Abstract
Patients affected by familial hypercholesterolemia possess elevated low-density lipoprotein cholesterol and therefore have greater risk for cardiovascular disease. About 90% of familial hypercholesterolemia cases are associated with aberrant LDLR. Over 3500 LDLR variants have been identified, 15% of which are considered “pathogenic.” Given the genetic diversity of LDLR variants, specific variants rarely receive attention. However, investigators have proposed the critical evaluation of individual variants as a method to clarify knowledge and to resolve discrepancies in the literature. This article reviews p.(Val429Met) (rs28942078) in the areas of pathology, epidemiology, lipid-lowering therapy, and genetic testing. The p.(Val429Met) variant is associated with a missense point substitution in exon 9 of chromosome 19. Biochemical studies have found severely reduced low-density lipoprotein receptor protein in autologous and heterologous expression systems. Additionally, there are inconsistencies regarding the functional classification of p.(Val429Met). Considered to be of European origin, p.(Val429Met) is found in extant populations due to founder effects. Evidence from clinical trials have also demonstrated variable responses to newer lipid-lowering therapies in patients with a p.(Val429Met) variant. Proper clinical detection and adequate genetic testing have been shown to greatly improve outcomes. Future research may be aimed at resolving discrepancies to better comprehend the implications of familial hypercholesterolemia.
1. Introduction
First described in 1938, familial hypercholesterolemia (FH) is a genetic disorder of cholesterol metabolism [1]. FH is caused by defective alleles in three regulatory genes (LDLR, ApoB, PCSK9). There are two disease states in FH. The most common is heterozygous familial hypercholesterolemia (HeFH), which is caused by one aberrant allele in the affected gene, whereas homozygous familial hypercholesterolemia (HoFH) is caused by two aberrant alleles in affected genes. Current diagnostic methods include physical examination, lipid-paneling, genetic testing, and cascade screening of family members [2]. It is estimated that HeFH occurs in 1 in 311 in the general population, while HoFH occurs in upwards of 1 in 300,000 in the general population [3,4]. Globally, FH is largely underdiagnosed and undertreated [5].
Variants in LDLR, the gene that encodes the low-density lipoprotein receptor (LDLR), are most associated with FH. It is estimated that around 90% of FH cases implicate variant LDLR [2]. As of August 2023, ClinVar (National Center for Biotechnology Information; NCBI) has identified more than 3500 LDLR variants, 15% of which are identified as “pathogenic,” based on guidelines set by the American College of Medical Genetics and Genomics (ACMG) [6]. Impaired LDLR limits the hepatic uptake of low-density lipoprotein cholesterol (LDL-C) from the bloodstream. Consequently, LDL-C is elevated by as much as two- to five-fold in FH patients, posing a serious risk for premature cardiovascular disease (CVD) [2,3]. Moreover, first-line statin therapy is ineffective at lowering LDL-C in FH patients due to statins’ dependence on functional LDLR. Fortunately, newer second-line therapies target alternative proteins involved in this pathway.
Given the diversity of LDLR variants, it is rare for a single variant to receive attention. However, critical evaluation of specific variants has been proposed as a method to clarify knowledge and to resolve discrepancies in the literature [7]. Additionally, focus on a single variant may expand the understanding of the functional class to which it belongs.
In this review, the LDLR variant of interest is p.(Val429Met) (p.V429M) (rs28942078), which has been associated with a severe form of FH. The function of p.V429M has been studied extensively. Investigators have also identified the variant globally and in ethnically diverse populations. This article reviews p.V429M in the areas of pathology, epidemiology, lipid-lowering therapy, and genetic testing as well as identifies disagreements in the literature.
2. Pathology
The p.V429M variant is associated with a missense point substitution in exon 9 of chromosome 19. In 1989, Leitersdorf et al. [8] first identified p.V429M in a population of Afrikaners. DNA sequencing revealed that adenine replaced guanine at nucleotide 1285 (1285G>A). As a result, methionine (Met, M) replaced valine (Val, V) at amino acid 429. Later studies by Pereira et al. (1995) [9] and by Fouchier et al. (2001) [10] confirmed the presence of p.V429M in Cuban and Dutch populations, respectively. The investigators all identified p.V429M as a cause of HoFH in the populations studied. Early on, genotype-phenotype relationships established p.V429M as a FH-associated variant of LDLR.
It is important to note that the nomenclature of this variant is inconsistent across the literature. The p.V429M variant was known as p.V408M when first discovered because the −21 position was labeled as the initiation codon [8,11]. Later, the initiation codon was revised to be at the +1 position [11]. Despite the change, studies from as late as 2018 still referred to the variant as p.V408M [12]. This disagreement may cause p.V429M and p.V408M to be misidentified as separate variants. Regardless, both refer to the same NCBI reference sequence: rs28942078. Furthermore, the allelic substitution of interest corresponds to transcript variant 1: NM_000527.5 [13]. In this review, p.(Val429Met) refers to rs28942078 but is abbreviated as p.V429M to follow recommendations from the Human Genome Variation Society [14].
2.1. Assessment of Disease
Evidence from biochemical assays have linked the p.V429M variant to significantly reduced concentrations of LDLR. In the first study of p.V429M, Leitersdorf et al. [8] transfected LDLR-naïve Chinese hamster ovary (CHO) cells with a plasmid that contained the p.V429M sequence. Immunoprecipitation followed by densitometry revealed that only 13% of LDLR remained after six hours of expression. A later study by Strøm et al. [15] measured the degradation of LDLR by serial immunoblotting. In that study, the investigators collected Western blots from p.V429M and wild-type expressing CHO cells. This method quantified LDLR expression by protein band opacity. After eight hours of expression, minimal LDLR was present on blots obtained from p.V429M expressing cells, compared to the amount found on blots obtained from wild-type expressing cells. Additional functional studies performed by Ranheim et al. [16] and Dušková et al. [17] found undetectable amounts of LDLR on the plasma membrane of p.V429M expressing CHO cells. As such, the p.V429M variant is classified as a “negative” or “null” variant because it produces LDLR with less than 2% of normal activity [18,19]. Overall, the literature agrees that p.V429M is associated with FH because of dysfunctional LDLR.
Elevated LDL-C is an independent risk factor for CVD [20]. As a result of reduced LDL-C uptake, patients with at least one p.V429M allele possess greater risk for CVD. A study by Umans-Eckenhausen et al. [21] found that carriers of p.V429M had a significant relative risk for CVD compared to noncarrier relatives (RR: 7.01; 95% CI: 2.18–22.59; p < 0.003). A similar study by Huijgen et al. [22] found that carriers of any FH-associated variant had a higher hazard ratio for CVD when compared to noncarrier relatives (HR: 3.64; 95% CI: 3.24–4.08; p < 0.001). Furthermore, a study by Bertolini et al. [23] found that the prevalence of CVD in heterozygotes with null variants was significantly greater than in heterozygotes with defective variants (i.e., variants with greater than 2% LDLR activity) (40.7% vs. 25.8%; p = 0.0001). The latter two studies did not evaluate p.V429M individually. However, both found that FH-associated variants significantly increased the risk for CVD. Additionally, the risk for CVD may have been underestimated because patients with FH-associated variants were more likely to receive lipid-lowering therapy [22]. Ultimately, carriers of p.V429M are at a potentially greater risk for CVD.
As described previously, biochemical assays are the standard method for functional determination. However, there is variation in the potential sources of DNA samples. Across multiple studies, variant p.V429M DNA has been isolated from skin fibroblasts, blood leukocytes, and saliva swabs [8,10,23,24,25]. Additionally, there is variation between heterologous and autologous expression systems. Heterologous expression systems have included ldlA-7 CHO (LDLR-naive) cells, tetracycline-induced CHO (T-Rex) cells, and human HepG2 cells [8,15,16,17]. Autologous expression systems have included human lymphocytes and skin fibroblasts [8,12,23]. Although saliva samples are easier to collect, there is evidence that blood samples provide more accurate copy number variant detection [26]. The choice of CHO cells or HepG2 cells is also debated. For instance, Banerjee et al. [27] noted that transfected CHO cells allowed for complete observation of the variant LDLR phenotype, while expression in wild-type HepG2 cells created a less severe heterozygous condition. Alternatively, Ranheim et al. [16] found that HepG2 cells better mimicked the in vivo environment due to presence of intracellular LDLR regulators, such as proprotein convertase subtilisin/kexin type 9 (PCSK9). In all, a variety of methods have been used to study the function and clinical consequences of p.V429M.
2.2. Variant Classification
In 1992, Hobbs et al. [19] described five functional classes of LDLR variants. Originally, p.V429M belonged to Class 5, defects in LDLR recycling. Early in vitro evidence of rapid LDLR degradation supported this classification [8,15,19]. Later studies identified defects in the epidermal growth factor (EGF) homology domain that were associated with poor recycling.
In Class 5 variants, impacted EGF homology domains fail to release LDL-C in the endosome [19]. An early study by Davis et al. [28] found that a conformational change that was required to release lipoprotein in acidic pH (i.e., the endosome) did not occur in variant LDLR with a deleted EGF homology domain. A later study by Rudenko et al. [29] identified the folding of the six-bladed β-propeller as the previously unknown conformational change. The EGF homology domain and the β-propeller are proximal to each other in the basic LDLR structure [30]. Thus, defects in the EGF homology domain spread to the β-propeller, affecting its release mechanism. In summary, failed release of LDL-C causes the LDLR complex to be completely digested by the endosome, preventing LDLR from being reused.
Recent evidence has supported the p.V429M variant as Class 2, defects in intracellular trafficking [19]. In 2020, Dušková et al. [17] found that p.V429M CHO cells accumulated immature LDLR in the endoplasmic reticulum (ER), as well as overexpressed stress-induced ER chaperonins. As a result, p.V429M cells failed to process LDLR into its mature form at the Golgi apparatus, which led to no LDL-C uptake at the plasma membrane. This finding conflicted with previous knowledge because it alternatively explained the lack of LDLR on the plasma membrane. As opposed to the degradation of matured LDLR, this proposed that LDLR never matured to begin with.
Interestingly, prior studies alluded to impaired intracellular processing. Although Leitersdorf et al. [8] concluded that p.V429M induced rapid degradation of LDLR, the investigators also identified the 120 kD (immature) and 160 kD (mature) forms within the immunoprecipitated LDLR. Additionally, Bertolini et al. [23] found the weight of LDLR in skin fibroblasts from p.V429M-affected individuals to range between 117.15 and 149.21 kD. Throughout its study, p.V429M has been associated with immature LDLR.
Furthermore, prior studies found that p.V429M deviated from other Class 5 variants. In a study that compared LDLR variants from different classes, Ranheim et al. [16] found that p.V429M had an LDL-C staining pattern that was similar to p.Gly565Val (p.G565V), a Class 2 variant. Additionally, measures of LDL-C uptake, binding, and degradation were most similar between these two variants. Notably, the investigators acknowledged the potential for misclassification but still assigned p.V429M to Class 5 because they observed some LDLR in the endosome, as evidenced by confocal laser imaging. In another study that only compared Class 5 variants, Strøm et al. [15] found that p.V429M had remarkably reduced LDLR when compared to p.Glu387Lys (p.E387K). Like Ranheim et al. [16], Strøm et al. [15] acknowledged the parallels between Class 2 and Class 5 variants but did not offer any reclassification. Due to the similarities between Class 2 and Class 5 variants, there is a crossover of evidence (Figure 1). This is not to say that impaired endosomal release and intracellular processing are mutually exclusive. It may be the case that p.V429M belongs in both classes due to the overlap between the EGF homology domain and the β-propeller. Alternatively, there may be more evidence favoring one class over another depending on the analytical methods used.
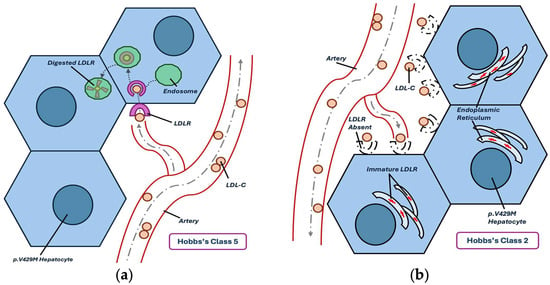
Figure 1.
Comparison of Hobbs’s functional classes, as relevant to p.V429M. (a) Visual representation of the effect of Class 5 LDLR variants; (b) visual representation of the effect of Class 2 LDLR variants.
In 2015, the ACMG published a five-tier classification system for genetic variants, regardless of disease. These guidelines classify variants as “pathogenic”, “likely pathogenic”, “uncertain significance”, “likely benign”, or “benign”. For pathogenic variants, the ACMG employs an algorithm that accounts for strength of evidence. Evidence may be considered “very strong”, “strong”, “moderate”, or “supporting”. For example, “very strong” evidence may be the presence of a known nonsense or frameshift variant, whereas “supporting” evidence may come from in silico analysis (e.g., SIFT, MutationTaster). An algorithm that combines evidence type and strength ultimately decides the variant’s pathogenicity [31]. Full description of individual ACMG criteria is beyond the scope of this paper. Instead, this discussion focuses on pathogenicity assessments that were reported to ClinVar.
According to ClinVar, 14 ACMG-guided assessments have been submitted for NM_000527.5. Moreover, “clinical testing” was the most common method for classification. Two submissions classified p.V429M as “likely pathogenic,” whereas the remainder classified p.V429M as “pathogenic.” The most recent ACMG-guided submission is from April 2024. The comments for this report cited inconclusive in silico data, individual variant rarity, and functional studies that demonstrated less than 2% functional activity in affected LDLR. As such, p.V429M was classified as “pathogenic.” Notably, a submission from December 2016 cited similar evidence, and yet classified p.V429M as “likely pathogenic.” All submissions were rated one star out of a possible four, which indicated that these reviews were from single submitters, as opposed to an expert panel [32]. The ACMG admits that there is no concrete method to quantify a “likely” rating, although they advise laboratories to use the term if there is 95% confidence of pathogenicity [31]. In general, these submissions somewhat conflict from lack of detail and source subjectivity.
The ACMG guidelines are more comprehensive than the classifications proposed by Hobbs. Nonetheless, the ACMG still regards validated functional studies as “strong” evidence for pathogenicity. This provides an opportunity to apply the antiquated functional classes to current practice. Previously, Chora et al. [33] performed a systematic review of FH-associated variants in the context of ACMG guidelines. The investigators found that functional studies were not widely performed on FH-associated variants. Additionally, they concluded that functional studies require more influence on pathogenicity determination than the ACMG currently assigns. This field may benefit from revised ACMG criteria that distinctly addresses unique factors of FH.
2.3. Additional Factors
Due to the presence of the wild-type allele in heterozygous patients, a less severe phenotype is observed. A study by Thedrez et al. [12] found that LDLR expression was two- to four-fold lower in heterozygous cells and three- to five-fold lower in homozygous cells with an FH-associated variant, compared to wild-type lymphocytes. Moreover, Ranheim et al. [16] and Chora et al. [33] proposed that homozygous patients with an FH-associated variant provide the best opportunity for study, as the wild-type allele cannot interfere. However, patients with the homozygous condition are very rare. As such, study in heterologous cells, such as LDLR-naïve CHO cells, is a popular alternative [8,17,33]. No matter the zygosity, the presence of one p.V429M allele is associated with markedly reduced LDLR on the plasma membrane [12]. Furthermore, the p.V429M variant appears to have an additive effect, as evidenced by the two-fold difference in LDLR concentration between heterozygous and homozygous lymphocytes. In sum, the heterozygous condition is one consideration that must be made when assessing phenotypes associated with LDLR variants.
Interestingly, acute infection from SARS-CoV-2 may reduce LDL-C to optimal levels in patients with an FH-associated variant. A case report by Bampatsias et al. [34] found that SARS-CoV-2 infection reduced LDL-C to 9 mg/dL in a HoFH patient with the p.V429M variant just 10 days after a positive test. After one month, LDL-C increased to 105 mg/dL on maximally tolerated lipid-lowering therapy, which consisted of rosuvastatin, ezetimibe, and lomitapide. The drastic decrease in LDL-C during acute infection may be attributed to the replication of SARS-CoV-2, which requires lipoprotein, whereas later increases in LDL-C may be attributed to impaired hepatic function caused by liver stiffness and steatosis following infection, in addition to the FH condition [35,36]. In a recent cohort study, general patients infected with SARS-CoV-2 had a 55.4% chance of having LDL-C levels greater than 130 mg/dL one year after infection (HR: 1.24; 95% CI: 1.20–1.29) [37]. Further research could elucidate the connections between lipid disorders and infectious disease as well as clarify the role of lipid disorders in the grand scheme of body systems.
There is some evidence that p.V429M is more harmful when inherited maternally. A study by Versmissen et al. [38] compared the standardized mortality ratio (SMR) between FH patients who inherited p.V429M maternally versus paternally. The SMR for maternal inheritance was significant (SMR: 2.49; 95% CI: 1.45–3.99; p = 0.001), whereas the SMR for paternal inheritance was insignificant (SMR: 1.30; 95% CI: 0.65–2.32; p = 0.234). Prior research found that a mother’s history of FH could induce aortic plaque formation in fetuses [39]. In this case, the risk for CVD is not due to differences in molecular architecture between maternally and paternally inherited p.V429M. Instead, epigenetic effects from the uterine environment explain the increased risk. In sum, maternal FH is a determinant for CVD prior to birth due to elevated LDL-C in mothers with identified FH-associated LDLR variants.
In addition to elevated LDL-C, elevated lipoprotein(a) (Lp(a)) is an independent risk factor for CVD [40]. Currently, the relationship between FH-associated variants and elevated Lp(a) is not resolved. An early study by Lingenhel et al. [41] found significantly elevated Lp(a) in carriers of FH-associated variants, including p.V429M, compared to unaffected relatives (p = 0.0047). However, a recent study by Marco-Benedi et al. [42] found that subjects without genetic hypercholesterolemia had greater mean Lp(a) levels than subjects with an LDLR variant (37.4 mg/dL vs. 21.7 mg/dL). Additionally, there was no significant difference between an LDLR variant’s functional class and associated Lp(a) concentration. Furthermore, a recent study by Medeiros et al. [43] found that the prevalence of elevated Lp(a), defined as >50 mg/dL, was significantly more prevalent in subjects without genetic hypercholesterolemia than in subjects with an FH-associated variant (38% vs. 32%; p = 0.035). Cohort heterogeneity may explain the conclusions of the latter two studies, as the polygenic variability (e.g., number of kringle IV2 repeats) between subjects could confound results [12], while a strength of Lingenhel et al. [41] was that analysis occurred in sibling pairs, which mitigated genetic differences. In all, conclusive research in this area is limited by the diverse genetic causes of hyper-Lp(a), as well as by differences in methodology.
3. Prevalence Across Populations
The p.V429M variant is one of only seven LDLR variants found on every populated continent [33]. Studies of FH-associated variants have identified populations in which p.V429M is more common. For example, Leitersdorf et al. [8] identified the p.V429M allele in 29.2% of LDLR samples from Afrikaners [8]. Additionally, Whitall et al. [44] identified p.V429M in 14.3% of Greek FH patients (n = 28). Despite the low sample size, an earlier study by Traeger-Synodinos et al. [45] identified p.V429M in a similar proportion of Greek FH patients (14.7%, n = 150). Furthermore, Pereira et al. [9] identified p.V429M in 10.7% of Cuban patients (n = 28). In an Asian population, Mak et al. [46] identified p.V429M in 6.7% of Chinese patients (n = 30). These results are not generalizable due to small population sizes. Additionally, the accuracy of prevalence estimates may be attenuated by varying clinical presentations, which impact rates of genetic testing. However, recognition of affected populations is important for awareness, which informs detection and treatment.
Other studies have found lower prevalences of p.V429M in certain European populations. For example, Huijgen et al. [11], Umans-Eckenhausen et al. [21], and Fouchier [10] identified p.V429M in 1.7% (n = 5467), 3.0% (n = 1695), and 3.1% (n = 1645) of Dutch FH patients, respectively. In addition, Bertolini et al. [23] identified p.V429M in 1.4% of Italian FH patients (n = 1018). Similarly, Brænne et al. [7] identified p.V429M in 1.6% of German FH patients (n = 255). These prevalences may be skewed by the characteristics of the populations sampled. For example, Brænne et al. included German patients with a history of premature myocardial infarction, a condition already associated with p.V429M [7], whereas Huijgen et al. sampled patients from a national cascade screening program in the Netherlands [11]. In all, there is a lack of standardized prevalence data across populations. For proper epidemiology, prevalence estimates may require data from each nation’s lipid clinic registry, proving a challenge in regions with low detection rates.
The p.V429M variant is thought to be of European origin [47]. As a result of global migration, a founder effect has been observed between related populations [8,46]. For instance, the prevalence of p.V429M in the Netherlands is between 1 and 3% [10,11,21], while the prevalence in Afrikaners approaches 30% [8]. This drastic difference indicates a greater concentration of p.V429M in the smaller Afrikaner population, as explained by the Dutch colonization of South Africa. A similar relationship can be seen between Spanish Caucasians and Cubans. For instance, Aparicio et al. [9] identified p.V429M in 6.0% of FH patients in mainland Spain (n = 182), compared to the 10.7% found in Havana [24]. Although the effect is observed to a lesser extent, this is an interesting finding within the context of genetic drift. The distinction between national identity and genetic identity is another important consideration, as nations with genetically heterogeneous populations may confound estimates. Additionally, allelic frequency may be a more comprehensive measure because it accounts for individual zygosity. Altogether, the p.V429M variant is of global interest due to its presence in diverse populations.
Despite the identification of populations in which p.V429M is more commonly found, small sample sizes and dated, single-country studies weaken the accuracy of prevalence estimates. More accurate allelic frequencies may be found in large, public genomic databases. For rs28942078, the global allelic frequencies are reported as 0.0013% in gnomAD v4.1.0 (n = 1614000) and as 0.0008334% in ExAC (n = 119988). In total, gnomAD reported 21 total alleles, whereas ExAC reported one allele. By genetic ancestry, European participants enrolled in gnomAD contributed the most alleles. The single allele identified in ExAC belonged to a South Asian participant [48,49]. Despite the contributions of these databases, they possess their own limitations. For instance, Chora et al. indicated a general lack of reporting outside of research studies. Additionally, most countries do not require reporting for FH variants [33]. There is great potential for these global databases. Yet, most countries do not have the infrastructure to realize this utility. As these databases currently stand, individual research studies provide more insight into prevalence, despite possessing significant limitations. Maintenance and synchronization of global prevalence data may fill these reporting gaps.
4. Studied Responses to Lipid-Lowering Therapy
Alirocumab (Praluent, Sanofi: Paris, France/Regeneron: Tarrytown, NY, USA) is a PCSK9 inhibitor. Overactive PCSK9 induces LDLR degradation. Therefore, inhibition of PCSK9 allows more LDLR for improved LDL-C clearance. ODYSSEY-HoFH, a randomized, placebo-controlled trial, investigated alirocumab as a lipid-lowering therapy in HoFH patients. Genetic testing identified three subjects who were homozygous for p.V429M. Notably, all p.V429M subjects had a prior history of CVD. In true homozygotes, baseline LDL-C ranged between 156 and 256 mg/dL. The primary endpoint was the percent reduction in LDL-C after 12 weeks, which ranged between −25.8% and +1.9%. All p.V429M homozygotes received 150 mg subcutaneous alirocumab every two weeks [50]. From this, a highly variable response to alirocumab was observed. Interestingly, the subject with the greatest LDL-C at baseline saw the 25.8% decrease, whereas the subject with lowest LDL-C at baseline saw the 1.9% increase [50]. Background lipid-lowering therapy may have not affected these results, as all subjects took concomitant rosuvastatin, ezetimibe, and lomitapide in consistent doses [50]. In p.V429M homozygotes, alirocumab did not reduce LDL-C by 50%, nor reduced LDL-C to less than 70 mg/dL, thereby underperforming American Heart Association (AHA) guidelines [51].
Evolocumab (Repatha, Amgen: Thousand Oaks, CA, USA) is another PCSK9 inhibitor. TAUSSIG, an open-label, single-arm trial, investigated evolocumab as a lipid-lowering therapy in HoFH patients. Genetic testing identified five subjects who were compound heterozygous for p.V429M and p.Asp206Glu (p.D296E), another FH-associated variant. Like Odyssey-HoFH, baseline LDL-C ranged between 173.2 and 273.9 mg/dL in these subjects. Percent reduction in LDL-C after 24 weeks ranged between −7% and −28%. All subjects received 420 mg subcutaneous evolocumab every two weeks for the duration of the trial [12,52]. As opposed to ODYSSEY-HoFH, the background lipid-lowering therapy that each subject took is not disclosed. Additionally, the investigators of TAUSSIG defined p.V429M as a “receptor-defective” variant, whereas the investigators of ODYSSEY-HoFH defined p.V429M as a “receptor-null” variant [12,50]. From this, clinical trials have inconsistently defined the function of p.V429M.
The functional definition of the variant is important because it guides the explanation of the lipid-lowering therapy’s efficacy. For example, “receptor-negative” (null) variants do not express enough functional LDLR for lipid-lowering therapy to have a benefit, as opposed to “receptor-defective” variants, which possess enough LDLR function for lipid-lowering therapy to reduce LDL-C [12]. The Leiden Open Variation Database (LOVD3) represents p.V429M as having less than 2% LDLR activity [53]. This agrees with the definition set by the investigators of ODYSSEY-HoFH [44]. If p.V429M were truly a “receptor-null” variant, the reductions in LDL-C observed in ODYSSEY-HoFH and TAUSSIG would not have occurred. This inconsistency may affect the reliability of data across different trials. Regardless of definition, however, alirocumab and evolocumab showed variable LDL-C reductions in HoFH subjects with at least one p.V429M variant.
Lomitapide (Juxtapid, Aegerion: Defunct 2019) inhibits the microsomal triglyceride transfer protein (MTP), which aids in the conversion of very low-density lipoprotein cholesterol (VLDL-C) to LDL-C. Therefore, inhibition of MTP reduces LDL-C concentration. A study by Kolovou et al. [54] investigated the efficacy of lomitapide in Indian HoFH patients. Genetic testing identified one p.V429M homozygote. This patient had a mean baseline LDL-C of 330 mg/dL. After 13 months of follow-up, this patient’s LDL-C decreased to 114 mg/dL (65% reduction). This patient received 10 mg lomitapide concomitant to maximally tolerated lipid-lowering therapy and apheresis treatment. Although the data on the efficacy of lipid-lowering therapy in p.V429M patients are scarce, this study found that lomitapide achieved a superior reduction in LDL-C. Furthermore, the investigators emphasized the influence of individual MTP variants on the efficacy of lipid-lowering therapy through genetic and environmental interactions [54]. Additionally, a cohort study that compared lomitapide to lipoprotein apheresis found that lomitapide significantly reduced the proportion of subjects who experienced major adverse cardiovascular events (13.3% vs. 55.2%; p < 0.001) [55], although in this study, none of the subjects possessed the p.V429M variant. The published LDL-C reductions, by subject, for alirocumab, evolocumab, and lomitapide are found in Table 1.

Table 1.
Summary of second-line LLT results.
Inclisiran (Leqvio, Novartis: Basel, Switzerland) is a small-interfering RNA (siRNA) that inhibits PCSK9. Inclsirian improves LDL-C metabolism by silencing the mRNA transcript that encodes the PCSK9 protein [56]. ORION-9 and ORION-5 were randomized, placebo-controlled trials that investigated the efficacy of inclisiran in HeFH and HoFH patients, respectively. Of note, the genotypes of individual subjects were not published. Therefore, the following results may not apply to subjects with the p.V429M variant. In HeFH patients (n = 482), inclisiran significantly reduced LDL-C by 39.7% after 24 months (between-group difference: 38.1%; 95% CI: −53.5% to −42.4%; p < 0.001) [57]. In HoFH patients (n = 56), however, inclisiran did not significantly reduce LDL-C (p = 0.90). The investigators of ORION-5 attributed the results to a small sample size and heterogeneity of FH genotypes [58]. An alternative explanation is that inclisiran was more effective for HeFH patients due to some functional LDLR activity. The study of inclisiran in HoFH patients, with consideration of individual genotypes, is an avenue for future research. Importantly, awareness of an individual LDLR variant’s function and its response to lipid-lowering therapy creates an opportunity for personalized medicine.
5. Genetic Testing
In addition to premature CVD, other common clinical signs of FH are arcus cornea and tendon xanthoma [19]. However, inconsistent phenotypic presentation limits diagnostic accuracy. For example, Pereira et al. observed a p.V429M heterozygous individual with premature CVD but without any history of arcus cornea nor tendon xanthoma [9]. Additionally, Brænne et al. [7] reported a pedigree of a German family affected by p.V429M, in which both children suffered premature myocardial infarction, as opposed to both parents who were unaffected. Reports such as these have demonstrated the incomplete penetrance of p.V429M.
Factors such as variable LDLR expression, polygenic mutations, and lifestyle may explain inconsistent clinical presentations. To illustrate, Thedrez et al. [12] found that maximal surface LDLR expression ranged from 113 to 195 among p.V429M compound heterozygotes. When extended to another FH-associated variant, p.Asp206Glu (p.D206E), maximal surface LDLR expression ranged from 184 to 607 in p.D206E homozygotes. Although p.V429M and p.D206E are separate variants, these results showed variable LDLR expression even within the same genotype. Variants in the PCSK9 and ApoB genes may also contribute to phenotypic variability, as these genes regulate LDLR activity [11,59]. Investigators have also attributed the variability to high-cholesterol diets observed across different regions [9,59]. Clinical features are important for recognizing index cases, but genetic testing is the most effective method for comprehensive diagnosis and treatment.
Cascade screening of p.V429M individuals has been reported in the literature since 2002 [21]. With this method, index cases of FH patients are identified, and the rest of the family is screened. Early on, cascade screening identified large yields of FH individuals. Genetic testing as part of cascade screening is cost-effective and has been correlated with better CVD risk detection and adherence to lipid-lowering therapy [24,59]. Although genetic testing is considered the best method for FH diagnosis, Reeskamp et al. [59] found that only 32.8% (n = 2320) of FH patients with a clinical diagnosis also had a confirmed genetic diagnosis. Genetic testing also allows for earlier detection and initiation of lipid-lowering therapy, which has been shown to significantly reduce the risk for CVD in FH patients [60].
Due to overreliance on clinical presentation and lack of provider knowledge, there have been reported cases of misidentified FH. For instance, Alnouri et al. [61] reported two HoFH patients who consulted dermatologists for tendon xanthomas. Both underwent surgical removal; however, the xanthomas eventually returned, and one patient suffered an ST-elevated myocardial infarction. After cardiology consultations, both patients received a clinical diagnosis of HoFH. Genetic testing revealed variants in the EGF-homology precursor domain, like p.V429M. In this case, lack of knowledge of FH across specialties led to a CVD event that may have been prevented if the condition was detected upon first observation.
A diagnosis of FH solely based on clinical features has also been shown to misclassify heterozygous and homozygous conditions. In an Italian cohort of clinically diagnosed HeFH patients, Bertolini et al. [23] found that 1.4% (n = 1018) truly possessed homozygous genotypes after genetic testing. Similarly, Chaudhry et al. [25] found that 0.9% (n = 705) of clinically diagnosed HeFH patients received a new diagnosis of true HoFH after genetic testing. Misdiagnosis attenuates care as CVD risk and treatment efficacy are different between HeFH and HoFH patients. Due to the diversity of FH-associated variants, genetic testing after clinical suspicion is a proper method for improved outcomes.
6. Conclusions
This review sought to determine the current knowledge of p.V429M. There are many qualities of this variant that may be resolved by future research, such as its functional classification, influence on other lipoprotein concentrations (e.g., lp(a)), and response to lipid-lowering therapy. Additionally, this review found that p.V429M experienced substantial migration throughout its history. Due to the individual rarity of LDLR variants, a significant limitation is the lack of standardized clinical and demographic data. All in all, this review brings awareness to the nuances of p.V429M, as well as identifies gaps in current reporting. This knowledge may be applied to other variants for greater comprehension of FH.
Author Contributions
Conceptualization, E.F.J. and M.S.K.; investigation, E.F.J.; writing—original draft preparation, E.F.J.; writing—review and editing, E.F.J. and M.S.K.; supervision, M.S.K.; project administration, E.F.J. and M.S.K. All authors have read and agreed to the published version of the manuscript.
Funding
Kindy is a Biomedical Laboratory Research & Development (BLR&D) Senior Research Career Scientist at the James A. Haley VA Medical Center.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
Yashwant Pathak and Kevin Sneed for their support from the Taneja College of Pharmacy at the University of South Florida, Tampa, Florida.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Müller, C. Xanthomata, Hypercholesterolemia, Angina Pectoris. Acta Med. Scand. 1938, 95, 75–84. [Google Scholar] [CrossRef]
- Brandts, R.; Ray, K.K. Familial Hypercholesterolemia. J. Am. Coll. Cardiol. 2021, 78, 1831–1843. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Dharmayat, K.I.; Stevens, C.A.; Sharabiani, M.T.; Jones, R.S.; Watts, G.F.; Genest, J.; Ray, K.K.; Vallejo-Vaz, A.J. Prevalence of Familial Hypercholesterolemia among the General Population and Patients with Atherosclerotic Cardiovascular Disease: A Systematic Review and Meta-Analysis. Circulation 2020, 141, 1742–1759. [Google Scholar] [CrossRef]
- Cuchel, M.; Bruckert, E.; Ginsberg, H.N.; Raal, F.J.; Santos, R.D.; Hegele, R.A.; Kuivenhoven, J.A.; Nordestgaard, B.G.; Descamps, O.S.; Steinhagen-Thiessen, E.; et al. Homozygous familial hypercholesterolaemia: New insights and guidance for clinicians to improve detection and clinical management. A position paper from the Consensus Panel on Familial Hypercholesterolaemia of the European Atherosclerosis Society. Eur. Heart J. 2014, 35, 2146–2157. [Google Scholar] [CrossRef]
- Nordestgaard, B.G.; Chapman, M.J.; Humphries, S.E.; Ginsberg, H.N.; Masana, L.; Descamps, O.S.; Wiklund, O.; Hegele, R.A.; Raal, F.J.; Defesche, J.C.; et al. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: Guidance for clinicians to prevent coronary heart disease: Consensus Statement of the European Atherosclerosis Society. Eur. Heart J. 2013, 34, 3478–3490. [Google Scholar] [CrossRef]
- Graça, R.; Zimon, M.; Alves, A.C.; Pepperkok, R.; Bourbon, M. High-Throughput Microscopy Characterization of Rare LDLR Variants. JACC Basic. Transl. Sci. 2023, 8, 1010–1021. [Google Scholar] [CrossRef]
- Brænne, I.; Kleinecke, M.; Reiz, B.; Graf, E.; Strom, T.; Wieland, T.; Fischer, M.; Kessler, T.; Hengstenberg, C.; Meitinger, T.; et al. Systematic analysis of variants related to familial hypercholesterolemia in families with premature myocardial infarction. Eur. J. Hum. Genet. 2016, 24, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Leitersdorf, E.; Van Der Westhuyzen, D.R.; Coetzee, G.A.; Hobbs, H.H. Two common low density lipoprotein receptor gene mutations cause familial hypercholesterolemia in Afrikaners. J. Clin. Investig. 1989, 84, 954–961. [Google Scholar] [CrossRef]
- Pereira, E.; Ferreira, R.; Hermelin, B.; Thomas, G.; Bernard, C.; Bertrand, V.; Nassiff, H.; Del Castillo, D.M.; Bereziat, G.; Benlian, P. Recurrent and novel LDL receptor gene mutations causing heterozygous familial hypercholesterolemia in La Habana. Hum. Genet. 1995, 96, 319–322. [Google Scholar] [CrossRef]
- Fouchier, S.W.; Defesche, J.C.; Umans-Eckenhausen, M.A.; Kastelein, J.J. The molecular basis of familial hypercholesterolemia in The Netherlands. Hum. Genet. 2001, 109, 602–615. [Google Scholar] [CrossRef]
- Huijgen, R.; Kindt, I.; Fouchier, S.W.; Defesche, J.C.; Hutten, B.A.; Kastelein, J.J.; Vissers, M.N. Functionality of sequence variants in the genes coding for the low-density lipoprotein receptor and apolipoprotein B in individuals with inherited hypercholesterolemia. Hum. Mutat. 2010, 31, 752–760. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thedrez, A.; Blom, D.J.; Ramin-Mangata, S.; Blanchard, V.; Croyal, M.; Chemello, K.; Nativel, B.; Pichelin, M.; Cariou, B.; Bourane, S.; et al. Homozygous Familial Hypercholesterolemia Patients with Identical Mutations Variably Express the LDLR (Low-Density Lipoprotein Receptor): Implications for the Efficacy of Evolocumab. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 592–598. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, 733–745. [Google Scholar] [CrossRef] [PubMed]
- Den Dunnen, J.T.; Dalgleish, R.; Maglott, D.R.; Hart, R.K.; Greenblatt, M.S.; McGowan-Jordan, J.; Roux, A.F.; Smith, T.; Antonarakis, S.E.; Taschner, P.E. HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Hum. Mutat. 2016, 37, 564–569. [Google Scholar] [CrossRef]
- Strøm, T.B.; Tveten, K.; Holla, Ø.L.; Cameron, J.; Berge, K.E.; Leren, T.P. The cytoplasmic domain is not involved in directing Class 5 mutant LDL receptors to lysosomal degradation. Biochem. Biophys. Res. Commun. 2011, 408, 642–646. [Google Scholar] [CrossRef]
- Ranheim, T.; Kulseth, M.A.; Berge, K.E.; Leren, T.P. Model System for Phenotypic Characterization of Sequence Variations in the LDL Receptor Gene. Clin. Chem. 2006, 52, 1469–1479. [Google Scholar] [CrossRef]
- Dušková, L.; Nohelová, L.; Loja, T.; Fialová, J.; Zapletalová, P.; Réblová, K.; Tichý, L.; Freiberger, T.; Fajkusová, L. Low Density Lipoprotein Receptor Variants in the Beta-Propeller Subdomain and Their Functional Impact. Front. Genet. 2020, 11, 691. [Google Scholar] [CrossRef]
- Raal, F.J.; Sjouke, B.; Hovingh, G.K.; Isaac, B.F. Phenotype diversity among patients with homozygous familial hypercholesterolemia: A cohort study. Atherosclerosis 2016, 248, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, H.H.; Brown, M.S.; Goldstein, J.L. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum. Mutat. 1992, 1, 445–466. [Google Scholar] [CrossRef]
- Gidding, S.S.; Ann Champagne, M.; De Ferranti, S.D.; Defesche, J.; Ito, M.K.; Knowles, J.W.; McCrindle, B.; Raal, F.; Rader, D.; Santos, R.D.; et al. The Agenda for Familial Hypercholesterolemia: A Scientific Statement From the American Heart Association. Circulation 2015, 132, 2167–2192. [Google Scholar] [CrossRef]
- Umans-Eckenhausen, M.A.; Sijbrands, E.J.; Kastelein, J.J.; Defesche, J.C. Low-Density Lipoprotein Receptor Gene Mutations and Cardiovascular Risk in a Large Genetic Cascade Screening Population. Circulation 2002, 106, 3031–3036. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huijgen, R.; Kindt, I.; Defesche, J.C.; Kastelein, J.J. Cardiovascular risk in relation to functionality of sequence variants in the gene coding for the low-density lipoprotein receptor: A study among 29,365 individuals tested for 64 specific low-density lipoprotein-receptor sequence variants. Eur. Heart J. 2012, 33, 2325–2330. [Google Scholar] [CrossRef] [PubMed]
- Bertolini, S.; Pisciotta, L.; Rabacchi, C.; Cefalù, A.B.; Noto, D.; Fasano, T.; Signori, A.; Fresa, R.; Averna, M.; Calandra, S. Spectrum of mutations and phenotypic expression in patients with autosomal dominant hypercholesterolemia identified in Italy. Atherosclerosis 2013, 227, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, A.; Villazón, F.; Suárez-Gutiérrez, L.; Gómez, J.; Martinez-Faedo, C.; Méndez-Torre, E.; Avanzas, P.; Álvarez-Velasco, R.; Cuesta-Llavona, E.; García-Lago, C.; et al. Clinical Evaluation of Patients with Genetically Confirmed Familial Hypercholesterolemia. J. Clin. Med. 2023, 12, 1030. [Google Scholar] [CrossRef]
- Chaudhry, A.; Trinder, M.; Vesely, K.; Cermakova, L.; Jackson, L.; Wang, J.; Hegele, R.A.; Brunham, L.R. Genetic Identification of Homozygous Familial Hypercholesterolemia by Long-Read Sequencing among Patients with Clinically Diagnosed Heterozygous Familial Hypercholesterolemia. Circ. Genom. Precis. Med. 2023, 16, e003887. [Google Scholar] [CrossRef]
- Yao, R.A.; Akinrinade, O.; Chaix, M.; Mital, S. Quality of whole genome sequencing from blood versus saliva derived DNA in cardiac patients. BMC Med. Genom. 2020, 13, 11. [Google Scholar] [CrossRef]
- Banerjee, P.; Chan, K.C.; Tarabocchia, M.; Benito-Vicente, A.; Alves, A.C.; Uribe, K.B.; Bourbon, M.; Skiba, P.J.; Pordy, R.; Gipe, D.A.; et al. Functional Analysis of LDLR (Low-Density Lipoprotein Receptor) Variants in Patient Lymphocytes to Assess the Effect of Evinacumab in Homozygous Familial Hypercholesterolemia Patients with a Spectrum of LDLR Activity. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 2248–2260. [Google Scholar] [CrossRef]
- Davis, C.G.; Goldstein, J.L.; Südhof, T.C.; Anderson, R.G.W.; Russell, D.W.; Brown, M.S. Acid-dependent ligand dissociation and recycling of LDL receptor mediated by growth factor homology region. Nature 1987, 326, 760–765. [Google Scholar] [CrossRef]
- Rudenko, G.; Henry, L.; Henderson, K.; Ichtchenko, K.; Brown, M.S.; Goldstein, J.L.; Deisenhofer, J. Structure of the LDL Receptor Extracellular Domain at Endosomal pH. Science 2002, 298, 2353–2358. [Google Scholar] [CrossRef]
- Andersen, O.M.; Dagil, R.; Kragelund, B.B. New horizons for lipoprotein receptors: Communication by β-propellers. J. Lipid Res. 2013, 54, 2763–2774. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hedge, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. ClinVar Accession Reference: NM_000527.5(LDLR):c.1285G>A (p.Val429Met). ClinVar; VCV000003694.60. Available online: https://www.ncbi.nlm.nih.gov/clinvar/variation/3694/ (accessed on 18 August 2024).
- Chora, J.R.; Medeiros, A.M.; Alves, A.C.; Bourbon, M. Analysis of publicly available LDLR, APOB, and PCSK9 variants associated with familial hypercholesterolemia: Application of ACMG guidelines and implications for familial hypercholesterolemia diagnosis. Genet. Med. 2018, 20, 591–598. [Google Scholar] [CrossRef]
- Bampatsias, D.; Dimopoulou, M.A.; Karagiannakis, D.; Sianis, A.; Korompoki, E.; Kantreva, K.; Psimenou, E.; Trakada, G.; Papatheodoridis, G.; Stamatelopoulos, K. SARS-CoV-2 infection-related deregulation of blood lipids in a patient with -/-LDLR familial homozygous hypercholesterolemia: A case report. J. Clin. Lipidol. 2023, 17, 219–224. [Google Scholar] [CrossRef]
- Kolesova, O.; Vanaga, I.; Laivacuma, S.; Dervos, A.; Kolesovs, A.; Radzina, M.; Platkajis, A.; Eglite, J.; Hagina, E.; Arutjunana, S.; et al. Intriguing findings of liver fibrosis following COVID-19. BMC Gastroenterol. 2021, 21, 370. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, M.; Anzali, B.C.; Mehryar, H.R. Liver enzymes as a predictor of mortality in patients with COVID-19? A cross-sectional study. Toxicol. Rep. 2024, 12, 266–270. [Google Scholar] [CrossRef]
- Xu, E.; Xie, Y.; Al-Aly, Z. Risks and burdens of incident dyslipidaemia in long COVID: A cohort study. Lancet Diabetes Endocrinol. 2023, 11, 120–128. [Google Scholar] [CrossRef]
- Versmissen, J.; Botden, I.P.G.; Huijgen, R.; Oosterveer, D.M.; Defesche, J.C.; Heil, T.C.; Muntz, A.; Langendonk, J.G.; Schinkel, A.F.; Kastelein, J.J.; et al. Maternal inheritance of familial hypercholesterolemia caused by the V408M low-density lipoprotein receptor mutation increases mortality. Atherosclerosis 2011, 219, 690–693. [Google Scholar] [CrossRef]
- Napoli, C.; D’Armiento, F.P.; Mancini, F.P.; Postiglione, A.; Witztum, J.L.; Palumbo, G.; Palinski, W. Fatty streak formation occurs in human fetal aortas and is greatly enhanced by maternal hypercholesterolemia. Intimal accumulation of low density lipoprotein and its oxidation precede monocyte recruitment into early atherosclerotic lesions. J. Clin. Investig. 1997, 100, 2680–2690. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Soffer, G.; Ginsberg, H.N.; Berglund, L.; Duell, P.B.; Heffron, S.P.; Kamstrup, P.R.; Lloyd-Jones, D.M.; Marcovina, S.M.; Yeang, C.; Koschinsky, M.L. Lipoprotein(a): A Genetically Determined, Causal, and Prevalent Risk Factor for Atherosclerotic Cardiovascular Disease: A Scientific Statement From the American Heart Association. Arterioscler. Thromb. Vasc. Biol. 2022, 42, e48–e60. [Google Scholar] [CrossRef]
- Lingenhel, A.; Kraft, H.G.; Kotze, M.; Peeters, A.V.; Kruse, R.; Utermann, G. Concentrations of the atherogenic Lp(a) are elevated in FH. Eur. J. Hum. Gen. 1998, 6, 50–60. [Google Scholar] [CrossRef]
- Marco-Benedí, V.; Cenarro, A.; Laclaustra, M.; Larrea-Sebal, A.; Jarauta, E.; Lamiquiz-Moneo, I.; Calmarza, P.; Bea, A.M.; Plana, N.; Pintó, X.; et al. Lipoprotein(a) in hereditary hypercholesterolemia: Influence of the genetic cause, defective gene and type of mutation. Atherosclerosis 2022, 349, 211–218. [Google Scholar] [CrossRef]
- Medeiros, A.M.; Alves, A.C.; Miranda, B.; Chora, J.R.; Bourbon, M. Unraveling the genetic background of individuals with a clinical familial hypercholesterolemia phenotype. J. Lipid Res. 2024, 65, 100490. [Google Scholar] [CrossRef]
- Whittall, R.A.; Scartezini, M.; Li, K.; Hubbart, C.; Reiner, Z.; Abraha, A.; Neil, H.A.; Dedoussis, G.; Humphries, S.E. Development of a high-resolution melting method for mutation detection in familial hypercholesterolaemia patients. Ann. Clin. Biochem. Int. J. Lab. Med. 2010, 47, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Traeger-Synodinos, J.; Mavroidis, N.; Kanavakis, E.; Eurydiki, D.; Humphries, S.E.; Day, I.N.; Kattamis, C.; Matsaniotis, N. Analysis of low density lipoprotein receptor gene mutations and microsatellite haplotypes in Greek FH heterozygous children: Six independent ancestors account for 60% of probands. Hum. Genet. 1998, 102, 343–347. [Google Scholar] [CrossRef] [PubMed]
- Mak, Y.T.; Pang, C.P.; Tomlinson, B.; Zhang, J.; Chan, Y.; Mak, T.W.; Masarei, J.R. Mutations in the Low-Density Lipoprotein Receptor Gene in Chinese Familial Hypercholesterolemia Patients. Arterioscler. Thromb. Vasc. Biol. 1998, 18, 1600–1605. [Google Scholar] [CrossRef] [PubMed]
- Schuster, H.; Fischer, H.J.; Keller, C.; Wolfram, G.; Zöllner, N. Identification of the 408 valine to methionine mutation in the low density lipoprotein receptor in a German family with familial hypercholesterolemia. Hum. Genet. 1993, 91, 287–289. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Weisburd, B.; Thomas, B.; Solomonson, M.; Ruderfer, D.M.; Kavanagh, D.; Hamamsy, T.; Lek, M.; Samocha, K.E.; Cummings, B.B.; et al. The ExAC browser: Displaying reference data information from over 60 000 exomes. Nucleic Acids Res. 2017, 45, 840–845. [Google Scholar] [CrossRef]
- Blom, D.J.; Harada-Shiba, M.; Rubba, P.; Gaudet, D.; Kastelein, J.J.; Charng, M.; Pordy, R.; Donahue, S.; Ali, S.; Dong, Y.; et al. Efficacy and Safety of Alirocumab in Adults with Homozygous Familial Hypercholesterolemia. J. Am. Coll. Cardiol. 2020, 76, 131–142. [Google Scholar] [CrossRef]
- Grundy, S.M.; Stone, N.J.; Bailey, A.L.; Beam, C.; Birtcher, K.K.; Blumenthal, R.S.; Braun, L.T.; De Ferranti, S.; Faiella-Tommasino, J.; Forman, D.E.; et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA Guideline on the Management of Blood Cholesterol: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation 2019, 139, e1046–e1081. [Google Scholar] [CrossRef]
- Santos, R.D.; Stein, E.A.; Hovingh, G.K.; Blom, D.J.; Soran, H.; Watts, G.F.; López, J.A.; Bray, S.; Kurtz, C.E.; Hamer, A.W.; et al. Long-Term Evolocumab in Patients with Familial Hypercholesterolemia. J. Am. Coll. Cardiol. 2020, 75, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Fokkema, I.F.A.C.; Taschner, P.E.M.; Schaafsma, G.C.P.; Celli, J.; Laros, J.F.J.; Den Dunnen, J.T. LOVD v.2.0: The next generation in gene variant databases. Hum. Mutat. 2011, 32, 557–563. [Google Scholar] [CrossRef]
- Kolovou, G.D.; Kolovou, V.; Papadopoulou, A.; Watts, G.F. MTP Gene Variants and Response to Lomitapide in Patients with Homozygous Familial Hypercholesterolemia. J. Atheroscler. Thromb. 2016, 23, 878–883. [Google Scholar] [CrossRef] [PubMed]
- D’Erasmo, L.; Gallo, A.; Cefalù, A.B.; Di Costanzo, A.; Saheb, S.; Glammanco, A.; Averna, M.; Buonaiuto, A.; Iannuzzo, G.; Fortunato, G.; et al. Long-term efficacy of lipoprotein apheresis and lomitapide in the treatment of homozygous familial hypercholesterolemia (HoFH): A cross-national retrospective survey. Orphanet J. Rare Dis. 2021, 16, 381. [Google Scholar] [CrossRef]
- Cesaro, A.; Fimiani, F.; Gragnano, F.; Moscarella, E.; Schiavo, A.; Vergara, A.; Akioyamen, L.; D’Erasmo, L.; Averna, M.; Arca, M.; et al. New Frontiers in the Treatment of Homozygous Familial Hypercholesterolemia. Heart Fail. Clin. 2022, 18, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Raal, F.J.; Kallend, D.; Ray, K.K.; Turner, T.; Koenig, W.; Wright, R.S.; Wijngaard, P.L.J.; Curcio, D.; Jaros, M.J.; Leiter, L.A.; et al. Inclisiran for the Treatment of Heterozygous Familial Hypercholesterolemia. N. Engl. J. Med. 2020, 382, 1520–1530. [Google Scholar] [CrossRef]
- Raal, F.; Durst, R.; Bi, R.; Talloczy, Z.; Maheux, P.; Lesogor, A.; Kastelein, J.J.P. fficacy, Safety, and Tolerability of Inclisiran in Patients with Homozygous Familial Hypercholesterolemia: Results From the ORION-5 Randomized Clinical Trial. Circulation 2024, 149, 354–362. [Google Scholar] [CrossRef]
- Reeskamp, L.F.; Tromp, T.R.; Defesche, J.C.; Grefhorst, A.; Stroes, E.S.; Hovingh, G.K.; Zuurbier, L. Next-generation sequencing to confirm clinical familial hypercholesterolemia. Eur. J. Prev. Cardiol. 2021, 28, 875–883. [Google Scholar] [CrossRef]
- Thompson, G.R.; Blom, D.J.; Marais, A.D.; Seed, M.; Pilcher, G.J.; Raal, F.J. Survival in homozygous familial hypercholesterolaemia is determined by the on-treatment level of serum cholesterol. Eur. Heart J. 2018, 39, 1162–1168. [Google Scholar] [CrossRef]
- Alnouri, F.; Al-Allaf, F.A.; Athar, M.; Abdulijaleel, Z.; Alabdullah, M.; Alammari, D.; Alanzi, M.; Alkaf, F.; Allehyani, A.; Alotaiby, M.A.; et al. Xanthomas Can Be Misdiagnosed and Mistreated in Homozygous Familial Hypercholesterolemia Patients: A Call for Increased Awareness among Dermatologists and Health Care Practitioners. Glob. Heart. 2020, 15, 19. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).