Effect of Oxidative Modification by Peroxyl Radical on the Characterization and Identification of Oxidative Aggregates and In Vitro Digestion Products of Walnut (Juglans regia L.) Protein Isolates
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Walnut Protein Isolates
2.3. Modification of Walnut Proteins with AAPH
2.4. Determination of Oxidation and Aggregation Formation in Walnut Proteins
2.4.1. Carbonyl Group
2.4.2. Free Sulfhydryl Group
2.4.3. Free Amino Group
2.4.4. Secondary Structure (FTIR)
2.4.5. Surface Hydrophobicity
2.4.6. UV Spectrum Analysis
2.4.7. Fluorescence Determination of Tryptophan, NFk, and Schiff Bases
2.5. Digestion In Vitro
2.5.1. Simulation of the Digestive Process
2.5.2. Gel Electrophoresis Analysis
2.5.3. Determination of the Protein Carbonyl Indexes in Gastrointestinal Digestion
2.5.4. Amino acid Decay Rate before and after Digestion of Oxidized Protein
2.5.5. Determination of Antioxidant Activity of Hydrolysates
2.6. Identification of Oxidized Protein Digestive Products
2.6.1. LC-MS/MS
In-Gel Digestion
2.6.2. Easy-n LC 1200 Combined with a Q-Exactive Mass Spectrometer
2.7. Statistical Analysis
3. Results
3.1. Effect of Oxidation on Protein Carbonyl Groups, Free Sulfhydryl Groups, and Free Amino Groups
3.2. Changes in Protein Structure after Oxidative Modifications
3.3. Effects of Oxidation on Protein Aggregation
3.4. Protein Oxidation and Digestibility during Digestion In Vitro Simulation
3.5. Dynamic Changes in Carbonyl Content during the Digestion of Walnut Protein
3.6. Antioxidant Capacity of Digestion Products
3.7. Mechanism of Selective Oxidation with Amino Acids
3.8. Antidigestive Protein Identification and Peptide Sequence Alignment
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sze-Tao, K.W.C.; Sathe, S.K. Walnuts (Juglans regia L): Proximate composition, protein solubility, protein amino acid composition and protein in vitro digestibility. J. Sci. Food Agric. 2000, 80, 1393–1401. [Google Scholar] [CrossRef]
- Oke, O.; Oyaniyi, T.; Adewumi, O.; Bamigboye, O.; Lawah, M.; Jatto, K.; Adara, C.; Marizu, J.; Ogunbela, A. Economic, nutritional and medicinal values of African walnut (Tetracarpidium conophorum) in Nigeria (Hutch. & Dalziel): A review. J. Res. For. Wildl. Environ. 2020, 12. [Google Scholar]
- Mao, X.-Y.; Hua, Y.-F. Chemical composition, molecular weight distribution, secondary structure and effect of NaCl on functional properties of walnut (Juglans regia L) protein isolates and concentrates. J. Food Sci. Technol.-Mysore 2014, 51, 1473–1482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Li, J.; Dong, N.; Zhang, Y.; Lu, X.; Hao, Y.; Qi, J. Separation and identification of ACE inhibitory peptides from defatted walnut meal. Eur. Food Res. Technol. 2020, 246, 2029–2038. [Google Scholar] [CrossRef]
- Mao, X.; Wang, D.; Sun, L.; Zhang, J.; Wu, Q. Effect of peroxyl-radicals-induced oxidative modification in the physicochemical and emulsifying properties of walnut protein. J. Am. Oil Chem. Soc. 2021, 98, 903–910. [Google Scholar] [CrossRef]
- Sun, L.; Wu, Q.; Mao, X. Effects of oxidation modification by malondialdehyde on the structure and functional properties of walnut protein. Foods 2022, 11, 2432. [Google Scholar] [CrossRef]
- Xi, J. Research progress of nutritional diet for the disabled elderly. Int. J. Front. Med. 2021, 3. [Google Scholar] [CrossRef]
- Li, F.; Wu, X.; Wu, W. Effects of oxidative modification by malondialdehyde on the in vitro digestion properties of rice bran protein. J. Cereal Sci. 2021, 97, 103158. [Google Scholar] [CrossRef]
- Zhao, J.; Su, G.; Zhao, M.; Sun, W. Physicochemical changes and in vitro gastric digestion of modified soybean protein induced by lipoxygenase catalyzed linoleic acid oxidation. J. Agric. Food Chem. 2019, 67, 13978–13985. [Google Scholar] [CrossRef]
- van Lieshout, G.A.A.; Lambers, T.T.; Bragt, M.C.E.; Hettinga, K.A. How processing may affect milk protein digestion and overall physiological outcomes: A systematic review. Crit. Rev. Food Sci. Nutr. 2020, 60, 2422–2445. [Google Scholar] [CrossRef] [Green Version]
- Goodman, B.E. Insights into digestion and absorption of major nutrients in humans. Adv. Physiol. Educ. 2010, 34, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Ren, S.; Shen, Q.; Ye, X.; Chen, J.; Ling, J. Protein oxidation and proteolysis during roasting and in vitro digestion of fish (Acipenser gueldenstaedtii). J. Sci. Food Agric. 2018, 98, 5344–5351. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Xiong, S.; Yin, T.; Hu, Y.; Liu, R.; Du, H.; Liu, Y.; You, J. Proteomic profiling and oxidation site analysis of gaseous ozone oxidized myosin from silver carp (Hypophthalmichthys molitrix) with different oxidation degrees. Food Chem. 2021, 363, 130307. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.J.; Tu, Z.C.; Wang, H.; Hu, Y.M.; Du, P.C.; Yang, Y.P. Mechanism of the effect of 2,2′-azobis (2-amidinopropane) dihydrochloride simulated lipid oxidation on the IgG/IgE binding ability of ovalbumin. Food Chem. 2020, 327, 127037. [Google Scholar] [CrossRef]
- Wang, C.; Zhao, F.; Bai, Y.; Li, C.; Xu, X.; Kristiansen, K.; Zhou, G. Effect of gastrointestinal alterations mimicking elderly conditions on in vitro digestion of meat and soy proteins. Food Chem. 2022, 383, 132465. [Google Scholar] [CrossRef]
- Yin, H.Y.; Xu, L.B.; Porter, N.A. Free radical lipid peroxidation: Mechanisms and analysis. Chem. Rev. 2011, 111, 5944–5972. [Google Scholar] [CrossRef]
- Zhu, Z.B.; Zhu, W.D.; Yi, J.H.; Liu, N.; Cao, Y.G.; Lu, J.L.; Decker, E.A.; McClements, D.J. Effects of sonication on the physicochemical and functional properties of walnut protein isolate. Food Res. Int. 2018, 106, 853–861. [Google Scholar] [CrossRef]
- Wang, Z.M.; He, Z.F.; Gan, X.; Li, H.J. Effect of peroxyl radicals on the structure and gel properties of isolated rabbit meat myofibrillar proteins. Int. J. Food Sci. Technol. 2018, 53, 2687–2696. [Google Scholar] [CrossRef]
- Wu, X.J.; Li, F.; Wu, W. Effects of rice bran rancidity on the oxidation and structural characteristics of rice bran protein. Lwt-Food Sci. Technol. 2020, 120, 108943. [Google Scholar] [CrossRef]
- Fu, Q.; Liu, R.; Wang, H.; Hua, C.; Song, S.; Zhou, G.; Zhang, W. Effects of oxidation in vitro on structures and functions of myofibrillar protein from beef muscles. J. Agric. Food Chem. 2019, 67, 5866–5873. [Google Scholar] [CrossRef]
- Chen, N.N.; Zhao, Q.Z.; Sun, W.Z.; Zhao, M.M. Effects of malondialdehyde modification on the in vitro digestibility of soy protein isolate. J. Agric. Food Chem. 2013, 61, 12139–12145. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.F.; Mao, X.Y.; Wu, Q.Z.; Zhang, J.; Deng, X.R. Effects of oxidative modification of peroxyl radicals on the structure and foamability of chickpea protein isolates. J. Food Sci. 2021, 86, 824–833. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Li, F.; Wu, X. Effects of rice bran rancidity on oxidation, structural characteristics and interfacial properties of rice bran globulin. Food Hydrocoll. 2021, 110, 106123. [Google Scholar] [CrossRef]
- Hellwig, M. Analysis of Protein oxidation in food and feed products. J. Agric. Food Chem. 2020, 68, 12870–12885. [Google Scholar] [CrossRef] [PubMed]
- Brodkorb, A.; Egger, L.; Alminger, M.; Alvito, P.; Assuncao, R.; Ballance, S.; Bohn, T.; Bourlieu-Lacanal, C.; Boutrou, R.; Carriere, F.; et al. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.C.; Chen, Y.M.; Zhang, C.M.; Kong, X.Z.; Hua, Y.F. The heat-induced protein aggregate correlated with trypsin inhibitor inactivation in soymilk processing. J. Agric. Food Chem. 2012, 60, 8012–8019. [Google Scholar] [CrossRef]
- Kuipers, B.J.H.; Gruppen, H. Prediction of molar extinction coefficients of proteins and peptides using UV absorption of the constituent amino acids at 214 nm to enable quantitative reverse phase high-performance liquid chromatography-mass spectrometry analysis. J. Agric. Food Chem. 2007, 55, 5445–5451. [Google Scholar] [CrossRef]
- Georgiou, C.D.; Zisimopoulos, D.; Argyropoulou, V.; Kalaitzopoulou, E.; Salachas, G.; Grune, T. Protein and cell wall polysaccharide carbonyl determination by a neutral pH 2,4-dinitrophenylhydrazine-based photometric assay. Redox Biol. 2018, 17, 128–142. [Google Scholar] [CrossRef]
- Soudani, N.; Ben Amara, I.; Troudi, A.; Hakim, A.; Bouaziz, H.; Makni, F.A.; Zeghal, K.M.; Zeghal, N. Oxidative damage induced by chromium (VI) in rat erythrocytes: Protective effect of selenium. J. Physiol. Biochem. 2011, 67, 577–588. [Google Scholar] [CrossRef]
- Werber, J.; Wang, Y.J.; Milligan, M.; Li, X.H.; Ji, J.A. Analysis of 2,2′-azobis (2-amidinopropane) dihydrochloride degradation and hydrolysis in aqueous solutions. J. Pharm. Sci. 2011, 100, 3307–3315. [Google Scholar] [CrossRef]
- Gomez-Pastor, R.; Perez-Torrado, R.; Cabiscol, E.; Ros, J.; Matallana, E. Engineered Trx2p industrial yeast strain protects glycolysis and fermentation proteins from oxidative carbonylation during biomass propagation. Microb. Cell Fact. 2012, 11, 4. [Google Scholar] [CrossRef] [PubMed]
- Kunath, S.; Schindeldecker, M.; De Giacomo, A.; Meyer, T.; Sohre, S.; Hajieva, P.; von Schacky, C.; Urban, J.; Moosmann, B. Prooxidative chain transfer activity by thiol groups in biological systems. Redox Biol. 2020, 36, 101628. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Wang, B.; Zhao, B.; Yang, F. Ultrasonic Assisted extraction of quinoa (Chenopodium quinoa Willd.) Protein and effect of heat treatment on its in vitro digestion characteristics. Foods 2022, 11, 771. [Google Scholar] [CrossRef] [PubMed]
- Moldogazieva, N.T.; Mokhosoev, I.M.; Mel’nikova, T.I.; Porozov, Y.B.; Terentiev, A.A. Oxidative Stress and advanced lipoxidation and glycation end products (ALEs and AGEs) in Aging and age-related diseases. Oxidative Med. Cell. Longev. 2019, 2019, 3085756. [Google Scholar] [CrossRef] [Green Version]
- Pan, J.; Lian, H.; Jia, H.; Hao, R.; Wang, Y.; Ju, H.; Li, S.; Dong, X. Dose affected the role of gallic acid on mediating gelling properties of oxidatively stressed Japanese seerfish myofibrillar protein. LWT-Food Sci. Technol. 2020, 118, 108849. [Google Scholar] [CrossRef]
- Wang, P.-Y.; Yang, C.-T.; Chu, L.-K. Differentiating the protein dynamics using fluorescence evolution of tryptophan residue(s): A comparative study of bovine and human serum albumins upon temperature jump. Chem. Phys. Lett. 2021, 781, 138998. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, C.M.; Kong, X.Z.; Hua, Y.F. Oxidative modification of soy protein by peroxyl radicals. Food Chem. 2009, 116, 295–301. [Google Scholar] [CrossRef]
- Wang, Z.; He, Z.; Gan, X.; Li, H. Interrelationship among ferrous myoglobin, lipid and protein oxidations in rabbit meat during refrigerated and superchilled storage. Meat Sci. 2018, 146, 131–139. [Google Scholar] [CrossRef]
- Wang, Q.X.; Liu, D.X.; Song, P.; Zou, M.H. Tryptophan-kynurenine pathway is dysregulated in inflammation, and immune activation. Front. Biosci.-Landmark 2015, 20, 1116–1143. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Zheng, J.; Ge, G.; Zhao, M.; Sun, W. Impact of heating treatments on physical stability and lipid-protein co-oxidation in oil-in-water emulsion prepared with soy protein isolates. Food Hydrocoll. 2020, 100, 105167. [Google Scholar] [CrossRef]
- Scheidegger, D.; Pecora, R.P.; Radici, P.M.; Kivatinitz, S.C. Protein oxidative changes in whole and skim milk after ultraviolet or fluorescent light exposure. J. Dairy Sci. 2010, 93, 5101–5109. [Google Scholar] [CrossRef] [PubMed]
- Xi, J.; Chen, Y. Analysis of the relationship between heterocyclic amines and the oxidation and thermal decomposition of protein using the dry heated soy protein isolate system. LWT 2021, 148, 111738. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, Z.; Wang, R.; Sui, X.; Qi, B.; Han, F.; Li, Y.; Jiang, L. Secondary structure and subunit composition of soy protein in vitro digested by pepsin and its relation with digestibility. BioMed. Res. Int. 2016, 2016, 5498639. [Google Scholar] [CrossRef] [Green Version]
- Sharma, N.; Sharma, R.; Rajput, Y.S.; Mann, B.; Singh, R.; Gandhi, K. Separation methods for milk proteins on polyacrylamide gel electrophoresis: Critical analysis and options for better resolution. Int. Dairy J. 2021, 114, 104920. [Google Scholar] [CrossRef]
- Feng, Y.L.; Wang, J.M.; Hu, H.Y.; Yang, C. Effect of oxidative modification by reactive oxygen species (ROS) on the aggregation of whey protein concentrate (WPC). Food Hydrocoll. 2022, 123, 107189. [Google Scholar] [CrossRef]
- Sante-Lhoutellier, V.; Aubry, L.; Gatellier, P. Effect of oxidation on in vitro digestibility of skeletal muscle myofibrillar proteins. J. Agric. Food Chem. 2007, 55, 5343–5348. [Google Scholar] [CrossRef] [PubMed]
- Dalle-Donne, I.; Rossi, R.; Giustarini, D.; Milzani, A.; Colombo, R. Protein carbonyl groups as biomarkers of oxidative stress. Clin. Chim. Acta 2003, 329, 23–38. [Google Scholar] [CrossRef]
- Van Hecke, T.; Vossen, E.; Bussche, J.V.; Raes, K.; Vanhaecke, L.; De Smet, S. Fat Content and nitrite-curing influence the formation of oxidation products and NOC-specific DNA adducts during in vitro digestion of meat. PLoS ONE 2014, 9, e0101122. [Google Scholar] [CrossRef] [Green Version]
- Karami, Z.; Akbari-adergani, B. Bioactive food derived peptides: A review on correlation between structure of bioactive peptides and their functional properties. J. Food Sci. Technol. 2019, 56, 535–547. [Google Scholar] [CrossRef]
- Stadtman, E.R. Oxidation of free amino acids and amino acid residues in proteins by radiolysis and by metal-catalyzed reactions. Annu. Rev. Biochem. 1993, 62, 797–821. [Google Scholar] [CrossRef]
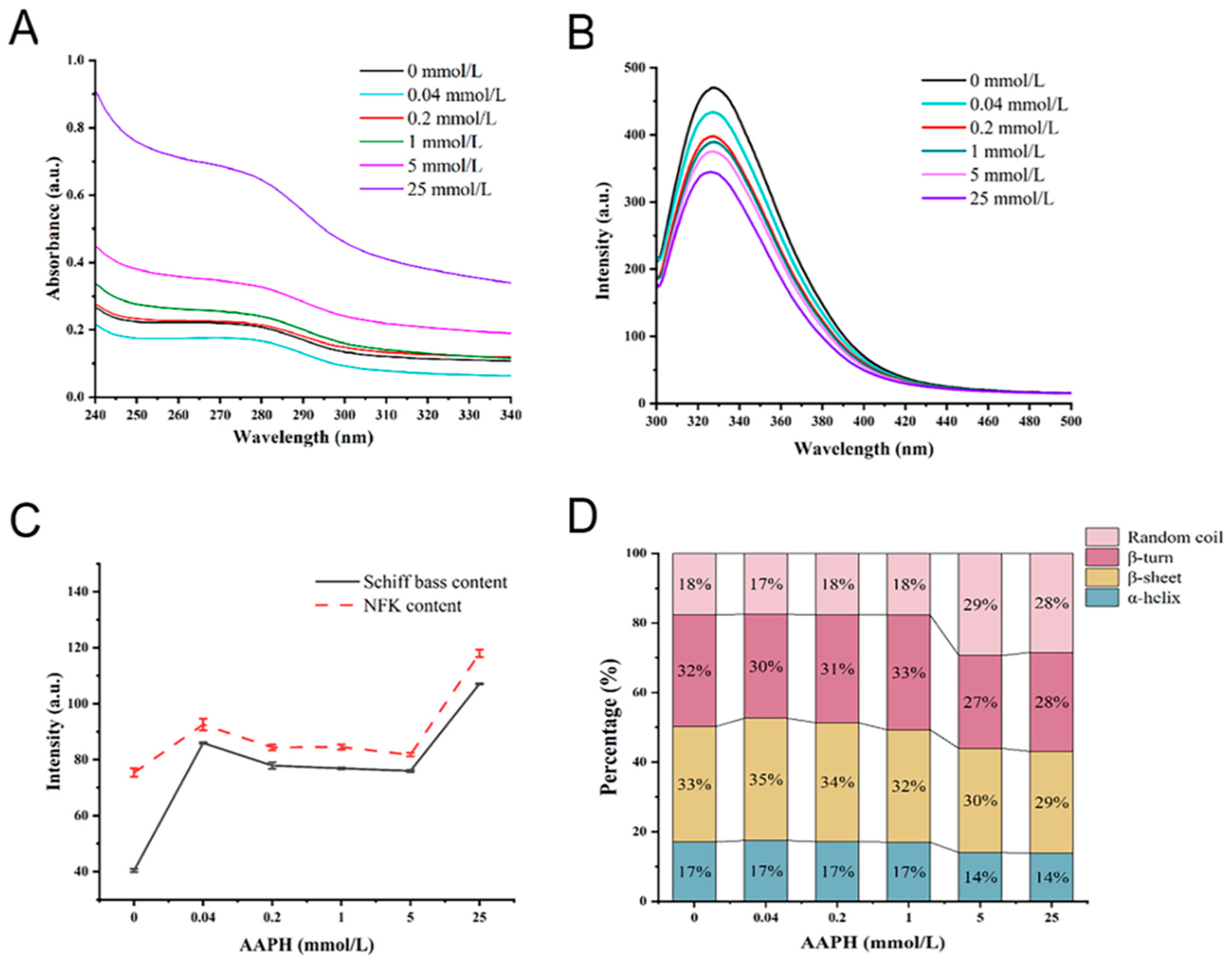

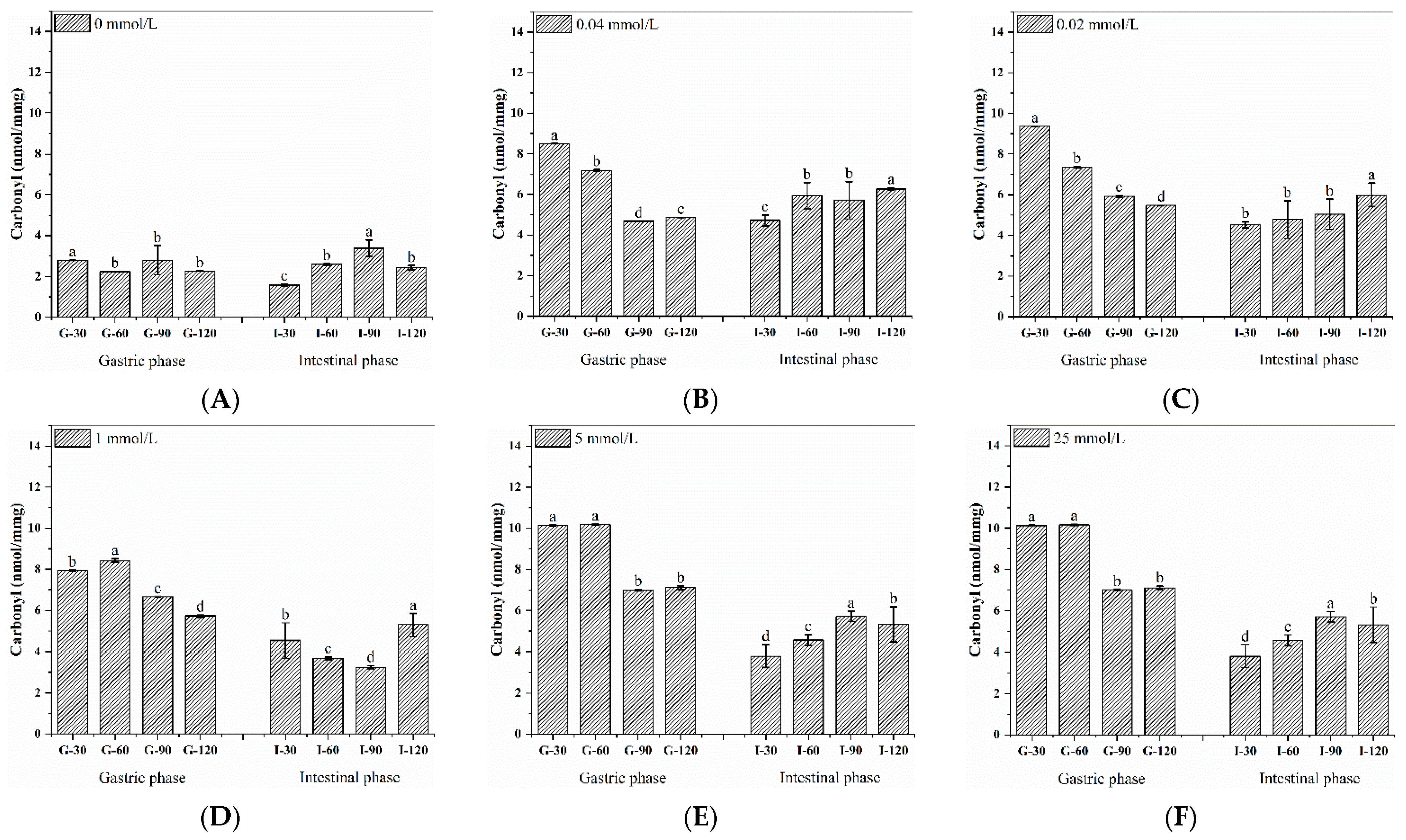




| AAPH (mmol/L) | Carbonyl (nmol/mg) | Free SH (nmol/mg) | Free Amino (μmol/mg) |
|---|---|---|---|
| 0 | 2.36 ± 0.09 f | 4.30 ± 0.03 a | 1.28 ± 0.03 c |
| 0.04 | 2.42 ± 0.11 e | 3.80 ± 0.05 b | 1.39 ± 0.01 b |
| 0.2 | 2.68 ± 0.05 d | 3.66 ± 0.02 c | 1.47 ± 0.02 a |
| 1 | 3.18 ± 0.02 c | 2.22 ± 0.01 d | 1.17 ± 0.01 d |
| 5 | 4.23 ± 0.07 b | 2.02 ± 0.01 e | 1.15 ± 0.02 d |
| 25 | 5.12 ± 0.10 a | 1.72 ± 0.08 f | 1.13 ± 0.02 d |
| ACCESSION | DESCRIPTION | MW | PI | G-0 | G-5 | I-5 | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| UP | SCORE | UP | SCORE | UP | SCORE | |||||
| 1 | XP_018818401.1 | 11S globulin seed storage protein 2-like | 54.3 | 6.81 | 11 | 40.43 | 62 | 9.91 | 63 | 965.24 |
| 2 | XP_018827137.1 | 11S globulin seed storage protein 2-like | 53.4 | 7.8 | 2 | 8.39 | 20 | 5.51 | 18 | 186.72 |
| 3 | XP_018827329.1 | 11S globulin seed storage protein Jug r 4 | 58.1 | 7.25 | 5 | 14.63 | 46 | 38.36 | 52 | 757.22 |
| 4 | XP_018827328.1 | 11S globulin-like | 58.3 | 7.77 | 13 | 45.10 | 66 | 119.56 | 59 | 908.08 |
| 5 | XP_018842296.1 | legumin B-like | 55.8 | 7.5 | 12 | 74.54 | 71 | 12.29 | 64 | 1504.61 |
| 6 | XP_018812171.1 | vicilin Car i 2.0101 | 94.3 | 6.83 | 9 | 24.86 | 23 | 1048.8 | 13 | 73.32 |
| 7 | XP_018814692.1 | Vicilin Jug r 6.0101 | 57.4 | 7.3 | 5 | 19.99 | 22 | 30.3 | 15 | 73.04 |
| 8 | XP_035546314.1 | vicilin-like seed storage protein At2g28490 | 59.6 | 6.92 | 2 | 5.38 | 6 | 199.66 | 5 | 17.19 |
| 9 | XP_018824007.1 | 2S sulfur-rich seed storage protein 2 | 17.1 | 6.52 | 5 | 27.03 | 3 | 2.7 | 3 | 65.45 |
| 10 | XP_018837699.1 | oleosin 18.2 kDa-like | 16.6 | 9.91 | 2 | 7.39 | 4 | 2.63 | 3 | 5.78 |
| 11 | XP_018842295.1 | legumin B-like | 55.8 | 8.18 | 1 | 28.82 | 2 | 21 | 2 | 399.8 |
| 12 | XP_018856265.1 | oleosin 1-like | 14.7 | 10.14 | 4 | 8.72 | 1 | 2.2 | 1 | 2.92 |
| 13 | XP_018842385.1 | oleosin 5-like | 16.3 | 10.08 | 2 | 5.3 | 5 | 0 | 2 | 2.46 |
| 14 | XP_018814488.1 | actin-97 | 41.7 | 5.49 | 3 | 6.52 | 5 | 7.3 | 3 | 8.54 |
| 15 | XP_018821252.1 | glutaredoxin-like | 13.7 | 6.09 | 1 | 2.05 | 2 | 2.15 | 1 | 1.73 |
| 16 | XP_035549483.1 | salicylate carboxymethyltransferase-like | 40.7 | 6.54 | 1 | 2.39 | 1 | 4.65 | 1 | 2.35 |
| 17 | XP_018805239.1 | ubiquitin-40S ribosomal protein S27a-like | 17.7 | 9.79 | 1 | 2.09 | 2 | 3.09 | 1 | 2.87 |
| 18 | XP_018810059.1 | peroxygenase-like | 26.9 | 5.87 | 2 | 2.6 | 2 | 2.17 | ||
| 19 | XP_018831031.1 | oleosin 1 | 14.8 | 9.63 | 1 | 0 | 1 | 2.62 | ||
| 20 | XP_018809740.1 | oleosin 18.2 kDa-like | 15.9 | 9.72 | 3 | 0 | 1 | 0 | ||
| 21 | XP_018818883.1 | haloacid dehalogenase-like hydrolase domain-containing protein Sgpp | 33.7 | 5.63 | 1 | 2.23 | 1 | 1.96 | ||
| 22 | XP_018841715.1 | histone H4-like | 14.2 | 10.84 | 2 | 6.31 | 1 | 2.17 | ||
| 23 | XP_018812582.1 | 40S ribosomal protein S8-like | 25.4 | 10.43 | 1 | 1.66 | ||||
| 24 | XP_018845470.1 | oleosin | 15.6 | 9.54 | 1 | 1.94 | ||||
| 25 | XP_018811862.1 | aspartic proteinase-like isoform X1 | 57.9 | 5.39 | 1 | 2.04 | ||||
| 26 | XP_018851532.1 | probable aquaporin TIP3-2 | 27.4 | 7.71 | 1 | 5.54 | ||||
| 27 | XP_018849277.1 | probable aquaporin TIP3-2 | 27.4 | 7.66 | 1 | 3.21 | ||||
| 28 | XP_018822092.1 | protein FAM135B-like | 91.8 | 8 | 1 | 874.88 | ||||
| 29 | XP_035550528.1 | putative glycine-rich cell wall structural protein 1 isoform X1 | 21.3 | 9.11 | 1 | 2.56 | ||||
| 30 | XP_018841706.1 | histone H2A | 15.9 | 10.67 | 1 | 1.74 | ||||
| 31 | XP_035543376.1 | glyceraldehyde-3-phosphate dehydrogenase GAPCP2, chloroplastic-like | 45.4 | 9.01 | 1 | 5.1 | ||||
| 32 | XP_018826340.2 | vicilin-like seed storage protein At2g18540 | 79 | 5.44 | 1 | 498.72 | ||||
| 33 | XP_018811454.1 | 11-beta-hydroxysteroid ehydrogenase B-like | 40.9 | 6.35 | 2 | 2.1 | ||||
| 34 | XP_018809799.1 | ankyrin repeat-containing protein ITN1-like | 25.6 | 7.46 | 1 | 2.79 | ||||
| 35 | XP_018821986.2 | malate synthase, glyoxysomal | 64.2 | 7.55 | 1 | 128.13 | ||||
| 36 | XP_018835846.1 | polygalacturonase QRT3 | 52.5 | 6.54 | 1 | 0 | ||||
| 37 | XP_018847613.1 | histone H2AX-like | 14.9 | 10.36 | 2 | 4.05 | ||||
| 38 | XP_018826287.1 | barwin-like | 20.4 | 7.62 | 1 | 0 | ||||
| 39 | XP_018828748.1 | elongation factor 1-alpha | 49.5 | 9.07 | 1 | 2.35 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, J.; Han, M.; Wu, Q.; Mao, X.; Zhang, J.; Lu, Z. Effect of Oxidative Modification by Peroxyl Radical on the Characterization and Identification of Oxidative Aggregates and In Vitro Digestion Products of Walnut (Juglans regia L.) Protein Isolates. Foods 2022, 11, 4104. https://doi.org/10.3390/foods11244104
Zhao J, Han M, Wu Q, Mao X, Zhang J, Lu Z. Effect of Oxidative Modification by Peroxyl Radical on the Characterization and Identification of Oxidative Aggregates and In Vitro Digestion Products of Walnut (Juglans regia L.) Protein Isolates. Foods. 2022; 11(24):4104. https://doi.org/10.3390/foods11244104
Chicago/Turabian StyleZhao, Jinjin, Miaomiao Han, Qingzhi Wu, Xiaoying Mao, Jian Zhang, and Zhenkang Lu. 2022. "Effect of Oxidative Modification by Peroxyl Radical on the Characterization and Identification of Oxidative Aggregates and In Vitro Digestion Products of Walnut (Juglans regia L.) Protein Isolates" Foods 11, no. 24: 4104. https://doi.org/10.3390/foods11244104
APA StyleZhao, J., Han, M., Wu, Q., Mao, X., Zhang, J., & Lu, Z. (2022). Effect of Oxidative Modification by Peroxyl Radical on the Characterization and Identification of Oxidative Aggregates and In Vitro Digestion Products of Walnut (Juglans regia L.) Protein Isolates. Foods, 11(24), 4104. https://doi.org/10.3390/foods11244104





