Abstract
Patients carrying the heterozygous A414G mutation in the HCN4 gene, which encodes the HCN4 protein, demonstrate moderate to severe bradycardia of the heart. Tetramers of HCN4 subunits compose the ion channels in the sinus node that carry the hyperpolarization-activated ‘funny’ current (If), also named the ‘pacemaker current’. If plays an essential modulating role in sinus node pacemaker activity. To assess the mechanism by which the A414G mutation results in sinus bradycardia, we first performed voltage clamp measurements on wild-type (WT) and heterozygous mutant HCN4 channels expressed in Chinese hamster ovary (CHO) cells. These experiments were performed at physiological temperature using the amphotericin-perforated patch-clamp technique. Next, we applied the experimentally observed mutation-induced changes in the HCN4 current of the CHO cells to If of the single human sinus node cell model developed by Fabbri and coworkers. The half-maximal activation voltage V1/2 of the heterozygous mutant HCN4 current was 19.9 mV more negative than that of the WT HCN4 current (p < 0.001). In addition, the voltage dependence of the heterozygous mutant HCN4 current (de)activation time constant showed a −11.9 mV shift (p < 0.001) compared to the WT HCN4 current. The fully-activated current density, the slope factor of the activation curve, and the reversal potential were not significantly affected by the heterozygous A414G mutation. In the human sinus node computer model, the cycle length was substantially increased, almost entirely due to the shift in the voltage dependence of steady-state activation, and this increase was more prominent under vagal tone. The introduction of a passive atrial load into the model sinus node cell further reduced the beating rate, demonstrating that the bradycardia of the sinus node was even more pronounced by interactions between the sinus node and atria. In conclusion, the experimentally identified A414G-induced changes in If can explain the clinically observed sinus bradycardia in patients carrying the A414G HCN4 gene mutation.
1. Introduction
The ‘funny’ current (If), also named the ‘pacemaker current’, is a key player in the pacemaker activity in the sinus node of the heart. If is a current of mixed ionic nature and is activated upon hyperpolarization. It is a (mainly) inwardly directed current and, consequently, it is important for the spontaneous depolarization that underlies sinus node pacemaker activity. Furthermore, cyclic AMP (cAMP), the (intracellular) level of which is lowered by acetylcholine (ACh), affects the current density and gating properties of If, and thus, If is an essential modulator of the cardiac pacing rate, as reviewed previously in detail [1,2,3,4,5,6]. The ion channel of If in the sinus node cell membrane is composed of four hyperpolarization-activated, cyclic-nucleotide-gated (HCN) subunits. The protein of HCN4, which is encoded by the HCN4 gene, is the main isoform of HCN in human and rabbit sinus node cells [7,8,9,10].
Two decades ago, Schulze-Bahr and colleagues [11] were the first to present a case of a patient with marked sinus bradycardia that could be attributed to a (heterozygous) loss-of-function mutation in HCN4. The loss-of-function phenotype of this particular mutation was demonstrated using patch-clamp methodology on COS-7 cells that were transiently transfected with wild-type (WT) and/or mutant HCN4 complementary DNA (cDNA) to make them express WT or mutant HCN4 channels. In the following years, a number of studies were published in which idiopathic or familial sinus bradycardia was associated with a (heterozygous) mutation in HCN4, as reviewed by us and by others [12,13,14,15,16]. Of note, the in vitro data on the mutation of interest, if provided, were regularly incomplete or not fully consistent with the clinical phenotype [14,15]. Generally speaking, HCN4 mutations associated with cardiac bradycardia are heterozygous dominant-negative (or dominant-negative-like) loss-of-function mutations.
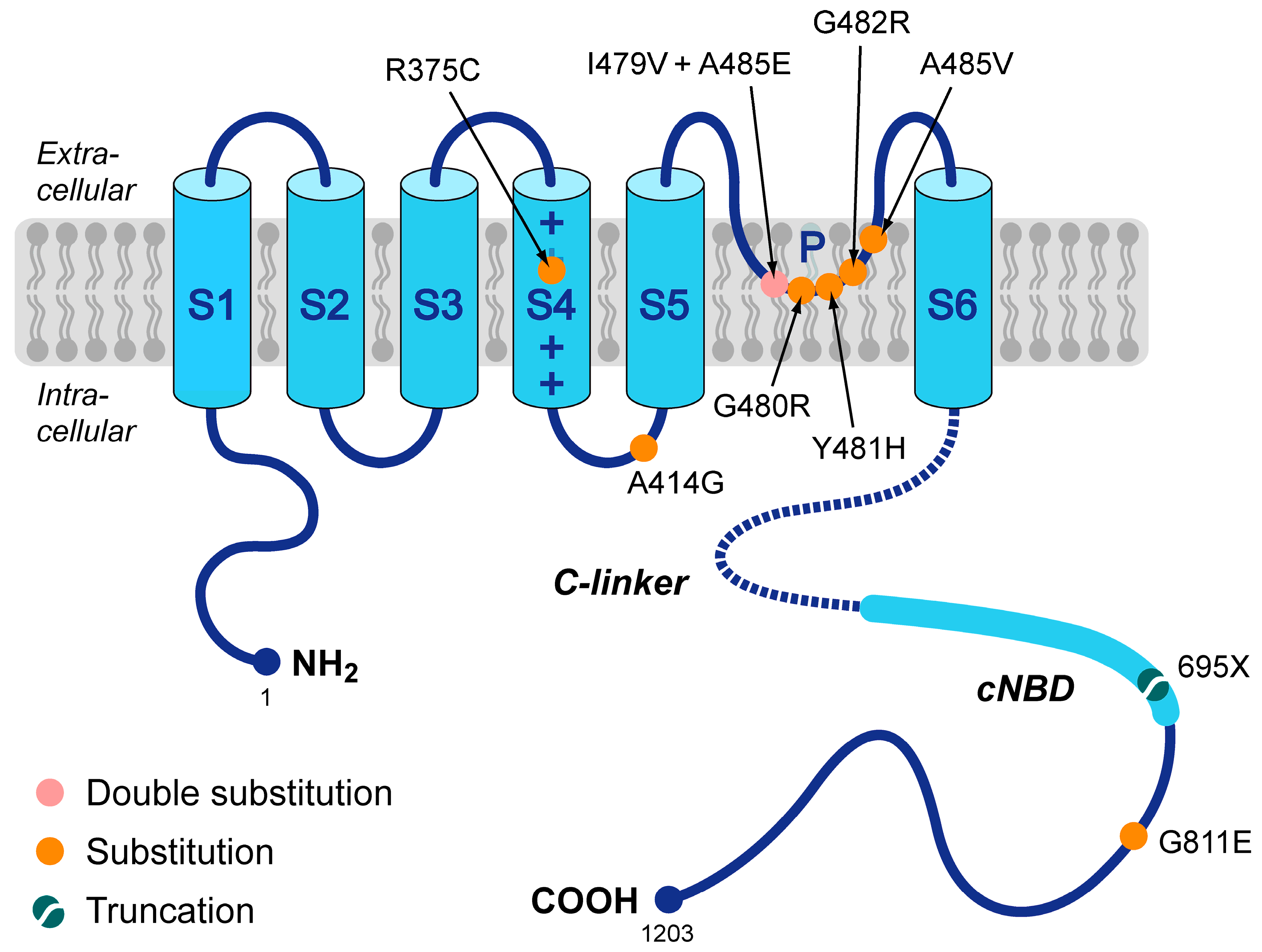
In 2014, Milano and coworkers [17] and Schweizer and coworkers [18] were the first to present a number of loss-of-function mutations in HCN4 that were not only associated with sinus bradycardia but also with mitral valve prolapse (MVP) and left ventricular non-compaction (LVNC). Since then, several more such mutations have been presented, together with more structural abnormalities, like right ventricular non-compaction, biventricular non-compaction, hypertrabeculation, left atrial dilatation, aortic dilation, and pulmonary artery dilation [19,20,21,22,23,24]. However, the exact mechanism by which these mutations in HCN4 lead to LVNC, MVP, and other structural abnormalities remains to be resolved. The mutations known today are summarized in Figure 1 and include the R375C [23,25,26], A414G [17], G480R [27], Y481H [17,20], G482R [17,18,19,22,24,25,26,27,28], A485V [25,27], and G811E [29] missense mutations, the I479V + A485E double mutation [21], and the 695X truncation mutation [18]. As can be directly inferred from Figure 1, many of these mutations are located in the pore-forming loop of the HCN4 channel. Of note, this figure is limited to mutations that have been classified as loss-of-function mutations, not taking into account that some of the loss-of-function effects may not be large enough to explain the bradycardic effects of the mutation, as in the case of the G811E mutation [29]. More mutations have been associated with the combined phenotype of LNVC and bradycardia of the heart, but in these cases, electrophysiological data are either lacking, like for the G480C missense mutation [26] and the R483_V487del deletion mutation [30], or gain-of-function characteristics rather than loss-of-function ones have been revealed, like for the P883R mutation [18,31], thus (partly) explaining the clinically observed tachycardia-bradycardia syndrome rather than the also observed sinus bradycardia.
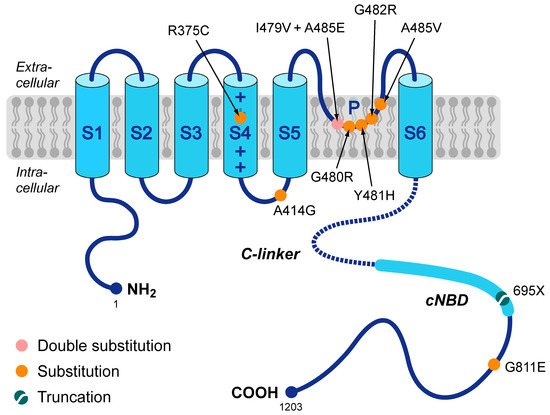
Figure 1.
Schematic topology of the HCN4 protein and loss-of-function mutations that have been associated with both sinus bradycardia and left ventricular non-compaction. Tetramers of HCN4 α-subunits constitute the cardiac ion channel that conducts the hyperpolarization-activated ‘funny’ current (If). The HCN4 protein possesses six transmembrane segments (S1–S6), of which the positively-charged S4 helix forms the voltage sensor of the channel. In addition, the HCN4 protein has a pore-forming loop (P) and intracellular N- and C-termini. The C-terminus comprises the C-linker (indicated by the dotted line) and the cyclic nucleotide-binding domain (cNBD), which mediates cyclic AMP (cAMP)-dependent changes in the gating properties of the HCN channel. Colored dots indicate the location of the currently known loss-of-function mutations in the HCN4 protein that are associated with both bradycardia of the sinus node and left ventricular non-compaction. These include seven substitutions (R375C, A414G, G480R, Y481H, G482R, A485V, and G811E), a double substitution (I479V + A485E), and a truncation (695X).
Once a specific mutation has been found, it is important to test its functional effects in extensive electrophysiological experiments to assess whether the mutation can explain the clinical phenotype, as demonstrated for the G811E and P883R mutations. Once electrophysiological data are available, these can be used in computer simulations to further determine the functional consequences of the mutation of interest. In this regard, the comprehensive single human sinus node cell model of Fabbri et al. [32] (known as the Fabbri–Severi model) can be a very helpful tool. Of note, this mathematical model allows different autonomic tone levels through the simulated administration of ACh (vagal tone).
Here, we characterized in detail the functional consequences of the A414G mutation in the S4-S5 linker of the HCN4 protein (Figure 1) that was previously found by Milano et al. [17] in a small family with sinus bradycardia and LVNC. Therefore, we first performed voltage-clamp experiments on the WT HCN4 channels and heterozygous HCN4 mutant channels expressed in Chinese hamster ovary (CHO) cells, importantly extending the electrophysiological data that were presented by Milano et al. [17]. Next, the experimentally identified mutation-induced changes in the kinetics of the HCN4 channels were incorporated into the Fabbri–Severi sinus node cell model. The computer simulations were performed with the default model as well as in the simulated presence of 10–20 nM ACh to test vagal effects. Also, we carried out computer simulations in the absence or presence of an additional outward current to test the effects of an atrial load. Our research had been presented at the 46th Computing in Cardiology Conference in Singapore and appeared in its conference proceedings [33].
2. Materials and Methods
2.1. Plasmid Construction and Transfection
The mutation of A414G was introduced in WT hHCN4 complementary DNA (cDNA) by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) [17]. CHO cells (Sigma-Aldrich, St. Louis, MO, USA) were cultured at 37 °C in Ham’s F-12 medium with 10% fetal bovine serum (Lonza, Basel, Switzerland), 2 mM glutamine, and 1% P/S (5000 U/mL penicillin and 5000 U/mL streptomycin sulfate, Lonza). The cells were transiently transfected with lipofectamine (Gibco BRL, Life Technologies, Grand Island, NY) using either 2 µg WT cDNA or—in order to recapitulate a heterozygous state—1 µg of WT and 1 µg of A414G mutant cDNA construct. Successfully transfected cells were visualized by co-expressed green fluorescent protein (GFP) (0.5 µg eGFP plasmid; pcDNA3-eGFP; Life Technologies, Carlsbad, CA, USA).
2.2. Patch-Clamp Experiments
The bath solution (36 ± 0.2 °C) contained (in mM): NaCl 140, KCl 5.4, CaCl2 1.8, MgCl2 1.0, glucose 5.5, and HEPES 5.0; pH 7.4 (NaOH). The patch-clamp experiments were performed two days after transfection using the amphotericin-perforated patch-clamp technique and an Axopatch 200B amplifier (Molecular Devices, Sunnyvale, CA, USA). The patch pipettes (2–2.5 MΩ) were pulled from borosilicate glass (GC100F-10; Harvard Apparatus, Waterbeach, UK) using a custom-made pipette puller. The pipette solution contained (in mM): K-gluconate 125, KCl 20, NaCl 10, amphotericin-B 0.88, and HEPES 10; pH 7.2 (KOH). Custom-made software was used for voltage control, data acquisition, and analysis. We corrected the potentials for the estimated liquid junction potential. Signals were low-pass filtered (cut-off frequency: 5 kHz) and digitized at 5 kHz. The cell membrane capacitance (Cm; 7.2 ± 0.8 pF (mean ± SEM, n = 24)) was determined by dividing the decay time constant of the capacitive transient in response to hyperpolarizing voltage-clamp steps of 5 mV (from a holding potential of −20 mV) by the series resistance. For proper voltage control, the series resistance was compensated by 70–80%.
HCN4 currents were measured using the voltage-clamp protocols as shown in the respective figures of Section 3.1.1 and 3.1.3. Currents analyzed at the end of the hyperpolarizing steps were normalized to Cm to obtain HCN4 current densities. To construct the steady-state activation curves, tail current amplitudes were normalized to the maximum tail current and plotted against the test voltage. The activation curves were fitted using the Boltzmann equation I/Imax = A/{1 + exp[(V½ − V)/k]}, where I is the HCN4 tail current at test potential V and Imax is the maximum HCN4 tail current, to determine the voltage of half-maximal activation V½ (in mV) and slope factor k (in mV). The HCN4 activation and deactivation time course was fitted by the mono-exponential functions I/Imax = A × [1 − exp(−t/τ)] and I/Imax = A × exp(−t/τ), respectively, to determine the time constant of (de)activation τ, ignoring the variable initial delay in (de)activation [34,35].
2.3. Computer Simulations
The electrical activity of a single human sinus node cell was mathematically simulated using the comprehensive Fabbri–Severi computer model of such a cell [32]. The heterozygous A414G mutation in HCN4 was implemented in the CellML code of the Fabbri–Severi model, which is available from the CellML Model Repository [36] at https://www.cellml.org/ (accessed on 16 April 2023), by a −19.9 mV shift in the voltage dependence of the steady-state activation curve as well as a −11.9 mV shift in the voltage dependence of its time constant of (de)activation. Either shift was based on the data from our patch-clamp experiments.
We introduced vagal tone into the Fabbri–Severi model by setting the model ACh concentration to 10 or 20 nM, thereby lowering the 74 beats/min beating rate of the default model (at zero ACh concentration) to 58 and 49 beats/min, respectively. The major consequences of the simulated ACh administration were the activation of the ACh-activated K+ current (IK,ACh), which was zero in the default model, and the If reduction through a negative shift in the If voltage dependence [32]. In order to assess the consequences of the sinus node–atrial interactions, we introduced a passive atrial load in the computer model through the addition of an ohmic outward current with a conductance of 0.2–0.4 pS/pF and a −80 mV reversal potential.
The CellML code [37] was edited and run in the Cellular Open Resource (COR) environment [38], version 0.9.31.1409. All simulations were run for 200 s, which proved to be a sufficiently long time to reach steady-state behavior. Data from the final 10 s were used for analysis.
2.4. Statistics
Data are presented as the mean ± SEM. We compared groups using an unpaired two-sided t-test or the two-way Repeated Measures (RM) ANOVA, which was followed by a pairwise comparison using the Student–Newman–Keuls test. p < 0.05 defined statistical significance.
3. Results
In the present study, we characterized the mechanism by which the HCN4 gene mutation A414G causes sinus bradycardia. Therefore, we performed voltage-clamp measurements on the WT and heterozygous mutant HCN4 channels expressed in CHO cells to determine in detail the changes in the biophysical properties of the HCN4 channels caused by the mutation, as set out in Section 3.1. Subsequently, the experimentally identified mutation-induced changes were incorporated into the Fabbri–Severi model of a human sinus node pacemaker cell to determine their functional effects. As described in Section 3.2, our computer simulations were carried out with the default model, in the presence of 10–20 nM Ach, to test the effects of vagal tone, and in the absence or presence of an extra outward current to test the effects of an atrial load.
3.1. Voltage-Clamp Experiments
3.1.1. Current Density
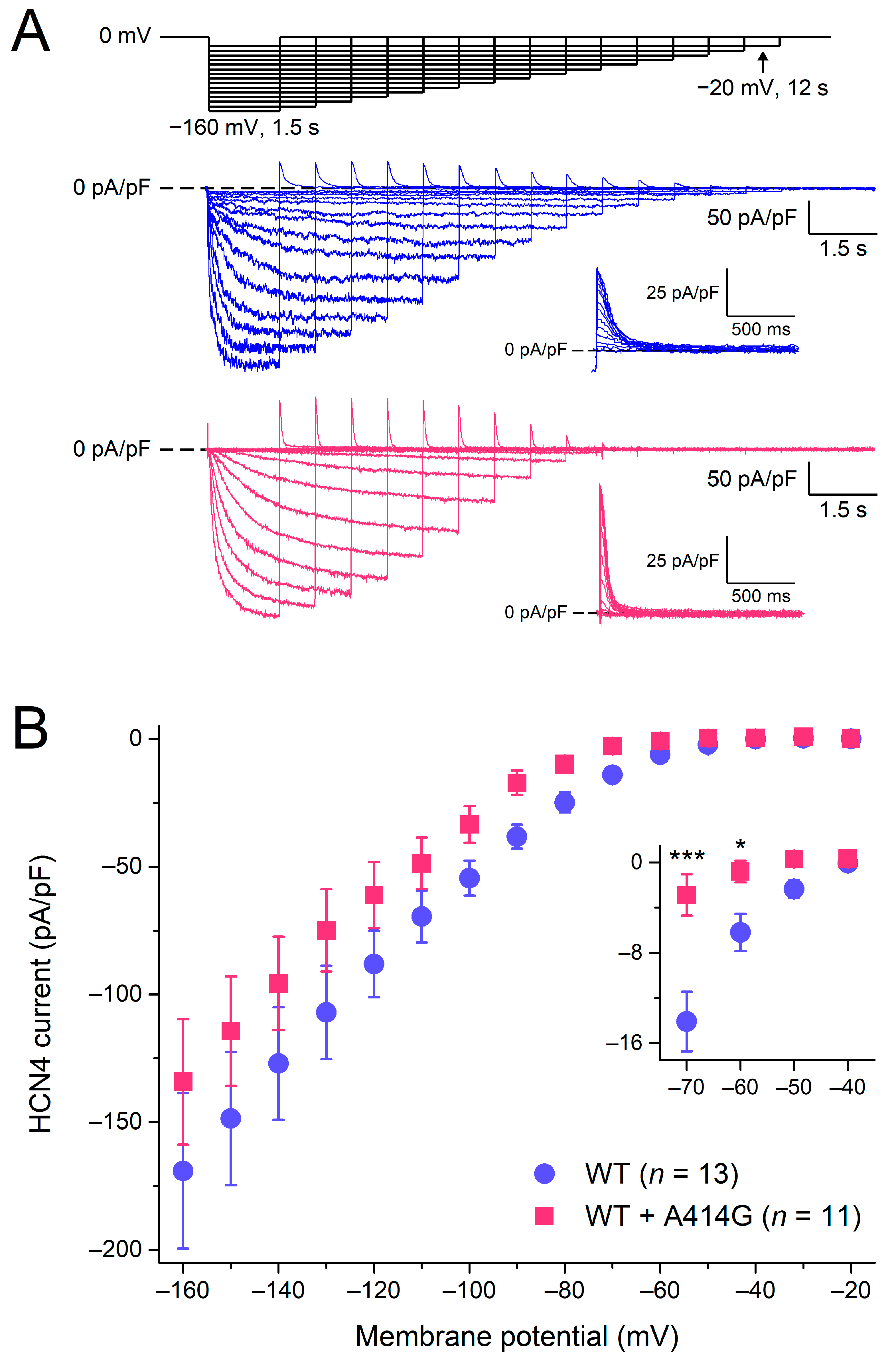
Figure 2A shows typical HCN4 currents in CHO cells transfected with WT (blue traces) or heterozygous mutant HCN4 channels (pink traces). To activate HCN4, the CHO cells were voltage-clamped from a holding potential of 0 mV to various test potentials (ranging from −160 to −20 mV, in 10 mV increments), as schematically shown in the protocol at the top of Figure 2A. The duration of the test step to −20 mV was 12 s to ensure full activation, and its duration was progressively reduced to 1.5 s at −160 mV to prevent membrane instability and cell death at very negative potentials. As typical for HCN4 currents, the activation threshold was in the −40 to −50 mV voltage range and the HCN4 current amplitude progressively increased with more hyperpolarized potentials. Figure 2B shows the current–voltage (I–V) relationships of the current densities in 13 WT and 11 heterozygous mutant HCN4 cells. Over the entire voltage range of the test potentials, the densities were not significantly different between the WT and heterozygous mutant HCN4 channels (two-way RM ANOVA with one-factor repetition). However, if the data analysis was limited to the voltage range of diastolic depolarization (roughly −70 to −40 mV), the current of the heterozygous mutant HCN4 channels seemed to activate at more negative potentials (for detailed activation kinetics, see Section 3.1.2), resulting in a lower current density during the diastolic depolarization of sinus node pacemaker cells (Figure 2B, inset).
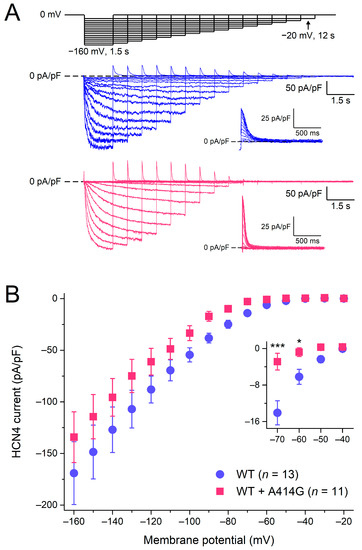
Figure 2.
Activation of wild-type (WT) and heterozygous A414G mutant (WT + A414G) HCN4 currents. (A) Voltage-clamp protocol (top panel) and typical WT (blue traces) and heterozygous mutant HCN4 currents (pink traces). Insets: superimposed tail currents at 0 mV. (B) Average densities of WT and heterozygous mutant HCN4 currents measured during the step potentials. The inset shows current densities on an enlarged current scale, focusing on the test potentials in the voltage range of diastolic depolarization of sinus node pacemaker cells and limiting data analysis to these potentials. * p < 0.05 and *** p < 0.001.
3.1.2. Activation Kinetics
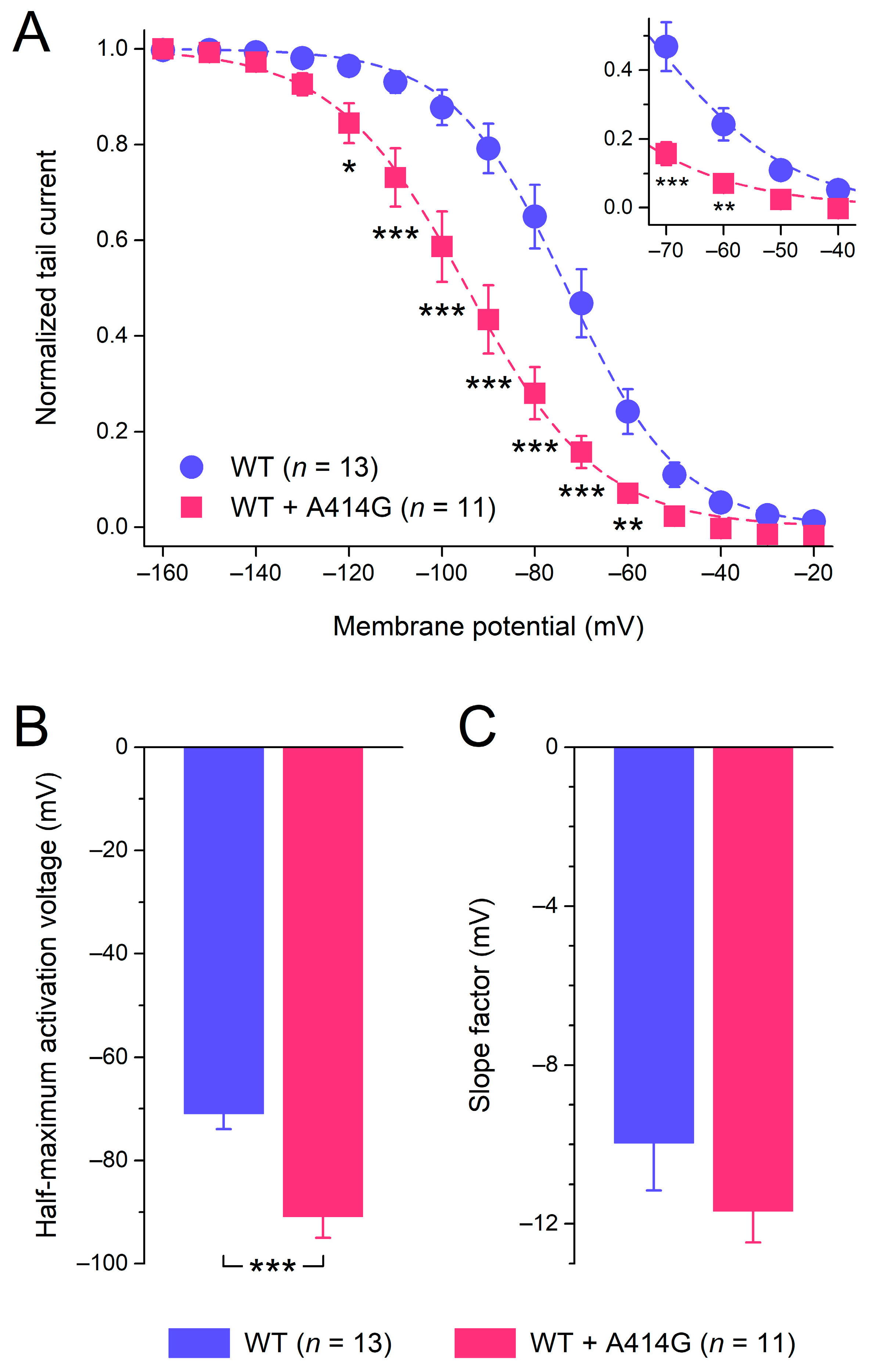
Figure 2B suggests that the heterozygous mutant HCN4 activates at more negative potentials than the WT. Therefore, we analyzed the voltage dependence of activation in more detail, using the HCN4 tail currents that could be recorded upon returning from the hyperpolarizing test potentials to the 0 mV holding potential, as illustrated in Figure 2A (insets). The tail currents were normalized to the maximum tail current and plotted against the preceding hyperpolarizing test potential to construct the WT and heterozygous mutant steady-state activation curves of Figure 3A. As can be inferred from Figure 3A, the heterozygous mutation shifted the steady-state activation curve by approximately −20 mV, thus decreasing the voltage range over which the HCN4 current is functionally active during a sinus node action potential (Figure 3A, inset). As a consequence of the shift, the half-maximal activation voltage V1/2 changed from −71.0 ± 3.1 mV for the WT (n = 13) to −90.9 ± 4.6 mV (n = 11) for the heterozygous mutation (Figure 3B; unpaired two-sided t-test, p < 0.001). With values of −10.0 ± 1.2 mV (WT) and −11.7 ± 0.9 mV (heterozygous mutation), the slope factor of the HCN4 steady-state activation curve was not significantly different between the WT and WT + A414G (Figure 3C; unpaired two-sided t-test).
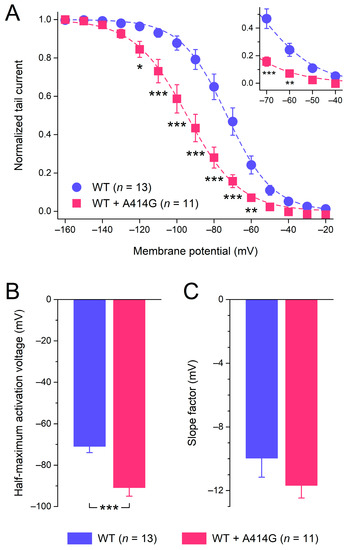
Figure 3.
Voltage dependence of HCN4 current activation. (A) Average tail current–voltage (I–V) relationships of the WT and the heterozygous A414G mutation. The dashed lines are the Boltzmann fits to the data, and the inset represents the voltage range of the diastolic depolarization of human sinus node pacemaker cells. (B) Average half-maximum activation voltage. (C) Average slope factor. * p < 0.05, ** p < 0.01, and *** p < 0.001.
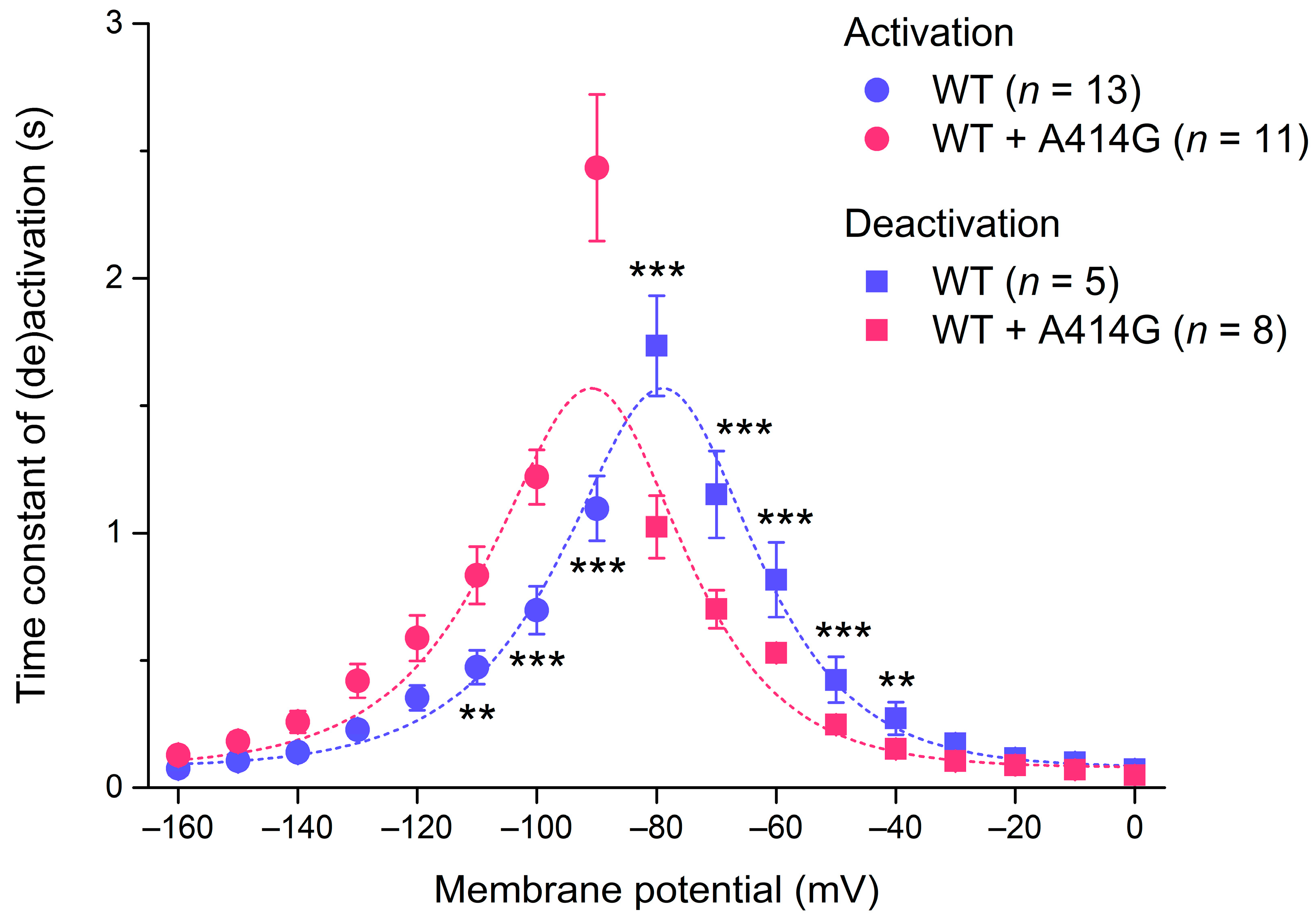
The native If and all HCN isoforms, including HCN4, activate faster at more hyperpolarized potentials [1], as can also be observed in the typical examples of Figure 2A. We fitted the time course of activation by a mono-exponential function to determine the time constant of activation. Figure 4 (filled circles) shows the thus obtained time constant of activation plotted against the hyperpolarizing test potential for the WT and the heterozygous mutation. The heterozygous mutation significantly decreased the activation rate of the HCN4 current at various test potentials (Figure 4; two-way RM ANOVA with one-factor repetition). For example, at −100 mV, the time constant of activation was 697 ± 98 ms for the WT (n = 13) and 1220 ± 126 ms (n = 11) for the heterozygous mutation (p < 0.001).
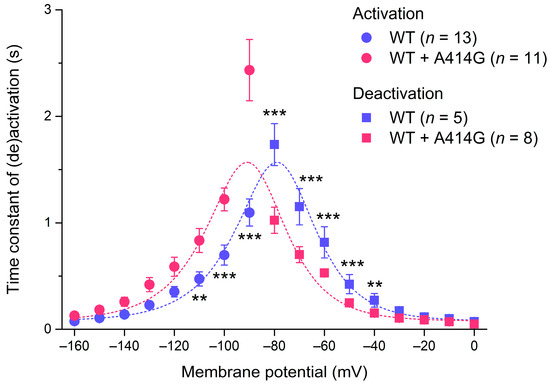
Figure 4.
Voltage dependence of the time constant of HCN4 current (de)activation. Bell-shaped curve: fit to the data according to τ = 1/[A1 × exp(–Vm/B1) + A2 × exp (Vm/B2)], where τ is the time constant of (de)activation, Vm is the membrane potential, and A1, A2, B1, and B2 are fitting parameters [39]. ** p < 0.01 and *** p < 0.001.
3.1.3. Deactivation Kinetics and Reversal Potentials
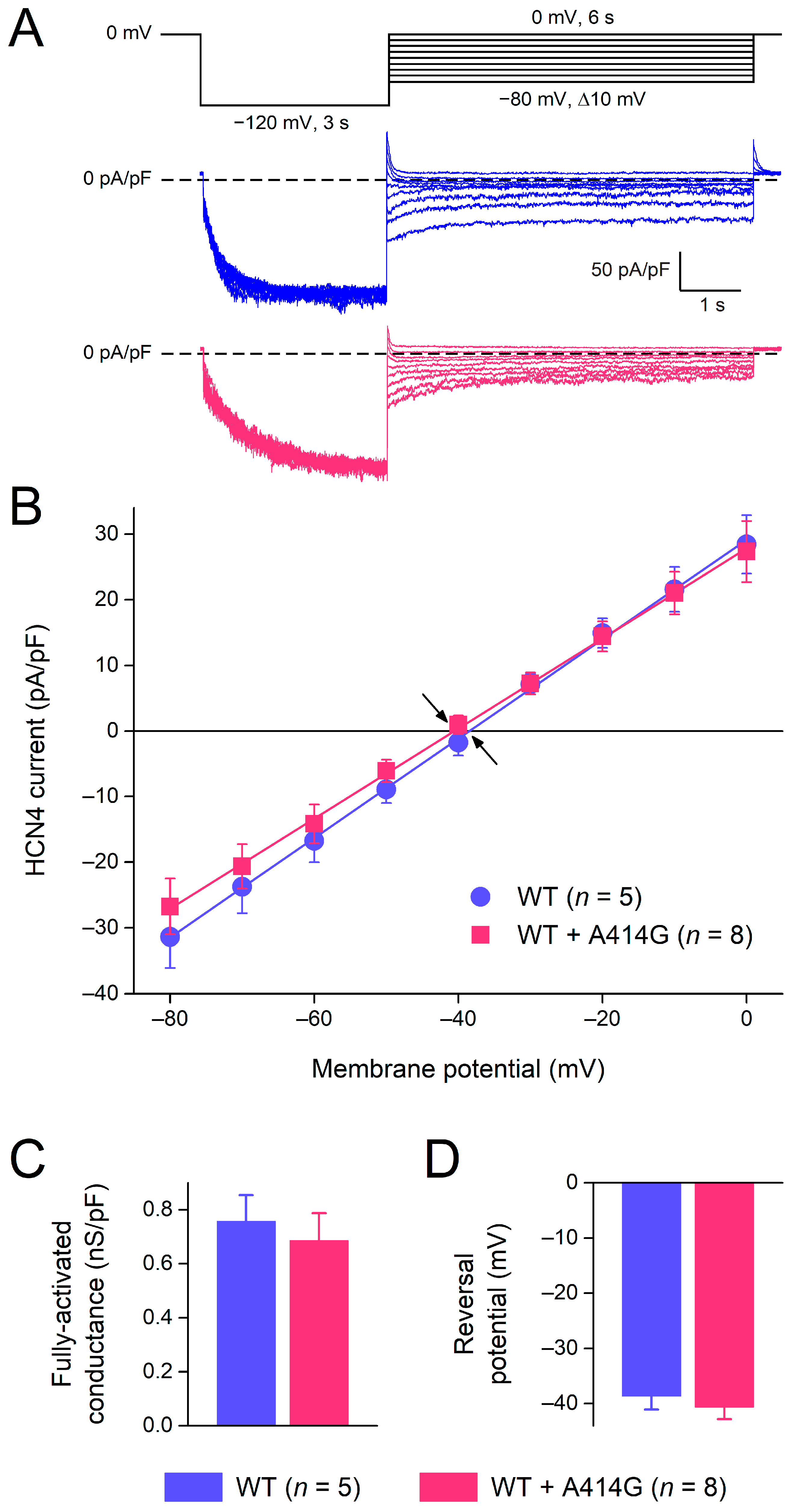
HCN4 currents activated during hyperpolarization demonstrate deactivation upon subsequent depolarization. Figure 5A shows typical examples of deactivation in a WT and heterozygous mutant cell. The cells were first voltage-clamped from a holding potential of 0 mV to −120 mV for 3 s, to ensure full activation, and this hyperpolarizing step was followed by a 6 s step to test potentials ranging from −80 to 0 mV (in 10 mV increments), as shown in the protocol at the top of Figure 5A. Typically, for HCN currents, deactivation becomes faster as test potentials become more depolarized. The time course of deactivation was fitted by a mono-exponential function to determine the time constant of deactivation. The thus obtained time constants of deactivation are plotted against the depolarizing test potentials in Figure 4 (filled squares). The heterozygous mutation significantly increased the deactivation rate of the HCN4 current at various test potentials. For example, at −60 mV, the time constant of deactivation was 816 ± 164 ms for the WT (n = 5) and 529 ± 26 ms (n = 8) for the heterozygous mutation (p < 0.001). The combination of the HCN4 activation and deactivation time constants usually constitutes a bell-shaped relationship [1], as is also the case in the present study, both for WT and WT + A414G (Figure 4, dashed lines). The heterozygous A414G mutation shifted the complete bell-shaped curve to the left by 11.9 mV.
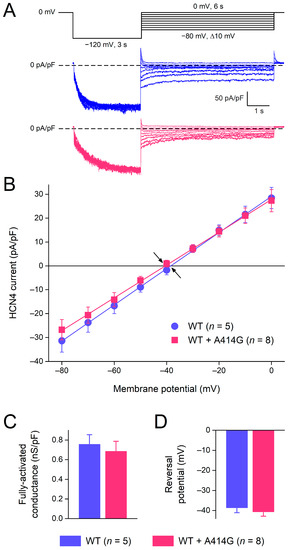
Figure 5.
Voltage dependence of HCN4 current deactivation and fully-activated I–V relationship. (A) Voltage-clamp protocol (top panel) and typical WT (blue traces) and heterozygous mutant HCN4 currents (pink traces). (B) The average I–V relationship of the fully-activated HCN4 current as determined from the deactivating tail currents at the test potentials ranging from −80 to 0 mV. Solid lines are the linear fits to the experimental data. Arrows indicate the reversal potential of the HCN4 current. (C) Fully-activated conductance and (D) reversal potential, as determined from five WT and eight heterozygous mutant I–V relationships.
The fully-activated I–V relationship was characterized by the determination of the peak of the deactivating tail current. The fully-activated I–V relationship was linear for the WT as well as for the mutation (Figure 5B). The fully-activated conductance, determined from the slopes of the individual linear fits, was not significantly different between the WT and the heterozygous mutation (Figure 5C). The reversal potential, determined from linear fits to the individual I–V relationships of the cells, was −38.7 ± 2.7 mV (n = 5) and −39.6 ± 2.3 mV (n = 8) for the WT and the heterozygous mutation, respectively (Figure 5D).
3.2. Computer Simulations
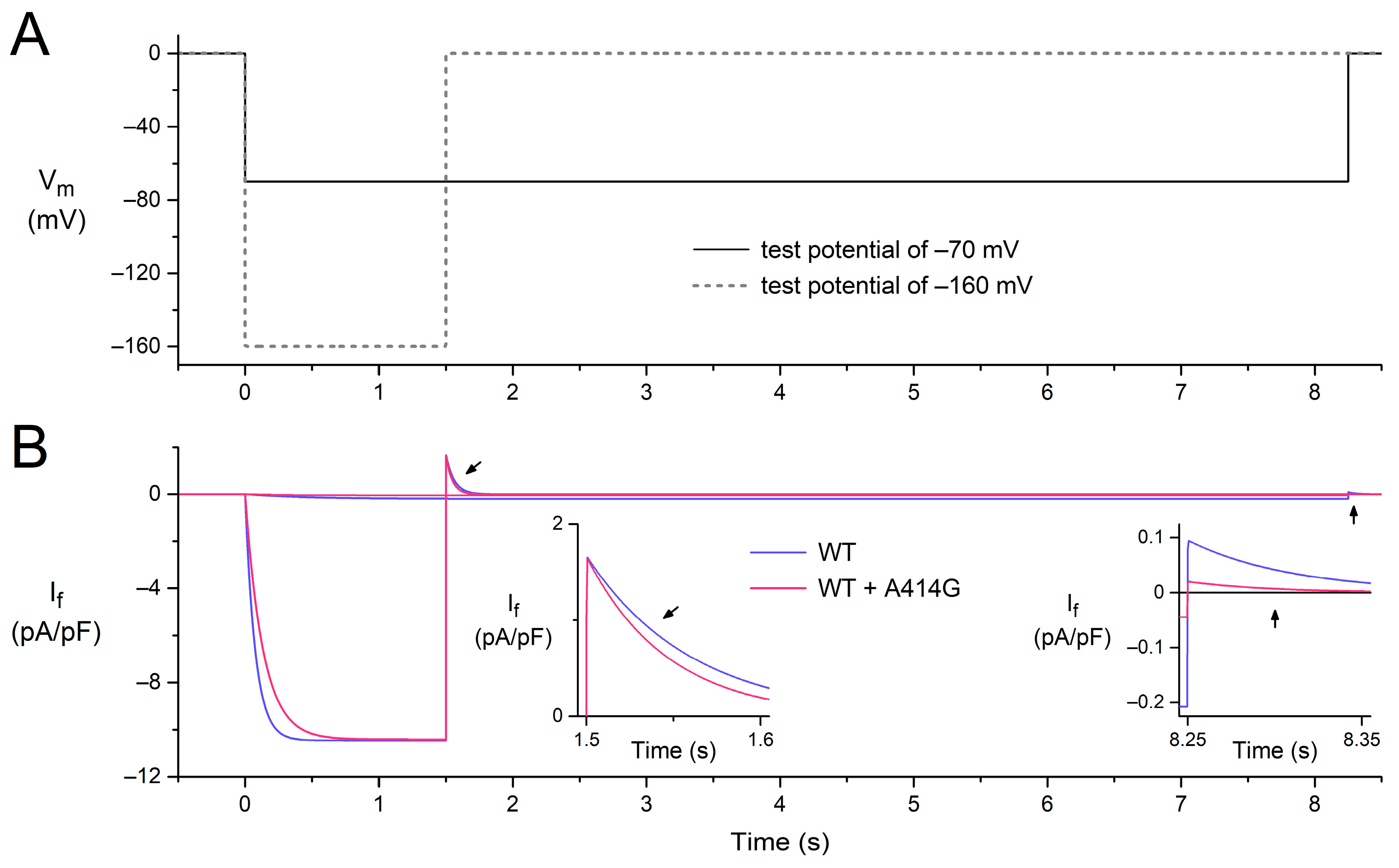
To assess the mechanism by which the heterozygous A414G mutation in HCN4 causes sinus bradycardia, we incorporated the experimentally observed mutation-induced changes in the HCN4 current into the comprehensive Fabbri–Severi model of a single human sinus node cell [32], assuming that the effects of the A414G mutation on the HCN4 current in CHO cells are similar to those on If in human sinus node cells. Accordingly, the voltage dependence of the steady-state activation curve of the model If was shifted by −19.9 mV and that of its time constant of (de)activation by −11.9 mV, whereas no changes were made to other If characteristics, like its fully-activated conductance and reversal potential. The effects of these shifts under voltage-clamp conditions are illustrated in Figure 6 for test potentials of −160 and −70 mV, following the voltage-clamp protocol of Figure 2A. At −160 mV, activation is slowed (Figure 2B, pink vs. blue traces), whereas deactivation upon stepping back to the holding potential of 0 mV is fastened (Figure 2B, left inset), which is in line with our experimental observations. At −70 mV, the shifts result in the activation of a substantially smaller amount of current, and its deactivation is fastened (Figure 2B, right inset, pink vs. blue traces), again in line with our experimental observations.
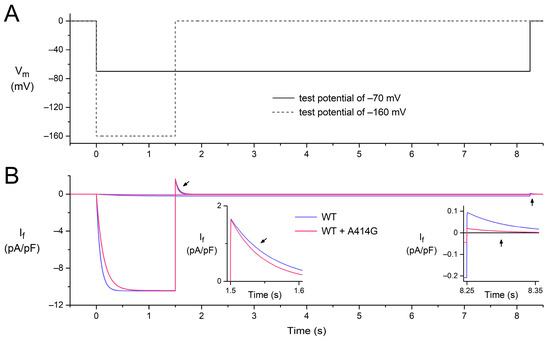
Figure 6.
Combined effects of a −19.9 mV shift in the voltage dependence of the steady-state activation of the model If and that of its time constant of (de)activation by −11.9 mV on the electrical activity of the Fabbri–Severi model of a single human sinus node cell under voltage-clamp conditions. (A) Voltage-clamp protocol with a 0 mV holding potential and test potentials of −70 and −160 mV (solid and dashed lines, respectively). Vm denotes the membrane potential of the model cell. (B) If under WT conditions (blue traces) and with changes induced by the WT + A414G heterozygous mutation in HCN4 (pink traces). Left inset: the deactivation of If in response to stepping back from the test potential of −160 mV to the holding potential of 0 mV (slanted arrows). Right inset: the fully-activated If during the test potential of −70 mV and deactivation in response to stepping back to the holding potential of 0 mV (vertical arrows).
3.2.1. Effects of Shifts in Voltage Dependence
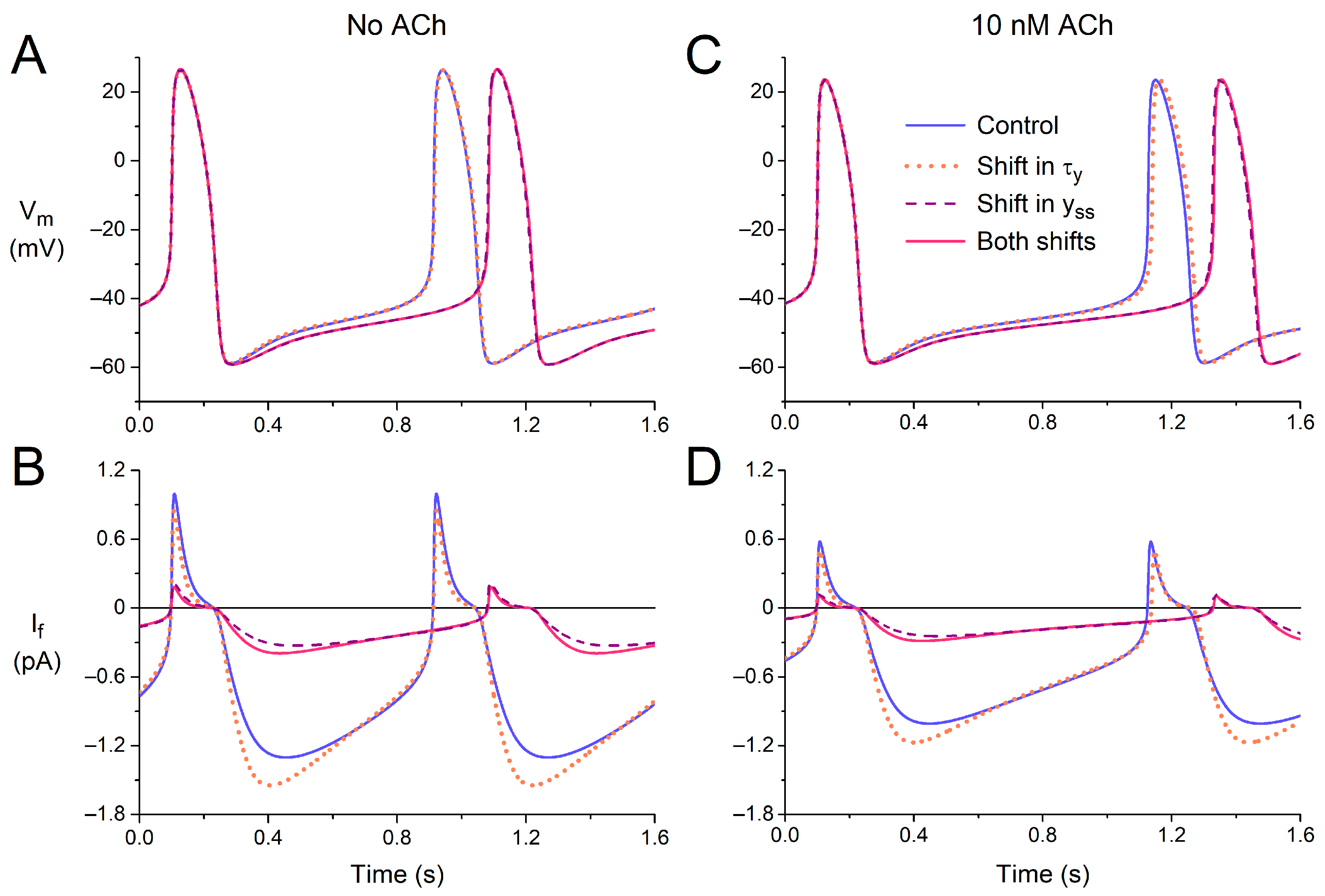
First, we tested the effects of the experimentally observed shift in the time constant of (de)activation of If (τy), both under control conditions (default model cell, no ACh) and under vagal tone (10 nM ACh). The −11.9 mV shift reduced τy in the voltage range of diastolic depolarization (Figure 4). As might be expected, the amplitude of If increased during the first half of diastolic depolarization (Figure 7A,B, blue solid and orange dotted lines) due to the increased rate of activation. However, the subsequent deactivation during the second half of diastolic depolarization was also fastened so that the amplitude of If was reduced during the second half. The overall effects of the changes in the time course of If on the action potential are small. The action potentials in the absence and presence of the shift in the voltage dependence of τy almost overlap (Figure 7A, blue solid and orange dotted lines), as illustrated by the negligible 2 ms increase in cycle length from 813 to 815 ms. Under vagal tone (10 nM ACh), the cycle length is prolonged, due to the ACh-induced inhibition of If and the concomitant activation of IK,ACh; the amplitude of If is reduced, but the effects of the −11.9 mV shift in the voltage dependence of τy are largely similar (Figure 7C,D). The cycle length now shows a 12 ms increase, from 1027 to 1039 ms (Figure 7C, blue solid and orange dotted lines).
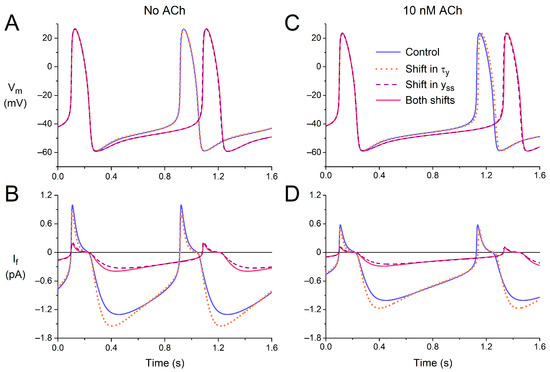
Figure 7.
Electrical activity of the Fabbri–Severi model of a single human sinus node cell in response to shifts in the voltage dependence of the steady-state activation (yss) or the (de)activation time constant (τy) of the hyperpolarization-activated current (If). (A) The membrane potential (Vm) and (B) If of the default model cell (no ACh) under control conditions (blue solid line) in response to a −11.9 mV shift in the voltage dependence of τy (orange dotted line), a −19.9 mV shift in the voltage dependence of yss (purple dashed line), or both shifts combined (pink solid line). (C) The Vm and (D) If under vagal tone (simulated administration of 10 nM ACh).
Next, we assessed the effects of the experimentally observed shift in the voltage dependence of the steady-state activation of If (yss). The −19.9 mV shift (Figure 3A,B) reduces yss in the voltage range of diastolic depolarization, thereby reducing the amount of If that can be maximally activated during diastolic depolarization (Figure 3A, inset). The amplitude of the simulated If is reduced approximately four-fold, both in the absence and presence of ACh (Figure 7B,D, purple dashed lines vs. blue solid lines), which is associated with a considerable increase in cycle length (Figure 7A,C), from 813 to 981 ms and from 1027 to 1227 ms in the absence and presence of ACh, respectively. We also tested the combined effects of the two shifts, thus assessing the functional effect of the heterozygous A414G mutation in HCN4 (Figure 7, pink vs. blue solid lines). The effects of the two shifts are near additive but determined almost entirely by the shift in the voltage dependence of yss, as demonstrated by the almost overlapping purple dashed and pink solid traces of Figure 7.
3.2.2. Effects of Atrial Load
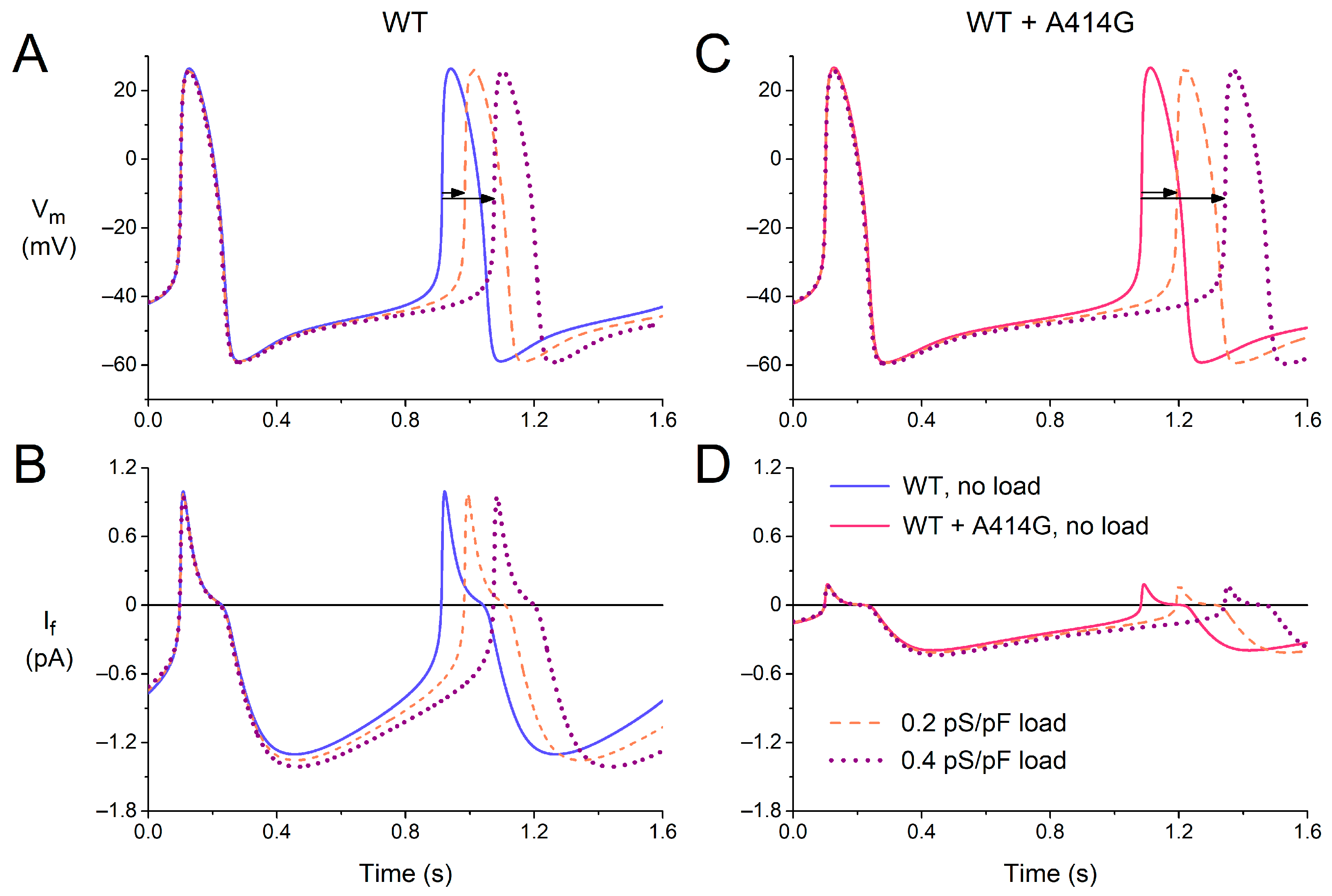
In the intact sinus node, cells in the peripheral area may have a more negative membrane potential and larger functional If than the central cells, both due to their intrinsic properties [40,41] and to the hyperpolarizing load imposed by the surrounding atrium [42]. The peripheral cells may protect themselves against this hyperpolarizing load by an increased activation of their If. To take the effects of such atrial load into account, at least in part, we introduced a passive load (simulated by an ohmic outward current with a conductance of 0.2–0.4 pS/pF and a reversal potential of −80 mV) into our cell model and tested whether the effects of the A414G mutation might be enhanced by the atrial load.
Figure 8 shows how the passive atrial load increases the cycle length of the model cell, which is only partly counteracted by an increase in If, both under the WT conditions (Figure 8A,B) and in the presence of the WT + A414G mutation in HCN4 (Figure 8C,D). Under the WT conditions, the cycle length increased by 9% and by 20% upon a passive load of 0.2 and 0.4 pS/pF, respectively (Figure 8A, horizontal arrows). In the presence of the mutation, this increase was noticeably larger, with values of 11% and 26%, respectively (Figure 8C, horizontal arrows). Apparently, the mutation-induced decrease in If makes the simulated cell less robust against the atrial load. Thus, the effects of the mutation are indeed enhanced by the atrial load. In the absence of atrial load, the mutation causes a 21% increase in cycle length (Figure 8A,C, solid lines). However, this increase becomes 24% (Figure 8A,C, dashed lines) and 27% (Figure 7A,C, dotted lines) in the presence of an atrial load of 0.2 and 0.4 pS/pF, respectively.
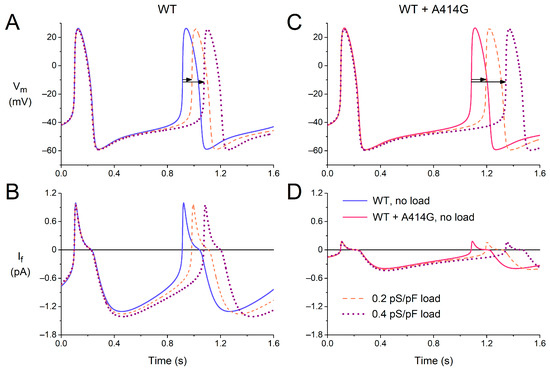
Figure 8.
Effects of a passive atrial load on the electrical activity of the Fabbri–Severi model of a single human sinus node cell under WT conditions and with changes in the characteristics of the hyperpolarization-activated current (If) induced by the WT + A414G heterozygous mutation in HCN4. (A) The membrane potential (Vm) and (B) If of the model cell under control conditions (no atrial load, blue solid line) and in response to an atrial load with a conductance of 0.2 pS/pF (orange dashed line) or 0.4 pS/pF (purple dotted line). (C) The Vm and (D) If in the presence of the mutation per se (pink solid line) or in combination with the atrial load. Horizontal arrows show the increase in cycle length induced by the atrial load.
3.2.3. Combined Effects of Atrial Load and Vagal Tone
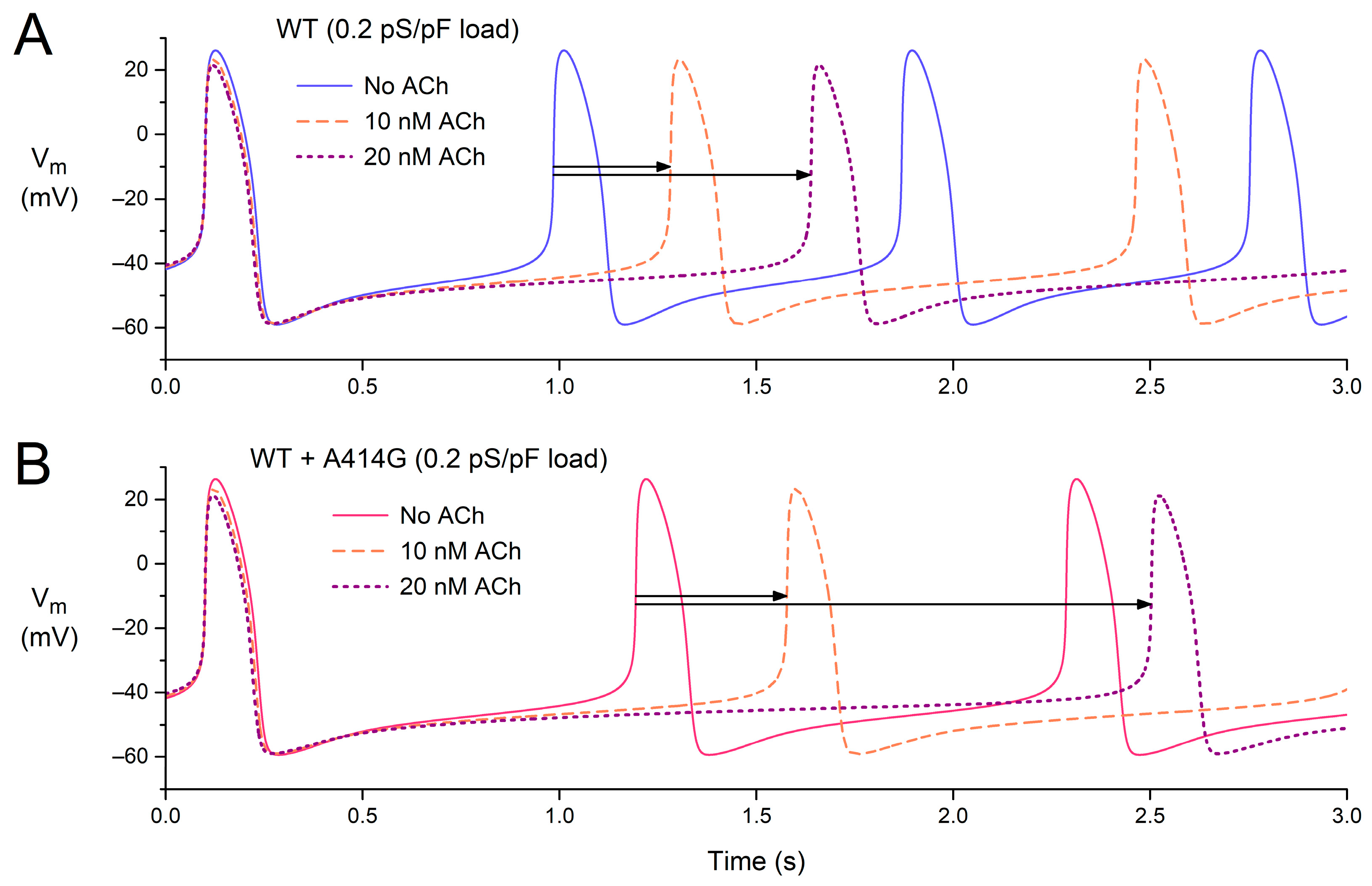
After studying the effects of vagal tone (Figure 7) and atrial load (Figure 8) separately, we also tested the effects of ACh in the presence of an atrial load. Figure 9 shows the effects of 0–20 nM ACh in the presence of an atrial load with a conductance of 0.2 pS/pF, both under WT conditions (Figure 9A) and in the presence of the WT + A414G mutation (Figure 9B).
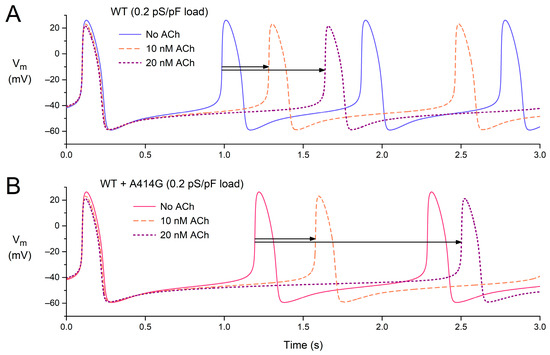
Figure 9.
Combined effects of atrial load and vagal tone on the electrical activity of the Fabbri–Severi model of a single human sinus node cell under WT conditions and with changes in the characteristics of the hyperpolarization-activated current (If) induced by the WT + A414G heterozygous mutation in HCN4. (A) The membrane potential (Vm) of the WT model cell in the presence of an atrial load at different levels of vagal tone (0, 10, and 20 nM ACh). (B) The Vm of the WT + A414G model cell in the presence of an atrial load at different levels of vagal tone. Horizontal arrows indicate the increase in cycle length in response to 10 or 20 nM ACh in the presence of the 0.2 pS/pF atrial load.
Under WT conditions, the cycle length is 885 ms in the absence of ACh (Figure 9A, blue solid line), which increases to 1182 ms (+34%) at 10 nM ACh and 1539 ms (+74%) at 20 nM ACh (Figure 9A, horizontal arrows). The percent increase is somewhat non-linear: it more than doubles with a doubling of the ACh concentration. In the presence of the WT + A414G mutation, the cycle length amounts to 1093 ms (Figure 9B, pink solid line). With 10 nM ACh, the cycle length becomes 1478 ms (+35%). Thus, the percent increase at 10 nM ACh is highly similar to that under WT conditions (Figure 9, two shortest horizontal arrows). With 20 nM ACh, however, the cycle length rises to 2403 ms (+120%). Thus, the response to ACh is highly non-linear in the case of the WT + A414G mutation, illustrating the increased sensitivity to ACh and potential vulnerability to sinus bradycardia.
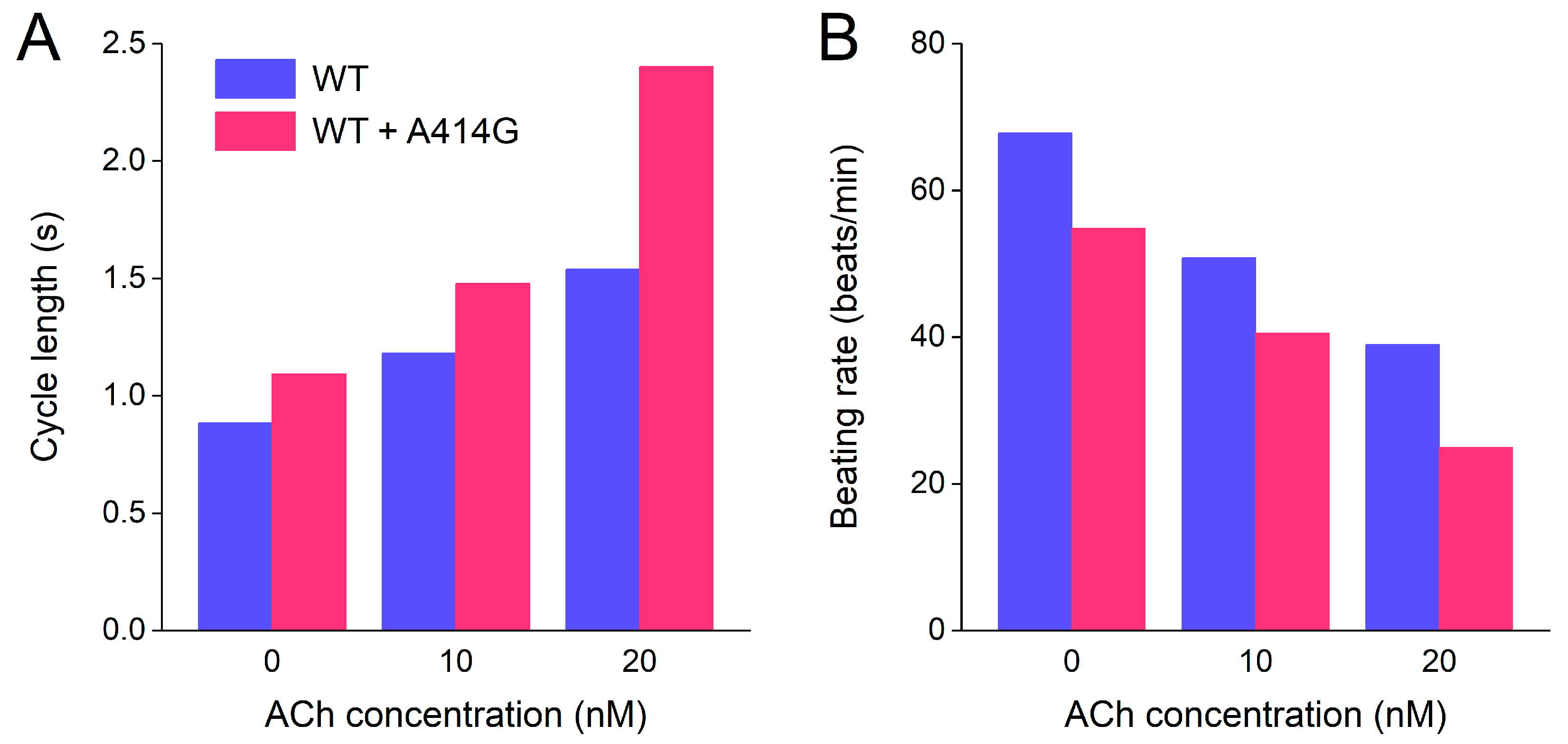
3.2.4. Beating Rate in the Presence of Atrial Load and Vagal Tone
Finally, to illustrate the effects of the WT + A414G mutation on beating rate at different levels of vagal tone, we converted the cycle lengths of Section 3.2.3, which are summarized in Figure 10A, to beating rate and created a simple bar graph (Figure 10B). Under WT conditions, the model cell fires at 68 beats/min at baseline (no ACh), which becomes 51 beats/min at 10 nM ACh and 39 beats/min at 20 nM ACh (Figure 10B, blue bars). In the case of the WT + A414G mutation, the beating rates are 55 beats/min at baseline, 41 beats/min at 10 nM ACh, and a poor 25 beats/min at 20 nM ACh (Figure 10B, pink bars). Thus, the mutant cell shows a remarkably slower beating rate at all levels of vagal tone, in particular at 20 nM ACh.
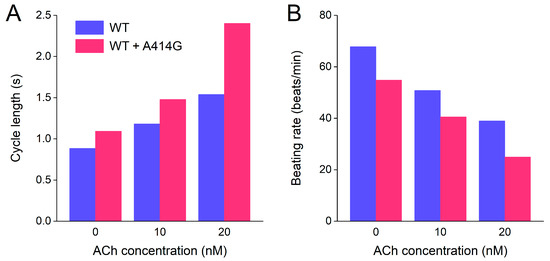
Figure 10.
Combined effects of atrial load and vagal tone on the cycle length and beating rate of the Fabbri–Severi model of a single human sinus node cell under WT conditions and with changes in the characteristics of the hyperpolarization-activated current (If) induced by the WT + A414G heterozygous mutation in HCN4. (A) Cycle length of the WT cell (blue bars) and the WT + A414G cell (pink bars) at different levels of vagal tone (0, 10, and 20 nM ACh) and in the presence of a 0.2 pS/pF atrial load. (B) Associated beating rate.
4. Discussion
4.1. Overview
We thoroughly characterized the biophysical properties of the A414G mutation in the HCN4 gene using patch-clamp methodology. We found that heterozygous mutant HCN4 channels expressed in CHO cells have a −19.9 mV shift in half-maximal activation voltage compared to WT channels. In addition, the A414G mutation shifted the voltage dependence of the time constant of (de)activation by −11.9 mV, whereas significant differences were not observed in the fully-activated current density, in the reversal potential or in the slope factor of the activation curve. Computer simulations of the effects of the experimentally identified mutation-induced changes on human sinus node pacemaker activity demonstrated a substantially increased cycle length, which was almost entirely due to the shift in the voltage dependence of steady-state activation. The cycle length increase was more prominent under vagal tone. Passive atrial load further reduced the beating rate, indicating a more prominent bradycardia in the presence of sinus node–atrial interactions. Thus, our experiments demonstrate that the altered gating properties of the HCN4-A414G channels are a valid explanation for the sinus bradycardia observed in the family described by Milano et al. [17].
4.2. Bradycardia and LVNC
The A414G mutation in HCN4 was found in 2014 by Milano et al. [17] in a small family with sinus bradycardia and LVNC. Since then, no further clinical data on HCN4-A414G mutation carriers have been published, in contrast to, for example, the G482R mutation, which was also originally presented in 2014 as the mutation in index families of Milano et al. [17] as well as Schweizer et al. [18]. Thereafter, further patient data on the latter mutation were presented by Millat et al. [19], Chanavat et al. [25], Ishikawa et al. [28], Richard et al. [26], Hanania et al. [22], Brunet-Garcia et al. [24], and Paszkowska et al. [27]. Starting with the findings by Milano et al. [17] and Schweizer et al. [18], the association of mutations in HCN4 and LVNC is now almost ten years old. LVNC is a genetically heterogeneous structural abnormality of the left ventricular myocardium [43,44]. Over one hundred genes have so far been reported in association with LVNC [45], including HCN4. Although outside the scope of the present study, it remains intriguing how mutations in HCN4 can lead to LVNC, or at least be associated with LVNC. Cannie and Elliott [44] suggested that non-compaction (or hypertrabeculation) may be a physiological response to bradycardia, as was also proposed by Milano et al. [17].
The intriguing association of functional mutations in a cardiac ion channel gene with LVNC is not limited to HCN4. In 2008, Shan et al. [46] screened patients with LVNC for genetic variants in the SCN5A gene, which encodes the pore-forming α-subunit of the cardiac fast sodium channel. They observed a high frequency of variants in SCN5A in their patients, which was significantly higher in the patients with cardiac arrhythmias, including sick sinus syndrome, than those without (50% vs. 7%: p < 0.001). On the one hand, this suggests that these variants represent a risk factor for the arrhythmias, and on the other hand, that mutations in SCN5A are somehow involved in LVNC [46].
Several studies have revealed an association between LVNC and mutations in the KCNH2 gene, which encodes the pore-forming α-subunit of the cardiac ion channel that conducts the rapid delayed rectifier potassium current (IKr). Ogawa et al. [47] reported two cases of young children with LVNC, in combination with long QT syndrome (LQTS), carrying the D501N or A561V missense mutation in KCNH2. The D501N mutation had already been identified in two unrelated patients with LQTS by Jongbloed et al. [48], whereas Bellocq et al. [49] identified the A561V mutation in an LQTS patient and demonstrated the loss-of-function effects of this mutation in patch-clamp experiments as well as in computer simulations. Recently, Xu et al. [50] and Caiffa et al. [51] each reported a small family in which a novel missense mutation in KCNH2 (T273M and V630G, respectively) was not only associated with LQTS but also with LVNC. Also recently, Maddali et al. [52] presented a case report of the well-known T613M mutation in KCNH2 in combination with LVNC. Interestingly, Sun et al. [53] recently reported a fetal case of a de novo mutation in KCNH2 (Y616C) that was not only associated with LQTS and biventricular non-compaction but also with sinus bradycardia.
In 2013, Nakashima et al. [54] presented a case report of the LQTS-related D611T mutation in the KCNQ1 gene, which encodes the pore-forming α-subunit of the cardiac ion channel that conducts the slow delayed rectifier potassium current (IKs), in combination with LVNC. A few years later, Kharbanda et al. [55] reported a family showing an association between LVNC and the LQTS-related L273F mutation in KCNQ1. Since IKs modulates pacemaker activity [56] and KCNQ1 loss-of-of function mutations may result in sinus bradycardia [57], the association between KCNQ1 mutations and LVNC may be somehow related to the bradycardia.
4.3. Limitations
In our study, we used the original If equations of the Fabbri–Severi model, thus describing the kinetics of If with a first-order Hodgkin and Huxley-type kinetic scheme [58], with identical time constants of activation and deactivation of If at a given voltage. Yet, experimental studies in which the activation and deactivation of (rabbit) If were measured at identical voltages demonstrated that If showed more complex kinetics, with differences in the time constants of activation and deactivation at a given voltage [35,59]. In the absence of sufficient quantitative data on the (de)activation of human If, we refrained from introducing the more complex kinetics of If into the Fabbri–Severi model. Also, we used recombinant heterologous expression systems for electrophysiological testing, and we simplified the complex interactions of the peripheral sinus node and the surrounding atrium to a passive atrial load. Although heterologous expression systems have been instrumental in the comparison of the electrical activity of the WT and mutant HCN4 channels [6], and the passive atrial load method has been applied for decades [60,61,62,63], the determined characteristics may not fully reflect the in vivo behavior underlying the clinical phenotype. The use of more advanced atrial cell models [64] or experimental studies using sinus node-like induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) [65,66,67,68] from patients with HCN4 mutations may prove helpful to better understand the mechanism underlying the clinically observed phenotype.
Author Contributions
Conceptualization, A.O.V. and R.W.; patch clamp experiments, A.O.V.; computer simulations, R.W.; all other aspects, A.O.V. and R.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are available to academic researchers upon request.
Acknowledgments
The authors thank Berend de Jonge for valuable biotechnical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Verkerk, A.O.; Van Ginneken, A.C.G.; Wilders, R. Pacemaker activity of the human sinoatrial node: Role of the hyperpolarization-activated current, If. Int. J. Cardiol. 2009, 132, 318–336. [Google Scholar] [CrossRef]
- Nof, E.; Antzelevitch, C.; Glikson, M. The contribution of HCN4 to normal sinus node function in humans and animal models. Pacing Clin. Electrophysiol. 2010, 33, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Baruscotti, M.; Barbuti, A.; Bucchi, A. The cardiac pacemaker current. J. Mol. Cell. Cardiol. 2010, 48, 55–64. [Google Scholar] [CrossRef] [PubMed]
- DiFrancesco, D. The role of the funny current in pacemaker activity. Circ. Res. 2010, 106, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Hennis, K.; Biel, M.; Wahl-Schott, C.; Fenske, S. Beyond pacemaking: HCN channels in sinoatrial node function. Prog. Biophys. Mol. Biol. 2021, 166, 51–60. [Google Scholar] [CrossRef]
- Hoekstra, M.; Van Ginneken, A.C.G.; Wilders, R.; Verkerk, A.O. HCN4 current during human sinoatrial node-like action potentials. Prog. Biophys. Mol. Biol. 2021, 166, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Wymore, R.; Yu, H.; Wu, J.; Wymore, R.T.; Pan, Z.; Robinson, R.B.; Dixon, J.E.; McKinnon, D.; Cohen, I.S. Distribution and prevalence of hyperpolarization-activated cation channel (HCN) mRNA expression in cardiac tissues. Circ. Res. 1999, 85, e1–e6. [Google Scholar] [CrossRef]
- Tellez, J.O.; Dobrzynski, H.; Greener, I.D.; Graham, G.M.; Laing, E.; Honjo, H.; Hubbard, S.J.; Boyett, M.R.; Billeter, R. Differential expression of ion channel transcripts in atrial muscle and sinoatrial node in rabbit. Circ. Res. 2006, 99, 1384–1393. [Google Scholar] [CrossRef]
- Brioschi, C.; Micheloni, S.; Tellez, J.O.; Pisoni, G.; Longhi, R.; Moroni, P.; Billeter, R.; Barbuti, A.; Dobrzynski, H.; Boyett, M.R.; et al. Distribution of the pacemaker HCN4 channel mRNA and protein in the rabbit sinoatrial node. J. Mol. Cell. Cardiol. 2009, 47, 221–227. [Google Scholar] [CrossRef]
- Chandler, N.J.; Greener, I.D.; Tellez, J.O.; Inada, S.; Musa, H.; Molenaar, P.; DiFrancesco, D.; Baruscotti, M.; Longhi, R.; Anderson, R.H.; et al. Molecular architecture of the human sinus node: Insights into the function of the cardiac pacemaker. Circulation 2009, 119, 1562–1575. [Google Scholar] [CrossRef]
- Schulze-Bahr, E.; Neu, A.; Friederich, P.; Kaupp, U.B.; Breithardt, G.; Pongs, O.; Isbrandt, D. Pacemaker channel dysfunction in a patient with sinus node disease. J. Clin. Investig. 2003, 111, 1537–1545. [Google Scholar] [CrossRef] [PubMed]
- Baruscotti, M.; Bottelli, G.; Milanesi, R.; DiFrancesco, J.C.; DiFrancesco, D. HCN-related channelopathies. Pflügers Arch. 2010, 460, 405–415. [Google Scholar] [CrossRef] [PubMed]
- DiFrancesco, D. Funny channel gene mutations associated with arrhythmias. J. Physiol. 2013, 591, 4117–4124. [Google Scholar] [CrossRef] [PubMed]
- Verkerk, A.O.; Wilders, R. Pacemaker activity of the human sinoatrial node: Effects of HCN4 mutations on the hyperpolarization-activated current. Europace 2014, 16, 384–395. [Google Scholar] [CrossRef]
- Verkerk, A.O.; Wilders, R. Pacemaker activity of the human sinoatrial node: An update on the effects of mutations in HCN4 on the hyperpolarization-activated current. Int. J. Mol. Sci. 2015, 16, 3071–3094. [Google Scholar] [CrossRef] [PubMed]
- DiFrancesco, D. HCN4, sinus bradycardia and atrial fibrillation. Arrhythmia Electrophysiol. Rev. 2015, 4, 9–13. [Google Scholar] [CrossRef]
- Milano, A.; Vermeer, A.M.C.; Lodder, E.M.; Barc, J.; Verkerk, A.O.; Postma, A.V.; Van der Bilt, I.A.C.; Baars, M.J.H.; Van Haelst, P.L.; Caliskan, K.; et al. HCN4 mutations in multiple families with bradycardia and left ventricular noncompaction cardiomyopathy. J. Am. Coll. Cardiol. 2014, 64, 745–756. [Google Scholar] [CrossRef]
- Schweizer, P.A.; Schröter, J.; Greiner, S.; Haas, J.; Yampolsky, P.; Mereles, D.; Buss, S.J.; Seyler, C.; Bruehl, C.; Draguhn, A.; et al. The symptom complex of familial sinus node dysfunction and myocardial noncompaction is associated with mutations in the HCN4 channel. J. Am. Coll. Cardiol. 2014, 64, 757–767. [Google Scholar] [CrossRef]
- Millat, G.; Janin, A.; De Tauriac, O.; Roux, A.; Dauphin, C. HCN4 mutation as a molecular explanation on patients with bradycardia and non-compaction cardiomyopathy. Eur. J. Med. Genet. 2015, 58, 439–442. [Google Scholar] [CrossRef]
- Vermeer, A.M.C.; Lodder, E.M.; Thomas, D.; Duijkers, F.A.M.; Marcelis, C.; Van Gorselen, E.O.F.; Fortner, P.; Buss, S.J.; Mereles, D.; Katus, H.A.; et al. Dilation of the aorta ascendens forms part of the clinical spectrum of HCN4 mutations. J. Am. Coll. Cardiol. 2016, 67, 2313–2315. [Google Scholar] [CrossRef]
- Servatius, H.; Porro, A.; Pless, S.A.; Schaller, A.; Asatryan, B.; Tanner, H.; De Marchi, S.F.; Roten, L.; Seiler, J.; Haeberlin, A.; et al. Phenotypic spectrum of HCN4 mutations: A clinical case. Circ. Genom. Precis. Med. 2018, 11, e002033. [Google Scholar] [CrossRef]
- Hanania, H.L.; Regalado, E.S.; Guo, D.-C.; Xu, L.; Demo, E.; Sallee, D.; Milewicz, D.M. Do HCN4 variants predispose to thoracic aortic aneurysms and dissections? Circ. Genom. Precis. Med. 2019, 12, e002626. [Google Scholar] [CrossRef]
- Alonso-Fernández-Gatta, M.; Gallego-Delgado, M.; Caballero, R.; Villacorta, E.; Díaz-Peláez, E.; García-Berrocal, B.; Crespo-García, T.; Plata-Izquierdo, B.; Marcos-Vadillo, E.; García-Cuenllas, L.; et al. A rare HCN4 variant with combined sinus bradycardia, left atrial dilatation, and hypertrabeculation/left ventricular noncompaction phenotype. Rev. Esp. Cardiol. 2021, 74, 781–789. [Google Scholar] [CrossRef]
- Brunet-Garcia, L.; Odori, A.; Fell, H.; Field, E.; Roberts, A.M.; Starling, L.; Kaski, J.P.; Cervi, E. Noncompaction cardiomyopathy, sick sinus disease, and aortic dilatation: Too much for a single diagnosis? JACC Case Rep. 2022, 4, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Chanavat, V.; Janin, A.; Millat, G. A fast and cost-effective molecular diagnostic tool for genetic diseases involved in sudden cardiac death. Clin. Chim. Acta 2016, 453, 80–85. [Google Scholar] [CrossRef]
- Richard, P.; Ader, F.; Roux, M.; Donal, E.; Eicher, J.C.; Aoutil, N.; Huttin, O.; Selton-Suty, C.; Coisne, D.; Jondeau, G.; et al. Targeted panel sequencing in adult patients with left ventricular non-compaction reveals a large genetic heterogeneity. Clin. Genet. 2019, 95, 356–367. [Google Scholar] [CrossRef]
- Paszkowska, A.; Piekutowska-Abramczuk, D.; Ciara, E.; Mirecka-Rola, A.; Brzezinska, M.; Wicher, D.; Kostrzewa, G.; Sarnecki, J.; Ziółkowska, L. Clinical presentation of left ventricular noncompaction cardiomyopathy and bradycardia in three families carrying HCN4 pathogenic variants. Genes 2022, 13, 477. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, T.; Ohno, S.; Murakami, T.; Yoshida, K.; Mishima, H.; Fukuoka, T.; Kimoto, H.; Sakamoto, R.; Ohkusa, T.; Aiba, T.; et al. Sick sinus syndrome with HCN4 mutations shows early onset and frequent association with atrial fibrillation and left ventricular noncompaction. Heart Rhythm 2017, 14, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, R.; Kinoshita, K.; Hata, Y.; Abe, M.; Matsuoka, K.; Hirono, K.; Kano, M.; Nakazawa, M.; Ichida, F.; Nishida, N.; et al. A mutant HCN4 channel in a family with bradycardia, left bundle branch block, and left ventricular noncompaction. Heart Vessel. 2018, 33, 802–819. [Google Scholar] [CrossRef]
- Ponińska, J.; Michalak, E.; Śpiewak, M.; Lutyńska, A.; Płoski, R.; Bilińska, Z.T. A novel HCN4 variant related to familial sinus bradycardia, left ventricular noncompaction, and thoracic aortic aneurysm. Pol. Arch. Intern. Med. 2021, 131, 70–72. [Google Scholar] [CrossRef]
- Weigl, I.; Geschwill, P.; Reiss, M.; Bruehl, C.; Draguhn, A.; Koenen, M.; Sedaghat-Hamedani, F.; Meder, B.; Thomas, D.; Katus, H.A.; et al. The C-terminal HCN4 variant P883R alters channel properties and acts as genetic modifier of atrial fibrillation and structural heart disease. Biochem. Biophys. Res. Commun. 2019, 519, 141–147. [Google Scholar] [CrossRef]
- Fabbri, A.; Fantini, M.; Wilders, R.; Severi, S. Computational analysis of the human sinus node action potential: Model development and effects of mutations. J. Physiol. 2017, 595, 2365–2396. [Google Scholar] [CrossRef] [PubMed]
- Verkerk, A.O.; Wilders, R. Mechanism of sinus bradycardia in carriers of the A414G mutation in the HCN4 gene. Comp. Cardiol. 2019, 46, 147. [Google Scholar] [CrossRef]
- DiFrancesco, D.; Ferroni, A.; Mazzanti, M.; Tromba, C. Properties of the hyperpolarizing-activated current (if) in cells isolated from the rabbit sino-atrial node. J. Physiol. 1986, 377, 61–88. [Google Scholar] [CrossRef] [PubMed]
- Van Ginneken, A.C.G.; Giles, W. Voltage clamp measurements of the hyperpolarization-activated inward current If in single cells from rabbit sino-atrial node. J. Physiol. 1991, 434, 57–83. [Google Scholar] [CrossRef]
- Lloyd, C.M.; Lawson, J.R.; Hunter, P.J.; Nielsen, P.F. The CellML model repository. Bioinformatics 2008, 24, 2122–2123. [Google Scholar] [CrossRef]
- Cuellar, A.A.; Lloyd, C.M.; Nielsen, P.F.; Bullivant, D.P.; Nickerson, D.P.; Hunter, P.J. An overview of CellML 1.1, a biological model description language. Simulation 2003, 79, 740–747. [Google Scholar] [CrossRef]
- Garny, A.; Kohl, P.; Noble, D. Cellular open resource (COR): A public CellML based environment for modelling biological function. Int. J. Bifurc. Chaos 2003, 13, 3579–3590. [Google Scholar] [CrossRef]
- Verkerk, A.O.; Den Ruijter, H.M.; Bourier, J.; Boukens, B.J.; Brouwer, I.A.; Wilders, R.; Coronel, R. Dietary fish oil reduces pacemaker current and heart rate in rabbit. Heart Rhythm 2009, 6, 1485–1492. [Google Scholar] [CrossRef]
- Kodama, I.; Boyett, M.R. Regional differences in the electrical activity of the rabbit sinus node. Pflügers Arch. 1985, 404, 214–226. [Google Scholar] [CrossRef]
- Boyett, M.R.; Honjo, H.; Kodama, I. The sinoatrial node, a heterogeneous pacemaker structure. Cardiovasc. Res. 2000, 47, 658–687. [Google Scholar] [CrossRef]
- Kirchhof, C.J.H.J.; Bonke, F.I.M.; Allessie, M.A.; Lammers, W.J.E.P. The influence of the atrial myocardium on impulse formation in the rabbit sinus node. Pflügers Arch. 1987, 410, 198–203. [Google Scholar] [CrossRef]
- Towbin, J.A.; Lorts, A.; Jefferies, J.L. Left ventricular non-compaction cardiomyopathy. Lancet 2015, 386, 813–825. [Google Scholar] [CrossRef]
- Cannie, D.; Elliott, P. The genetics of left ventricular noncompaction. Curr. Opin. Cardiol. 2021, 36, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Finsterer, J.; Stöllberger, C. Left ventricular noncompaction syndrome: Genetic insights and therapeutic perspectives. Curr. Cardiol. Rep. 2020, 22, 84. [Google Scholar] [CrossRef] [PubMed]
- Shan, L.; Makita, N.; Xing, Y.; Watanabe, S.; Futatani, T.; Ye, F.; Saito, K.; Ibuki, K.; Watanabe, K.; Hirono, K.; et al. SCN5A variants in Japanese patients with left ventricular noncompaction and arrhythmia. Mol. Genet. Metab. 2008, 93, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, K.; Nakamura, Y.; Terano, K.; Ando, T.; Hishitani, T.; Hoshino, K. Isolated non-compaction of the ventricular myocardium associated with long QT syndrome: A report of 2 cases. Circ. J. 2009, 73, 2169–2172. [Google Scholar] [CrossRef]
- Jongbloed, R.; Marcelis, C.; Velter, C.; Doevendans, P.; Geraedts, J.; Smeets, H. DHPLC analysis of potassium ion channel genes in congenital long QT syndrome. Hum. Mutat. 2002, 20, 382–391. [Google Scholar] [CrossRef]
- Bellocq, C.; Wilders, R.; Schott, J.-J.; Louérat-Oriou, B.; Boisseau, P.; Le Marec, H.; Escande, D.; Baró, I. A common antitussive drug, clobutinol, precipitates the long QT syndrome 2. Mol. Pharmacol. 2004, 66, 1093–1102. [Google Scholar] [CrossRef]
- Xu, B.; Li, K.; Liu, F.; Kong, L.; Yang, J.; Zhou, B.; Lv, T.; Liu, Y.; She, F.; He, R.; et al. Mexiletine shortened QT interval and reduced ventricular arrhythmias in a pedigree of type 2 long QT syndrome combined with left ventricular non-compaction. Int. Heart J. 2021, 62, 427–431. [Google Scholar] [CrossRef]
- Caiffa, T.; Tessitore, A.; Leoni, L.; Reffo, E.; Chicco, D.; D’Agata Mottolese, B.; Rubinato, E.; Girotto, G.; Lenarduzzi, S.; Barbi, E.; et al. Long QT syndrome and left ventricular non-compaction in a family with KCNH2 mutation: A case report. Front. Pediatr. 2022, 10, 970240. [Google Scholar] [CrossRef] [PubMed]
- Maddali, M.M.; Thomas, E.; Al Abri, I.A.; Patel, M.H.; Al Maskari, S.N.; Al Yamani, M.I. Dilated cardiomyopathy phenotype-associated left ventricular noncompaction and congenital long QT syndrome type-2 in infants with KCNH2 gene mutation: Anesthetic considerations. J. Cardiothorac. Vasc. Anesth. 2022, 36, 3662–3667. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Liu, X.; Hao, X.; Zhou, X.; Wang, J.; Han, J.; Liang, M.; Zhang, H.; He, Y. Case report: Biventricular noncompaction cardiomyopathy with pulmonary stenosis and bradycardia in a fetus with KCNH2 mutation. Front. Genet. 2022, 13, 821226. [Google Scholar] [CrossRef]
- Nakashima, K.; Kusakawa, I.; Yamamoto, T.; Hirabayashi, S.; Hosoya, R.; Shimizu, W.; Sumitomo, N. A left ventricular noncompaction in a patient with long QT syndrome caused by a KCNQ1 mutation: A case report. Heart Vessel. 2013, 28, 126–129. [Google Scholar] [CrossRef]
- Kharbanda, M.; Hunter, A.; Tennant, S.; Moore, D.; Curtis, S.; Hancox, J.C.; Murday, V. Long QT syndrome and left ventricular noncompaction in 4 family members across 2 generations with KCNQ1 mutation. Eur. J. Med. Genet. 2017, 60, 233–238. [Google Scholar] [CrossRef]
- Verkerk, A.O.; Wilders, R. Human sinoatrial node pacemaker activity: Role of the slow component of the delayed rectifier K+ current, IKs. Int. J. Mol. Sci. 2023, 24, 7264. [Google Scholar] [CrossRef]
- Wilders, R.; Verkerk, A.O. Long QT syndrome and sinus bradycardia–A mini review. Front. Cardiovasc. Med. 2018, 5, 106. [Google Scholar] [CrossRef]
- Hodgkin, A.L.; Huxley, A.F. A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. 1952, 117, 500–544. [Google Scholar] [CrossRef] [PubMed]
- DiFrancesco, D. The cardiac hyperpolarizing-activated current, if. Origins and developments. Prog. Biophys. Mol. Biol. 1985, 46, 163–183. [Google Scholar] [CrossRef] [PubMed]
- Tan, R.C.; Joyner, R.W. Electrotonic influences on action potentials from isolated ventricular cells. Circ. Res. 1990, 67, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, E.I.; Honjo, H.; Anno, T.; Boyett, M.R.; Kodama, I.; Toyama, J. Modulation of pacemaker activity of sinoatrial node cells by electrical load imposed by an atrial cell model. Am. J. Physiol. 1995, 269, H1735–H1742. [Google Scholar] [CrossRef] [PubMed]
- Kohl, P.; Noble, D.; Winslow, R.L.; Hunter, P.J. Computational modelling of biological systems: Tools and visions. Philos. Trans. A Math. Phys. Eng. Sci. 2000, 358, 579–610. [Google Scholar] [CrossRef]
- Clayton, R.H.; Bernus, O.; Cherry, E.M.; Dierckx, H.; Fenton, F.H.; Mirabella, L.; Panfilov, A.V.; Sachse, F.B.; Seemann, G.; Zhang, H. Models of cardiac tissue electrophysiology: Progress, challenges and open questions. Prog. Biophys. Mol. Biol. 2011, 104, 22–48. [Google Scholar] [CrossRef]
- Maleckar, M.M.; Greenstein, J.L.; Trayanova, N.A.; Giles, W.R. Mathematical simulations of ligand-gated and cell-type specific effects on the action potential of human atrium. Prog. Biophys. Mol. Biol. 2008, 98, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Peischard, S.; Möller, M.; Disse, P.; Ho, H.T.; Verkerk, A.O.; Strutz-Seebohm, N.; Budde, T.; Meuth, S.G.; Schweizer, P.A.; Morris, S.; et al. Virus-induced inhibition of cardiac pacemaker channel HCN4 triggers bradycardia in human-induced stem cell system. Cell. Mol. Life Sci. 2022, 79, 440. [Google Scholar] [CrossRef]
- Wiesinger, A.; Li, J.; Fokkert, L.; Bakker, P.; Verkerk, A.O.; Christoffels, V.M.; Boink, G.J.J.; Devalla, H.D. A single cell transcriptional roadmap of human pacemaker cell differentiation. Elife 2022, 11, e76781. [Google Scholar] [CrossRef]
- Li, J.; Wiesinger, A.; Fokkert, L.; Boukens, B.J.; Verkerk, A.O.; Christoffels, V.M.; Boink, G.J.J.; Devalla, H.D. Molecular and electrophysiological evaluation of human cardiomyocyte subtypes to facilitate generation of composite cardiac models. J. Tissue Eng. 2022, 13, 20417314221127908. [Google Scholar] [CrossRef]
- Li, J.; Wiesinger, A.; Fokkert, L.; Boukens, B.; Verkerk, A.; Christoffels, V.; Boink, G.; Devalla, H. Comparative characterization of single cells and engineered heart tissues from hiPSC-derived cardiomyocyte subtypes. Cardiovasc. Res. 2022, 118 (Suppl. S1), cvac066.024. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).