Lower Circulating Cell-Free Mitochondrial DNA Is Associated with Heart Failure in Type 2 Diabetes Mellitus Patients
Highlights
- We found that bidirectional changes (an increase in the nuclear fraction and a decrease in the mitochondrial fraction) in patients with type 2 diabetes mellitus are associated with adverse cardiac remodeling, renal damage and inflammation.
- We found a superior discriminatory ability in cell-free nuclear DNA compared with N-terminal brain natriuretic peptide for heart failure in patients with type 2 diabetes mellitus.
- They may improve the conventional predictive models for the development of any phenotype of chronic heart failure in diabetics.
- They suggest a new methodology for the identification of patients and the application of early treatment responses through serial measuring of circulating levels of cell-free-DNA.
Abstract
1. Introduction
2. Materials and Methods
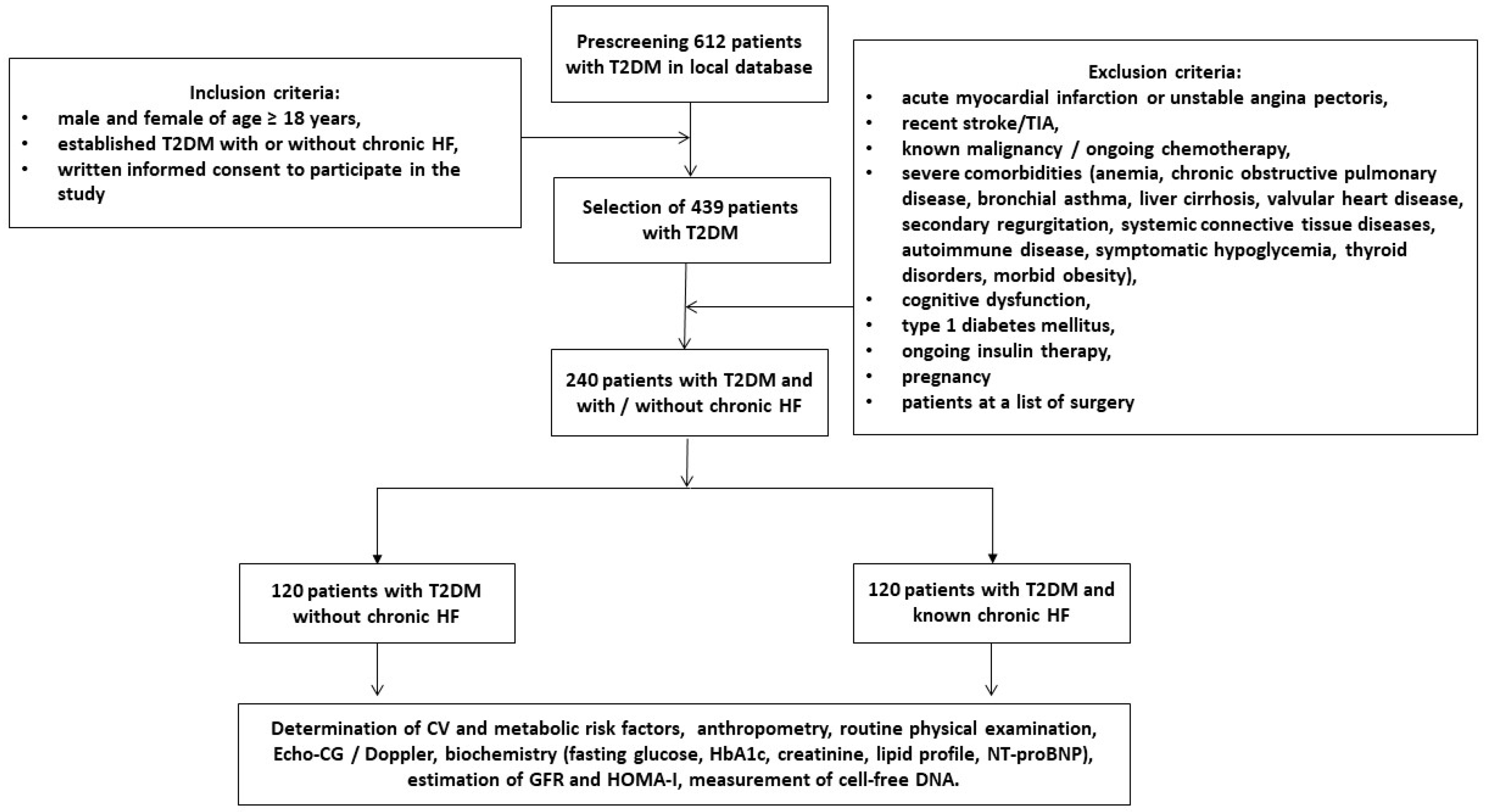
2.1. Study Patients
2.2. Medical Information Collection
2.3. Examination of Hemodynamics
2.4. Blood Sampling and Biomarker Measures
2.5. Cell-Free DNA Extraction
2.6. Measurement of Cell-Free DNAs in Plasma Samples
2.7. Statistical Analysis
3. Results
3.1. General Patient Characteristics
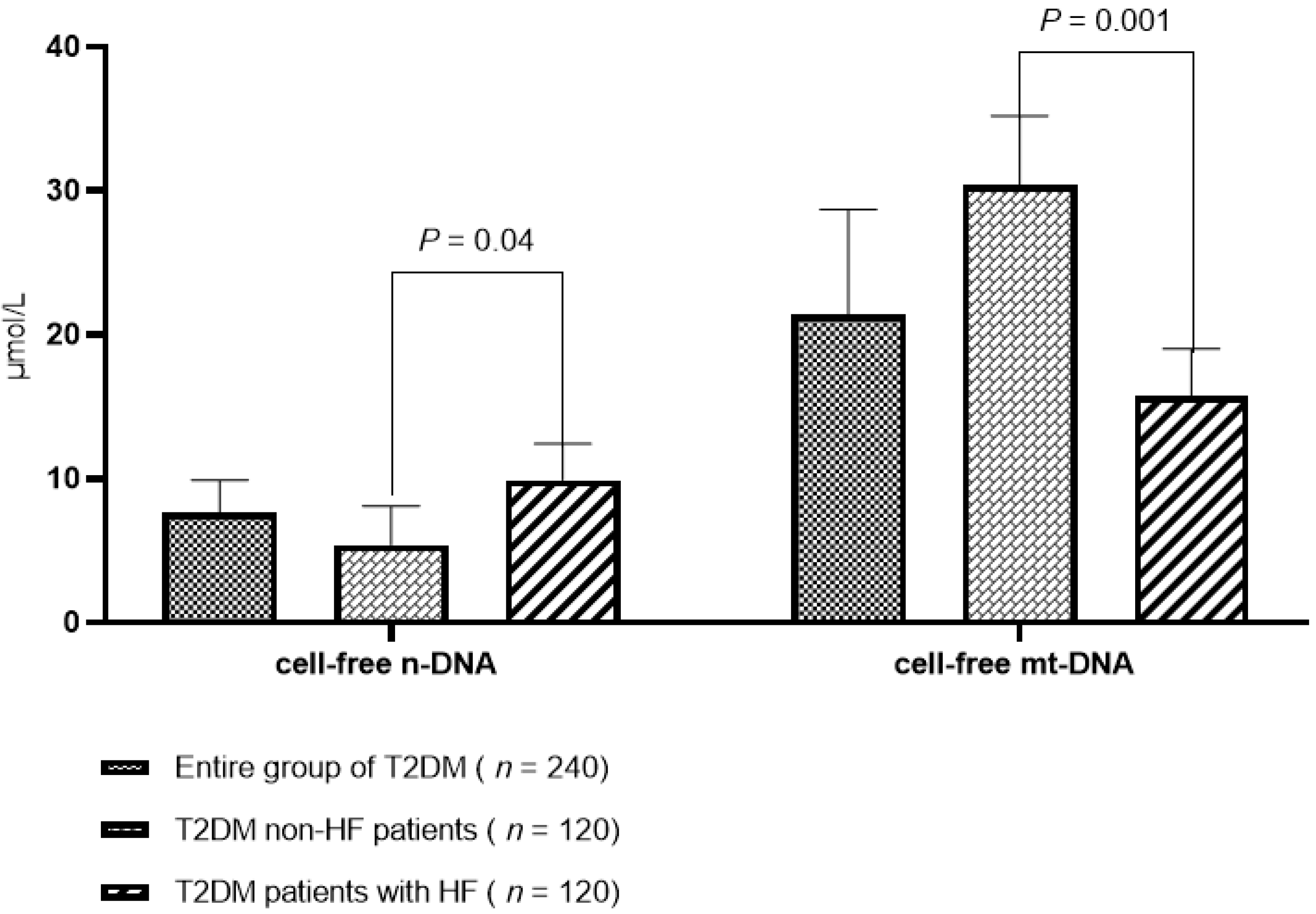
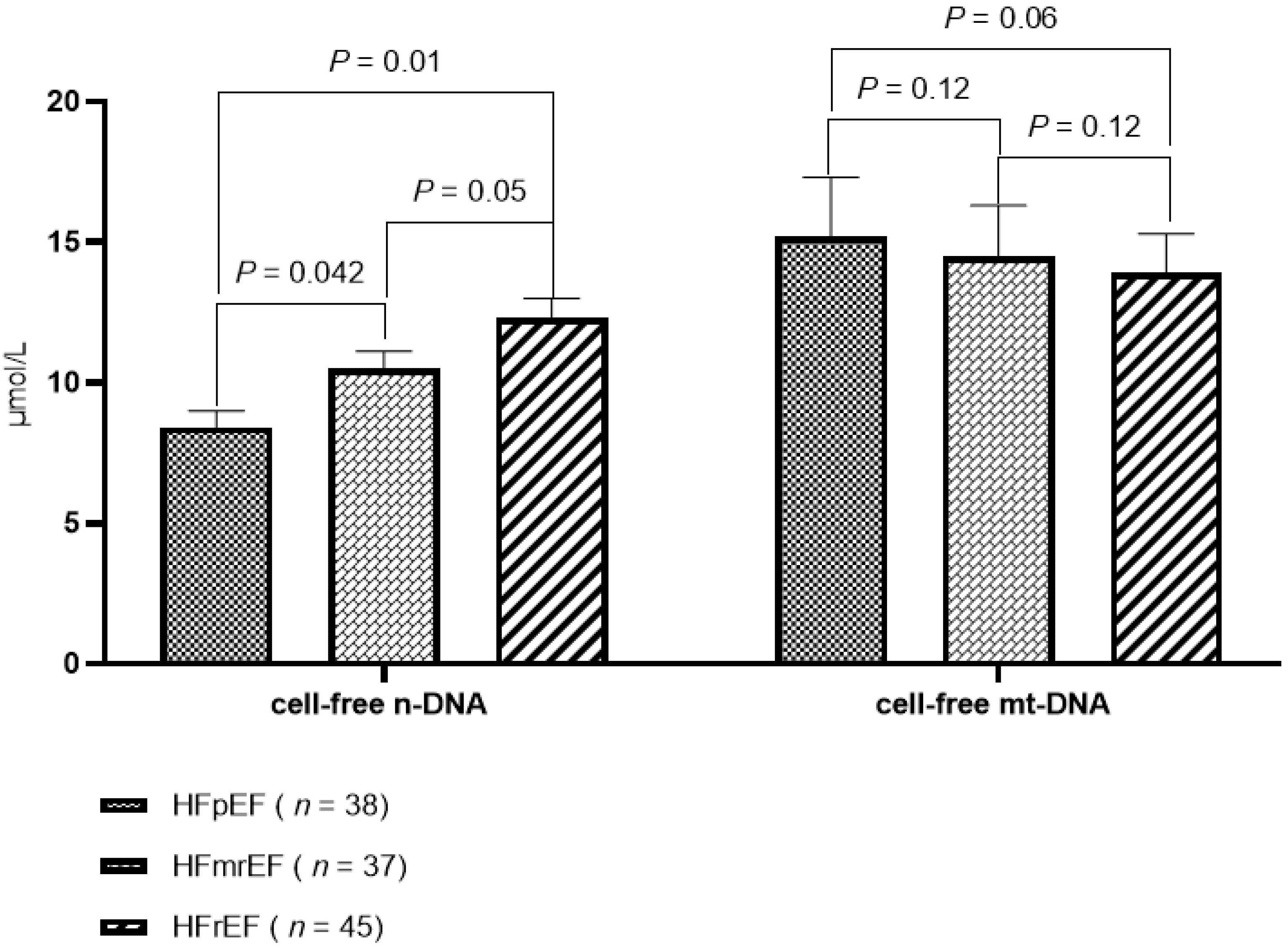
3.2. The Levels of Circulating Cell-Free DNAs
3.3. Spearman’s Correlation between the Circulating Levels of Cell-Free DNA and Other Parameters
3.4. The Factors Associated with HF in T2DM Patients: The Univariate and Multivariate Logistic Regression
3.5. Comparison of the Models
4. Discussion
5. Study Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yap, J.; Tay, W.T.; Teng, T.K.; Anand, I.; Richards, A.M.; Ling, L.H.; MacDonald, M.R.; Chandramouli, C.; Tromp, J.; Siswanto, B.B.; et al. ASIAN-HF (Asian Sudden Cardiac Death in Heart Failure) Registry Investigators. Association of Diabetes Mellitus on Cardiac Remodeling, Quality of Life, and Clinical Outcomes in Heart Failure With Reduced and Preserved Ejection Fraction. J. Am. Heart Assoc. 2019, 8, e013114. [Google Scholar] [CrossRef] [PubMed]
- Kannel, W.B.; McGee, D.L. Diabetes and cardiovascular disease. The Framingham study. JAMA 1979, 241, 2035–2038. [Google Scholar] [CrossRef] [PubMed]
- Kenny, H.C.; Abel, E.D. Heart Failure in Type 2 Diabetes Mellitus. Circ. Res. 2019, 124, 121–141. [Google Scholar] [CrossRef]
- Tromp, J.; Paniagua, S.M.A.; Lau, E.S.; Allen, N.B.; Blaha, M.J.; Gansevoort, R.T.; Hillege, H.L.; Lee, D.E.; Levy, D.; Vasan, R.S.; et al. Age dependent associations of risk factors with heart failure: Pooled population based cohort study. BMJ 2021, 372, n461. [Google Scholar] [CrossRef]
- Randhawa, V.K.; Dhanvantari, S.; Connelly, K.A. How Diabetes and Heart Failure Modulate Each Other and Condition Management. Can. J. Cardiol. 2021, 37, 595–608. [Google Scholar] [CrossRef]
- Park, J.J. Epidemiology, Pathophysiology, Diagnosis and Treatment of Heart Failure in Diabetes. Diabetes Metab. J. 2021, 45, 146–157. [Google Scholar] [CrossRef]
- Pop-Busui, R.; Januzzi, J.L.; Bruemmer, D.; Butalia, S.; Green, J.B.; Horton, W.B.; Knight, C.; Levi, M.; Rasouli, N.; Richardson, C.R. Heart Failure: An Underappreciated Complication of Diabetes. A Consensus Report of the American Diabetes Association. Diabetes Care 2022, 45, 1670–1690. [Google Scholar] [CrossRef]
- Berezin, A. Metabolic memory phenomenon in diabetes mellitus: Achieving and perspectives. Diabetes Metab. Syndr. 2016, 10 (Suppl. S1), S176–S183. [Google Scholar] [CrossRef]
- Caprnda, M.; Mesarosova, D.; Ortega, P.F.; Krahulec, B.; Egom, E.; Rodrigo, L.; Kruzliak, P.; Mozos, I.; Gaspar, L. Glycemic Variability and Vascular Complications in Patients with Type 2 Diabetes Mellitus. Folia Med. 2017, 59, 270–278. [Google Scholar] [CrossRef]
- Cardoso, C.R.L.; Leite, N.C.; Moram, C.B.M.; Salles, G.F. Long-term visit-to-visit glycemic variability as predictor of micro- and macrovascular complications in patients with type 2 diabetes: The Rio de Janeiro Type 2 Diabetes Cohort Study. Cardiovasc. Diabetol. 2018, 17, 33. [Google Scholar] [CrossRef]
- Berezin, A. Neutrophil extracellular traps: The core player in vascular complications of diabetes mellitus. Diabetes Metab. Syndr. 2019, 13, 3017–3023. [Google Scholar] [CrossRef] [PubMed]
- Del Re, D.P.; Amgalan, D.; Linkermann, A.; Liu, Q.; Kitsis, R.N. Fundamental Mechanisms of Regulated Cell Death and Implications for Heart Disease. Physiol. Rev. 2019, 99, 1765–1817. [Google Scholar] [CrossRef] [PubMed]
- De Miranda, F.S.; Barauna, V.G.; Dos Santos, L.; Costa, G.; Vassallo, P.F.; Campos, L.C.G. Properties and Application of Cell-Free DNA as a Clinical Biomarker. Int. J. Mol. Sci. 2021, 22, 9110. [Google Scholar] [CrossRef] [PubMed]
- Bronkhorst, A.J.; Ungerer, V.; Diehl, F.; Anker, P.; Dor, Y.; Fleischhacker, M.; Gahan, P.B.; Hui, L.; Holdenrieder, S.; Thierry, A.R. Towards systematic nomenclature for cell-free DNA. Hum. Genet. 2021, 140, 565–578. [Google Scholar] [CrossRef] [PubMed]
- De Bont, C.M.; Boelens, W.C.; Pruijn, G.J.M. NETosis, complement, and coagulation: A triangular relationship. Cell Mol. Immunol. 2019, 16, 19–27. [Google Scholar] [CrossRef] [PubMed]
- De Vlaminck, I.; Valantine, H.A.; Snyder, T.M.; Strehl, C.; Cohen, G.; Luikart, H.; Neff, N.F.; Okamoto, J.; Bernstein, D.; Weisshaar, D.; et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci. Transl. Med. 2014, 6, 241ra77. [Google Scholar] [CrossRef]
- Dawson, S.-J.; Tsui, D.W.; Murtaza, M.; Biggs, H.; Rueda, O.M.; Chin, S.-F.; Dunning, M.J.; Gale, D.; Forshew, T.; Mahler-Araujo, B.; et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013, 368, 1199–1209. [Google Scholar] [CrossRef]
- Antonatos, D.; Patsilinakos, S.; Spanodimos, S.; Korkonikitas, P.; Tsigas, D. Cell-free DNA levels as a prognostic marker in acute myocardial infarction. Ann. N. Y. Acad. Sci. 2006, 1075, 278–281. [Google Scholar] [CrossRef]
- Cheng, Z.; Abrams, S.T.; Toh, J.; Wang, S.S.; Wang, Z.; Yu, Q.; Yu, W.; Toh, C.H.; Wang, G. The Critical Roles and Mechanisms of Immune Cell Death in Sepsis. Front. Immunol. 2020, 11, 1918. [Google Scholar] [CrossRef]
- Menegazzo, L.; Ciciliot, S.; Poncina, N.; Mazzucato, M.; Persano, M.; Bonora, B.; Albiero, M.; Vigili de Kreutzenberg, S.; Avogaro, A.; Fadini, G.P. NETosis is induced by high glucose and associated with type 2 diabetes. Acta Diabetol. 2015, 52, 497–503. [Google Scholar] [CrossRef]
- Dou, H.; Kotini, A.; Liu, W.; Fidler, T.; Endo-Umeda, K.; Sun, X.; Olszewska, M.; Xiao, T.; Abramowicz, S.; Yalcinkaya, M.; et al. Oxidized Phospholipids Promote NETosis and Arterial Thrombosis in LNK(SH2B3) Deficiency. Circulation 2021, 144, 1940–1954. [Google Scholar] [CrossRef]
- Njeim, R.; Azar, W.S.; Fares, A.H.; Azar, S.T.; Kfoury Kassouf, H.; Eid, A.A. NETosis contributes to the pathogenesis of diabetes and its complications. J. Mol. Endocrinol. 2020, 65, R65–R76. [Google Scholar] [CrossRef] [PubMed]
- Ling, S.; Xu, J.W. NETosis as a Pathogenic Factor for Heart Failure. Oxid. Med. Cell. Longev. 2021, 2021, 6687096. [Google Scholar] [CrossRef] [PubMed]
- Yokokawa, T.; Misaka, T.; Kimishima, Y.; Shimizu, T.; Kaneshiro, T.; Takeishi, Y. Clinical Significance of Circulating Cardiomyocyte-Specific Cell-Free DNA in Patients With Heart Failure: A Proof-of-Concept Study. Can. J. Cardiol. 2020, 36, 931–935. [Google Scholar] [CrossRef]
- Ren, J.; Jiang, L.; Liu, X.; Liao, Y.; Zhao, X.; Tang, F.; Yu, H.; Shao, Y.; Wang, J.; Wen, L.; et al. Heart-specific DNA methylation analysis in plasma for the investigation of myocardial damage. J. Transl. Med. 2022, 20, 36. [Google Scholar] [CrossRef]
- Toto, R.D. Microalbuminuria: Definition, detection, and clinical significance. J. Clin. Hypertens. 2004, 6 (Suppl. S3), 2–7. [Google Scholar] [CrossRef]
- American Diabetes Association. 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes—2021. Diabetes Care 2021, 44 (Suppl. S1), S15–S33. [Google Scholar] [CrossRef] [PubMed]
- McDonagh, T.A.; Metra, M.; Adamo, M.; Gardner, R.S.; Baumbach, A.; Bohm, M.; Burri, H.; Butler, J.; Čelutkienė, J.; Chioncel, O.; et al. 2021 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur. Heart J. 2021, 42, 3599–3726. [Google Scholar] [CrossRef]
- Williams, B.; Mancia, G.; Spiering, W.; Agabiti Rosei, E.; Azizi, M.; Burnier, M.; Burnier, M.; Clement, D.L.; Coca, A.; de Simone, G.; et al. 2018 ESC/ESH Guidelines for the management of arterial hypertension. Eur. Heart J. 2018, 39, 3021–3104. [Google Scholar] [CrossRef]
- Mach, F.; Baigent, C.; Catapano, A.L.; Koskinas, K.C.; Casula, M.; Badimon, L.; Chapman, M.J.; De Backer, G.G.; Delgado, V.; Ference, B.A.; et al. 2019 ESC/EAS Guidelines for the management of dyslipidaemias: Lipid modification to reduce cardiovascular risk. Eur. Heart J. 2020, 41, 111–188. [Google Scholar] [CrossRef]
- Knuuti, J.; Wijns, W.; Saraste, A.; Capodanno, D.; Barbato, E.; Funck-Brentano, C.; Prescott, E.; Storey, R.F.; Deaton, C.; Cuisset, T.; et al. 2019 ESC Guidelines for the diagnosis and management of chronic coronary syndromes. Eur. Heart J. 2020, 41, 407–477. [Google Scholar] [CrossRef] [PubMed]
- De Boer, I.H.; Khunti, K.; Sadusky, T.; Tuttle, K.R.; Neumiller, J.J.; Rhee, C.M.; Rosas, S.E.; Rossing, P.; Bakris, G. Diabetes Management in Chronic Kidney Disease: A Consensus Report by the American Diabetes Association (ADA) and Kidney Disease: Improving Global Outcomes (KDIGO). Diabetes Care 2022, 45, 3075–3090. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, J.; Wakami, K.; Muto, K.; Kikuchi, S.; Goto, T.; Fukuta, H.; Seo, Y.; Ohte, N. Verification of Echocardiographic Assessment of Left Ventricular Diastolic Dysfunction in Patients With Preserved Left Ventricular Ejection Fraction Using the American Society of Echocardiography and European Association of Cardiovascular Imaging 2016 Recommendations. Circ. Rep. 2019, 1, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Nagueh, S.F.; Smiseth, O.A.; Appleton, C.P.; Byrd, B.F., 3rd; Dokainish, H.; Edvardsen, T.; Flachskampf, F.A.; Gillebert, T.C.; Klein, A.L.; Lancellotti, P.; et al. Recommendations for the Evaluation of Left Ventricular Diastolic Function by Echocardiography: An Update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. J. Am. Soc. Echocardiogr. 2016, 29, 277–314. [Google Scholar] [CrossRef] [PubMed]
- Levey, A.S.; Stevens, L.A.; Schmid, C.H.; Zhang, Y.L.; Castro, A.F., 3rd; Feldman, H.I.; Kusek, J.W.; Eggers, P.; Van Lente, F.; Greene, T.; et al. CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration). A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 2009, 150, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Matthews, D.R.; Hosker, J.P.; Rudenski, A.S.; Naylor, B.A.; Treacher, D.F.; Turner, R.C. Homeostasis model assessment: Insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985, 28, 412–419. [Google Scholar] [CrossRef]
- Hasenleithner, S.O.; Speicher, M.R. A clinician’s handbook for using ctDNA throughout the patient journey. Mol. Cancer 2022, 21, 81. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Chen, C.; Lu, X.; Song, Y.; Zhang, Z.; Zeng, C.; Chiu, R.; Li, L.; Xu, M.; He, C.; et al. Alterations of 5-hydroxymethylcytosines in circulating cell-free DNA reflect retinopathy in type 2 diabetes. Genomics 2021, 113 Pt 1, 79–87. [Google Scholar] [CrossRef]
- Yuzefovych, L.V.; Pastukh, V.M.; Ruchko, M.V.; Simmons, J.D.; Richards, W.O.; Rachek, L.I. Plasma mitochondrial DNA is elevated in obese type 2 diabetes mellitus patients and correlates positively with insulin resistance. PLoS ONE 2019, 14, e0222278. [Google Scholar] [CrossRef]
- Bryk, A.H.; Prior, S.M.; Plens, K.; Konieczynska, M.; Hohendorff, J.; Malecki, M.T.; Butenas, S.; Undas, A. Predictors of neutrophil extracellular traps markers in type 2 diabetes mellitus: Associations with a prothrombotic state and hypofibrinolysis. Cardiovasc. Diabetol. 2019, 18, 49. [Google Scholar] [CrossRef]
- Belcher, D.J.; Sousa, C.A.; Carzoli, J.P.; Johnson, T.K.; Helms, E.R.; Visavadiya, N.P.; Zoeller, R.F.; Whitehurst, M.; Zourdos, M.C. Time course of recovery is similar for the back squat, bench press, and deadlift in well-trained males. Appl. Physiol. Nutr. Metab. 2019, 44, 1033–1042. [Google Scholar] [CrossRef] [PubMed]
- Camuzi Zovico, P.V.; Gasparini Neto, V.H.; Venâncio, F.A.; Soares Miguel, G.P.; Graça Pedrosa, R.; Kenji Haraguchi, F.; Barauna, V.G. Cell-free DNA as an obesity biomarker. Physiol Res. 2020, 69, 515–520. [Google Scholar] [CrossRef]
- Martinod, K.; Witsch, T.; Erpenbeck, L.; Savchenko, A.; Hayashi, H.; Cherpokova, D.; Gallant, M.; Mauler, M.; Cifuni, S.M.; Wagner, D.D. Peptidylarginine deiminase 4 promotes age-related organ fibrosis. J. Exp. Med. 2017, 214, 439–458. [Google Scholar] [CrossRef] [PubMed]
- Devaux, Y. Cardiomyocyte-Specific Cell-Free DNA as a Heart Failure Biomarker? Can. J. Cardiol. 2020, 36, 807–808. [Google Scholar] [CrossRef] [PubMed]
- Buckley, L.F.; Canada, J.M.; Del Buono, M.G.; Carbone, S.; Trankle, C.R.; Billingsley, H.; Kadariya, D.; Arena, R.; Van Tassell, B.W.; Abbate, A. Low NT-proBNP levels in overweight and obese patients do not rule out a diagnosis of heart failure with preserved ejection fraction. ESC Heart Fail. 2018, 5, 372–378. [Google Scholar] [CrossRef]
- Nah, E.H.; Kim, S.Y.; Cho, S.; Kim, S.; Cho, H.I. Plasma NT-proBNP levels associated with cardiac structural abnormalities in asymptomatic health examinees with preserved ejection fraction: A retrospective cross-sectional study. BMJ Open 2019, 9, e026030. [Google Scholar] [CrossRef]
- Oremus, M.; McKelvie, R.; Don-Wauchope, A.; Santaguida, P.L.; Ali, U.; Balion, C.; Hill, S.; Booth, R.; Brown, J.A.; Bustamam, A.; et al. A systematic review of BNP and NT-proBNP in the management of heart failure: Overview and methods. Heart Fail. Rev. 2014, 19, 413–419. [Google Scholar] [CrossRef]
- Jhund, P.S.; Anand, I.S.; Komajda, M.; Claggett, B.L.; McKelvie, R.S.; Zile, M.R.; Carson, P.E.; McMurray, J.J. Changes in N-terminal pro-B-type natriuretic peptide levels and outcomes in heart failure with preserved ejection fraction: An analysis of the I-Preserve study. Eur. J. Heart Fail. 2015, 17, 809–817. [Google Scholar] [CrossRef]
- Rørth, R.; Jhund, P.S.; Yilmaz, M.B.; Kristensen, S.L.; Welsh, P.; Desai, A.S.; Køber, L.; Prescott, M.F.; Rouleau, J.L.; Solomon, S.D.; et al. Comparison of BNP and NT-proBNP in Patients With Heart Failure and Reduced Ejection Fraction. Circ. Heart Fail. 2020, 13, e006541. [Google Scholar] [CrossRef]
- Kwak, S.H.; Park, K.S. Role of mitochondrial DNA variation in the pathogenesis of diabetes mellitus. Front. Biosci. 2016, 21, 1151–1167. [Google Scholar] [CrossRef]
- Walczak, K.; Stawski, R.; Perdas, E.; Brzezinska, O.; Kosielski, P.; Galczynski, S.; Budlewski, T.; Padula, G.; Nowak, D. Circulating cell free DNA response to exhaustive exercise in average trained men with type I diabetes mellitus. Sci. Rep. 2021, 11, 4639. [Google Scholar] [CrossRef] [PubMed]
- Rautenberg, E.K.; Hamzaoui, Y.; Coletta, D.K. Mini-review: Mitochondrial DNA methylation in type 2 diabetes and obesity. Front. Endocrinol. 2022, 13, 968268. [Google Scholar] [CrossRef]
- Wang, W.; Luo, J.; Willems van Dijk, K.; Hägg, S.; Grassmann, F.; T Hart, L.M.; van Heemst, D.; Noordam, R. Assessment of the bi-directional relationship between blood mitochondrial DNA copy number and type 2 diabetes mellitus: A multivariable-adjusted regression and Mendelian randomisation study. Diabetologia 2022, 65, 1676–1686. [Google Scholar] [CrossRef]
- Li, X.; Hu, R.; Luo, T.; Peng, C.; Gong, L.; Hu, J.; Yang, S.; Li, Q. Serum cell-free DNA and progression of diabetic kidney disease: A prospective study. BMJ Open Diabetes Res. Care 2020, 8, e001078. [Google Scholar] [CrossRef] [PubMed]
- Ishida, M.; Ueda, K.; Sakai, C.; Ishida, T.; Morita, K.; Kobayashi, Y.; Okazaki, Y.; Baba, A.; Horikoshi, Y.; Yoshizumi, M.; et al. Cigarette smoke induces mitochondrial DNA damage and activates cGAS-STING pathway -Application to a biomarker for atherosclerosis. Clin. Sci. 2023, 137, CS20220525. [Google Scholar] [CrossRef]
- Park, J.H.; Kim, J.E.; Gu, J.Y.; Yoo, H.J.; Park, S.H.; Kim, Y.I.; Nam-Goong, I.S.; Kim, E.S.; Kim, H.K. Evaluation of Circulating Markers of Neutrophil Extracellular Trap (NET) Formation as Risk Factors for Diabetic Retinopathy in a Case-Control Association Study. Exp. Clin. Endocrinol Diabetes 2016, 124, 557–561. [Google Scholar] [CrossRef]
- Vulesevic, B.; Lavoie, S.S.; Neagoe, P.E.; Dumas, E.; Räkel, A.; White, M.; Sirois, M.G. CRP Induces NETosis in Heart Failure Patients with or without Diabetes. Immunohorizons 2019, 3, 378–388. [Google Scholar] [CrossRef]
- Carestia, A.; Frechtel, G.; Cerrone, G.; Linari, M.A.; Gonzalez, C.D.; Casais, P.; Schattner, M. NETosis before and after Hyperglycemic Control in Type 2 Diabetes Mellitus Patients. PLoS ONE 2016, 11, e0168647. [Google Scholar] [CrossRef]
- Thiam, H.R.; Wong, S.L.; Wagner, D.D.; Waterman, C.M. Cellular Mechanisms of NETosis. Annu. Rev. Cell Dev. Biol. 2020, 36, 191–218. [Google Scholar] [CrossRef]
- Miyoshi, A.; Yamada, M.; Shida, H.; Nakazawa, D.; Kusunoki, Y.; Nakamura, A.; Miyoshi, H.; Tomaru, U.; Atsumi, T.; Ishizu, A. Circulating Neutrophil Extracellular Trap Levels in Well-Controlled Type 2 Diabetes and Pathway Involved in Their Formation Induced by High-Dose Glucose. Pathobiology 2016, 83, 243–251. [Google Scholar] [CrossRef]
- Liu, J.; Zou, Y.; Tang, Y.; Xi, M.; Xie, L.; Zhang, Q.; Gong, J. Circulating cell-free mitochondrial deoxyribonucleic acid is increased in coronary heart disease patients with diabetes mellitus. J. Diabetes Investig. 2016, 7, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.P.; Cheng, K.; Ning, M.A.; Li, H.H.; Wang, H.C.; Li, F.; Chen, S.Y.; Qu, F.L.; Guo, W.Y. Association between peripheral blood cells mitochondrial DNA content and severity of coronary heart disease. Atherosclerosis 2017, 261, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Berezin, A.E. Circulating Cell-Free Mitochondrial DNA as Biomarker of Cardiovascular risk: New Challenges of Old Findings. Angiology 2015, 3, 161. [Google Scholar] [CrossRef]
| Variables | Entire Patient Cohort (n = 240) | Patients without HF (n = 120) | Patients with HF (n = 120) | p Value |
|---|---|---|---|---|
| Demographics and anthropomorphic parameters | ||||
| Age, years | 52 (40–65) | 50 (40–61) | 53 (42–66) | 0.44 |
| Male/female, n (%) | 136 (56.7)/104 (43.3) | 69 (57.5)/51 (42.5) | 67 (55.8)/53 (44.2) | 0.12 |
| BMI, kg/m2 | 25.2 ± 2.9 | 25.8 ± 2.5 | 23.9 ± 2.0 | 0.26 |
| Waist circumference, cm | 96.4 ± 3.7 | 97.9 ± 3.8 | 95.3 ± 3.3 | 0.20 |
| Male, cm | 98.6 ± 3.0 | 99.0 ± 2.7 | 97.2 ± 2.6 | 0.78 |
| Female, cm | 95.1 ± 3.5 | 95.7 ± 3.0 | 94.5 ± 2.4 | 0.84 |
| WHR, units | 0.86 ± 0.06 | 0.86 ± 0.06 | 0.85 ± 0.04 | 0.82 |
| Male, cm | 0.87 ± 0.04 | 0.88 ± 0.05 | 0.87 ± 0.03 | 0.90 |
| Female, cm | 0.84 ± 0.05 | 0.84 ± 0.03 | 0.85 ± 0.04 | 0.90 |
| Comorbidities and CV risk factors | ||||
| Dyslipidemia, n (%) | 196 (81.7) | 99 (82.5) | 97 (80.8) | 0.78 |
| Hypertension, n (%) | 152 (63.3) | 78 (65.0) | 74 (61.7) | 0.78 |
| Stable CAD, n (%) | 81 (33.8) | 32 (26.7) | 49 (40.8) | 0.04 |
| Paroxysmal/persistent AF, n (%) | 17 (7.1) | 4 (3.3) | 13 (10.8) | 0.01 |
| Smoking, n (%) | 98 (40.8) | 50 (41.6) | 48 (40.0) | 0.68 |
| Abdominal obesity, n (%) | 109 (45.4) | 57 (47.5) | 52 (43.3) | 0.56 |
| LV hypertrophy, n (%) | 194 (80.8) | 95 (79.1) | 99 (82.5) | 0.16 |
| CKD 1–3 grades, n (%) | 62 (25.8) | 25 (20.8) | 37 (30.8) | 0.02 |
| Microalbuminuria, n (%) | 45 (18.8) | 23 (19.2) | 22 (18.3) | 0.88 |
| HF phenotypes and functional classification | ||||
| HFpEF, n (%) | 38 (15.8) | - | 38 (32.0) | 0.001 |
| HFmrEF, n (%) | 37 (15.4) | - | 37 (30.8) | 0.001 |
| HFrEF, n (%) | 45 (18.8) | - | 45 (37.5) | 0.001 |
| I/II HF NYHA class, n (%) | 81 (33.8) | - | 81 (67.5) | 0.001 |
| III HF NYHA class, n (%) | 39 (16.6) | - | 39 (32.5) | 0.001 |
| Hemodynamic performance | ||||
| SBP, mm Hg | 131 ± 6 | 129 ± 7 | 132 ± 5 | 0.22 |
| DBP, mm Hg | 75 ± 6 | 76 ± 5 | 74 ± 6 | 0.64 |
| LVEDV, mL | 155 (129–168) | 144 (126–158) | 162 (154–170) | 0.04 |
| LVESV, mL | 69 (49-87) | 55 (47–64) | 86 (80–93) | 0.01 |
| LVEF, % | 55 (46–65) | 62 (56–67) | 46 (37–55) | 0.01 |
| LVMMI, g/m2 | 126 ± 9 | 108 ± 5 | 154 ± 5 | 0.001 |
| LAVI, mL/m2 | 39 (33–47) | 34 (30–36) | 43 (37–52) | 0.01 |
| E/è, unit | 10.3 ± 2.1 | 6.51 ± 1.1 | 13.5 ± 1.3 | 0.01 |
| Biochemistry parameters | ||||
| eGFR, mL/min/1.73 m2 | 74 ± 7 | 81 ± 5 | 67 ± 6 | 0.02 |
| HOMA-IR | 6.87 ± 1.1 | 6.10± 0.9 | 7.95 ± 2.3 | 0.16 |
| Fasting glucose, mmol/L | 6.09 ± 1.2 | 6.08 ± 0.8 | 6.12 ± 1.3 | 0.62 |
| HbA1c, % | 6.40 ± 0.10 | 6.20 ± 0.05 | 6.59 ± 0.02 | 0.58 |
| Creatinine, µmol/L | 92.8 ± 9.7 | 77.4 ± 8.0 | 108.6 ± 8.5 | 0.04 |
| TC, mmol/L | 5.92 ± 0.90 | 5.48 ± 0.40 | 6.43 ± 0.60 | 0.05 |
| HDL-C, mmol/L | 0.98 ± 0.16 | 1.01 ± 0.15 | 0.97 ± 0.17 | 0.48 |
| LDL-C, mmol/L | 3.63 ± 0.19 | 3.10 ± 0.14 | 4.38 ± 0.10 | 0.01 |
| TG, mmol/L | 1.92 ± 0.16 | 1.80 ± 0.12 | 2.21 ± 0.17 | 0.04 |
| hs-CRP, mg/L | 4.06 (2.51–6.90) | 2.94 (1.88–3.76) | 5.83 (3.12–7.22) | 0.02 |
| NT-proBNP, pmol/mL | 1043 (0–2155) | 56 (0–102) | 2615 (1380–3750) | 0.001 |
| Concomitant medications | ||||
| ACEI, n (%) | 117 (48.8) | 53 (44.2) | 64 (53.3) | 0.04 |
| ARB, n (%) | 20 (16.7) | 11 (9.2) | 9 (7.5) | 0.22 |
| ARNI, n (%) | 47 (19.6) | - | 47 (39.2) | 0.001 |
| Beta-blocker, n (%) | 132 (55.0) | 14 (11.7) | 118 (98.3) | 0.001 |
| Ivabradine, n (%) | 13 (5.4) | - | 13 (10.8) | 0.001 |
| Calcium channel blocker, n (%) | 37 (15.4) | 17 (14.2) | 24 (20.0) | 0.04 |
| MRA, n (%) | 45 (18.8) | - | 45 (37.5) | 0.001 |
| Loop diuretic, n (%) | 123 (51.3) | 5 (4.2) | 118 (98.3) | 0.001 |
| Antiplatelet, n (%) | 81 (33.8) | 32 (26.7) | 49 (40.8) | 0.04 |
| Anticoagulants, n (%) | 17 (7.1) | 4 (3.3) | 13 (10.8) | 0.01 |
| Metformin, n (%) | 240 (100) | 120 (100) | 120 (100) | 1.0 |
| SGLT2 inhibitors, n (%) | 177 (73.8) | 57 (47.5) | 120 (100) | 0.001 |
| Statins, n (%) | 222 (92.5) | 109 (90.8) | 113 (94.2) | 0.88 |
| Dependent Variable: HF | ||||
|---|---|---|---|---|
| Variables | Univariate Logistic Regression | Multivariate Logistic Regression | ||
| OR (95% CI) | p-Value | OR (95% CI) | p-Value | |
| NT-proBNP | 1.09 (1.05–1.16) | 0.001 | 1.10 (1.04–1.19) | 0.001 |
| cell-free nDNA (>7.6 μmol/L vs. ≤7.6 μmol/L) | 1.05 (1.02–1.08) | 0.02 | 1.07 (1.03–1.12) | 0.01 |
| cell-free mtDNA (<21.4 μmol/L vs. ≥21.4 μmol/L) | 1.03 (1.01–1.06) | 0.04 | 1.02 (1.00–1.05) | 0.16 |
| LAVI | 1.05 (1.02–1.09) | 0.02 | 1.03 (1.00–1.07) | 0.050 |
| E/è | 1.02 (0.98–1.05) | 0.86 | - | |
| Predictive Models | AUC | NRI | IDI | |||
|---|---|---|---|---|---|---|
| M (95% CI) | p-Value | M (95% CI) | p-Value | M (95% CI) | p-Value | |
| NT-proBNP | 0.67 (0.60–0.74) | - | Reference | - | Reference | - |
| cf n-DNA | 0.80 (0.75–0.88) | 0.001 | 0.36 (0.30–0.43) | 0.001 | 0.48 (0.43–0.54) | 0.001 |
| NT-proBNP + cf n-DNA | 0.83 (0.74–0.92) | 0.001 | 0.38 (0.35–0.42) | 0.001 | 0.48 (0.42–0.55) | 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berezina, T.A.; Kopytsya, M.P.; Petyunina, O.V.; Berezin, A.A.; Obradovic, Z.; Schmidbauer, L.; Lichtenauer, M.; Berezin, A.E. Lower Circulating Cell-Free Mitochondrial DNA Is Associated with Heart Failure in Type 2 Diabetes Mellitus Patients. Cardiogenetics 2023, 13, 15-30. https://doi.org/10.3390/cardiogenetics13010003
Berezina TA, Kopytsya MP, Petyunina OV, Berezin AA, Obradovic Z, Schmidbauer L, Lichtenauer M, Berezin AE. Lower Circulating Cell-Free Mitochondrial DNA Is Associated with Heart Failure in Type 2 Diabetes Mellitus Patients. Cardiogenetics. 2023; 13(1):15-30. https://doi.org/10.3390/cardiogenetics13010003
Chicago/Turabian StyleBerezina, Tetiana A., Mykola P. Kopytsya, Olga V. Petyunina, Alexander A. Berezin, Zeljko Obradovic, Lukas Schmidbauer, Michael Lichtenauer, and Alexander E. Berezin. 2023. "Lower Circulating Cell-Free Mitochondrial DNA Is Associated with Heart Failure in Type 2 Diabetes Mellitus Patients" Cardiogenetics 13, no. 1: 15-30. https://doi.org/10.3390/cardiogenetics13010003
APA StyleBerezina, T. A., Kopytsya, M. P., Petyunina, O. V., Berezin, A. A., Obradovic, Z., Schmidbauer, L., Lichtenauer, M., & Berezin, A. E. (2023). Lower Circulating Cell-Free Mitochondrial DNA Is Associated with Heart Failure in Type 2 Diabetes Mellitus Patients. Cardiogenetics, 13(1), 15-30. https://doi.org/10.3390/cardiogenetics13010003







