Cdk5-mediated phosphorylation of Prx2 in MPTP toxicity and Parkinson’s disease. Neuron. 2007, 55, 37–52. [Google Scholar] [CrossRef]
- Gan, Y.; Ji, X.; Hu, X.; Luo, Y.; Zhang, L.; Li, P.; Liu, X.; Yan, F.; Vosler, P.; Gao, Y.; et al. Transgenic overexpression of peroxiredoxin-2 attenuates ischemic neuronal injury via suppression of a redox-sensitive pro-death signaling pathway. Antioxid. Redox Signal. 2012, 17, 719–732. [Google Scholar] [CrossRef]
- Park, J.; Choi, H.; Kim, B.; Chae, U.; Gil Lee, D.; Lee, S.-R.; Lee, H.J.; Lee, H.-S.; Lee, H.J. Peroxiredoxin 5 (Prx5) decreases LPS-induced microglial activation through regulation of Ca2+/calcineurin-Drp1-dependent mitochondrial fission. Free Radic. Biol. Med. 2016, 99, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Sekeljic, V.; Bataveljic, D.; Stamenkovic, S.; Ułamek, M.; Jabłoński, M.; Radenovic, L.; Pluta, R.; Andjus, P.R. Cellular markers of neuroinflammation and neurogenesis after ischemic brain injury in the long-term survival rat model. Brain Struct. Funct. 2012, 217, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Radenovic, L.; Nenadic, M.; Ułamek-Kozioł, M.; Januszewski, S.; Czuczwar, S.J.; Andjus, P.R.; Pluta, R. Heterogeneity in brain distribution of activated microglia and astrocytes in a rat ischemic model of Alzheimer’s disease after 2 years of survival. Aging 2020, 12, 12251–12267. [Google Scholar] [CrossRef] [PubMed]
- Power, J.H.T.; Asad, S.; Chataway, T.K.; Chegini, F.; Manavis, J.; Temlett, J.A.; Jensen, P.H.; Blumbergs, P.C.; Gai, W.-P. Peroxiredoxin 6 in human brain: Molecular forms, cellular distribution and association with Alzheimer’s disease pathology. Acta Neuropathol. 2008, 115, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Manevich, Y.; Hutchens, S.; Halushka, P.; Tew, K.; Townsend, D.M.; Jauch, E.; Borg, K. Peroxiredoxin VI oxidation in cerebrospinal fluid correlates with traumatic brain injury outcome. Free Radic. Biol. Med. 2014, 72, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.Y.; Zhou, X.L.; Wang, X.Y.; Liang, J.; Xue, Q. Peroxiredoxin-6 Released by Astrocytes Contributes to Neuroapoptosis During Ischemia. Neuroscience 2023, 512, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Yun, H.-M.; Choi, D.Y.; Oh, K.W.; Hong, J.T. PRDX6 Exacerbates Dopaminergic Neurodegeneration in a MPTP Mouse Model of Parkinson’s Disease. Mol. Neurobiol. 2014, 52, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Fisher, A.B. The phospholipase A2 activity of peroxiredoxin 6. J. Lipid Res. 2018, 59, 1132–1147. [Google Scholar] [CrossRef]
- Zhou, S.; Dodia, C.; Feinstein, S.I.; Harper, S.; Forman, H.J.; Speicher, D.W.; Fisher, A.B. Oxidation of Peroxiredoxin 6 in the Presence of GSH Increases its Phospholipase A₂ Activity at Cytoplasmic pH. Antioxidants 2018, 8, 4. [Google Scholar] [CrossRef]
- Chou, J.L.; Wu, C.H.; Tsai, C.Y.; Chang, A.Y.; Chan, S.H. Proteomic investigation of a neural substrate intimately related to brain death. Proteomics 2011, 11, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Dayon, L.; Turck, N.; Garcí-Berrocoso, T.; Walter, N.; Burkhard, P.R.; Vilalta, A.; Sahuquillo, J.; Montaner, J.; Sanchez, J.C. Brain extracellular fluid protein changes in acute stroke patients. J. Proteome Res. 2011, 10, 1043–1051. [Google Scholar] [CrossRef] [PubMed]
- Pankiewicz, J.E.; Diaz, J.R.; Martá-Ariza, M.; Lizińczyk, A.M.; Franco, L.A.; Sadowski, M.J. Peroxiredoxin 6 mediates protective function of astrocytes in Aβ proteostasis. Mol. Neurodegener. 2020, 15, 50. [Google Scholar] [CrossRef]
- Ma, S.; Zhang, X.; Zheng, L.; Li, Z.; Zhao, X.; Lai, W.; Shen, H.; Lv, J.; Yang, G.; Wang, Q.; et al. Peroxiredoxin 6 Is a Crucial Factor in the Initial Step of Mitochondrial Clearance and Is Upstream of the PINK1-Parkin Pathway. Antioxid. Redox Signal. 2016, 24, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Yoo, K.Y.; Park, O.K.; Yu, J.; Yan, B.; Li, H.; Lee, C.H.; Choi, J.H.; Kim, D.W.; Hwang, I.K.; Won, M.H. Expression and changes of hyperoxidized peroxiredoxins in non-pyramidal and polymorphic cells in the gerbil hippocampus during normal aging. Cell. Mol. Neurobiol. 2009, 29, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.K.; Yoo, K.Y.; Kim, D.W.; Choi, J.H.; Lee, I.S.; Won, M.H. Hyperoxidized peroxiredoxins and glyceraldehyde-3-phosphate dehydrogenase immunoreactivity and protein levels are changed in the gerbil hippocampal CA1 region after transient forebrain ischemia. Neurochem. Res. 2007, 32, 1530–1538. [Google Scholar] [CrossRef]



| Antioxidant/Group | CON | IR | IPC |
|---|---|---|---|
| SOD (U/mg protein) | 65.59 ± 0.94 | 66.12 ± 0.40 | 66.52 ± 1.07 |
| GR (μmol/min/mg protein) | 3.83 ± 1.99 | 5.51 ± 1.18 | 14.05 ± 1.61 *** |
| GPx (μmol/min/mg protein) | 0.15 ± 0.03 | 0.19 ± 0.03 | 0.18 ± 0.02 |
| Total glutathione (nmol/mg protein) | 106.57 ± 8.57 | 103.19 ± 46.04 | 88.65 ± 20.72 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lysikova, T.; Tomascova, A.; Kovalska, M.; Lehotsky, J.; Leskova Majdova, K.; Kaplan, P.; Tatarkova, Z. Dynamics in Redox-Active Molecules Following Ischemic Preconditioning in the Brain. Neurol. Int. 2024, 16, 533-550. https://doi.org/10.3390/neurolint16030040
Lysikova T, Tomascova A, Kovalska M, Lehotsky J, Leskova Majdova K, Kaplan P, Tatarkova Z. Dynamics in Redox-Active Molecules Following Ischemic Preconditioning in the Brain. Neurology International. 2024; 16(3):533-550. https://doi.org/10.3390/neurolint16030040
Chicago/Turabian StyleLysikova, Terezia, Anna Tomascova, Maria Kovalska, Jan Lehotsky, Katarina Leskova Majdova, Peter Kaplan, and Zuzana Tatarkova. 2024. "Dynamics in Redox-Active Molecules Following Ischemic Preconditioning in the Brain" Neurology International 16, no. 3: 533-550. https://doi.org/10.3390/neurolint16030040
APA StyleLysikova, T., Tomascova, A., Kovalska, M., Lehotsky, J., Leskova Majdova, K., Kaplan, P., & Tatarkova, Z. (2024). Dynamics in Redox-Active Molecules Following Ischemic Preconditioning in the Brain. Neurology International, 16(3), 533-550. https://doi.org/10.3390/neurolint16030040






