Sparse Regularized Autoencoders-Based Radiomics Data Augmentation for Improved EGFR Mutation Prediction in NSCLC
Abstract
1. Introduction
Main Contributions
- Introducing a scalable kl_weight hyperparameter into the GSRA-KL loss function to adaptively balance sparse regularization and reconstruction fidelity.
- Enhancing synthetic data faithfulness through systematic generation across mutation subtypes by jointly adjusting GSRA-KL hyperparameters, particularly tuning kl_weight values relative to hidden-layer sizes.
- Integrating an Optuna-based Bayesian optimization framework for hyperparameter tuning, which significantly reduces computational cost compared to the PSO-based approach used previously.
- Conducting a comprehensive evaluation across 63 synthetic datasets for EGFR gene mutation using resemblance-dimension score (RDS), the novel utility-dimension score (UDS), and t-SNE visualizations.
- Performing additional quantitative validation using non-parametric Friedman and Spearman rank-based statistical analyses to assess consistency across resemblance and computational cost dimensions.
- Comparing the effectiveness of GSRA-KL, GSRA, and TVAE models to address prediction enhancement challenges in small, imbalanced CT-based radiomics datasets.
- Performing an ablation-like analysis demonstrating that setting kl_weight = 0.0 reverts GSRA-KL to the baseline GSRA model, enabling an isolated evaluation of KL divergence regularization effects on model performance and robustness.
2. Materials and Methods
2.1. Data-Preprocessing Phase
2.2. Synthetic Data Generation
| Algorithm 1 Pseudocode for GSRA–KL-Based Synthetic Radiomics Data Generation with Custom Loss and Optuna Optimization |
| Input: , , , , , , , , , epochs E, batch size B, random seed. Initialize: Fix random seed; set ; preprocess ; define . Output: Trained encoder/decoder, best hyperparameters, synthetic datasets , and resource metrics. Process:
|
2.3. Synthetic Data Evaluation
2.4. Resemblance-Dimension Evaluation
2.4.1. Univariate Resemblance Analysis
2.4.2. Bivariate Effectiveness (BE)
2.4.3. Multivariate Resemblance
2.4.4. Resemblance Score (RS)
2.4.5. Data-Labeling Analysis (DLA)
- Logistic Regression—Default;
- Decision Tree—random_state = 9;
- Random Forest—n_estimators = 100, random_state = 9;
- Support Vector Machine—C = 100, max_iter = 300, kernel = ’linear’, probability = True, random_state = 9;
- K-Nearest Neighbors—n_neighbors = 10;
- Multi-layer Perceptron—hidden_layer_sizes = (128, 64, 32), max_iter = 300, random_state = 9;
- Naive Bayes—Default (GaussianNB);
- Gaussian Process—Default;
- XGBoost—Default;
- LightGBM—Default.
2.4.6. Computation of Resemblance-Dimension Score (RDS)
2.5. Utility-Dimension Evaluation
2.5.1. Evaluation Strategy
2.5.2. Training and Testing Protocol
2.5.3. Utility Score Interpretation
3. Results
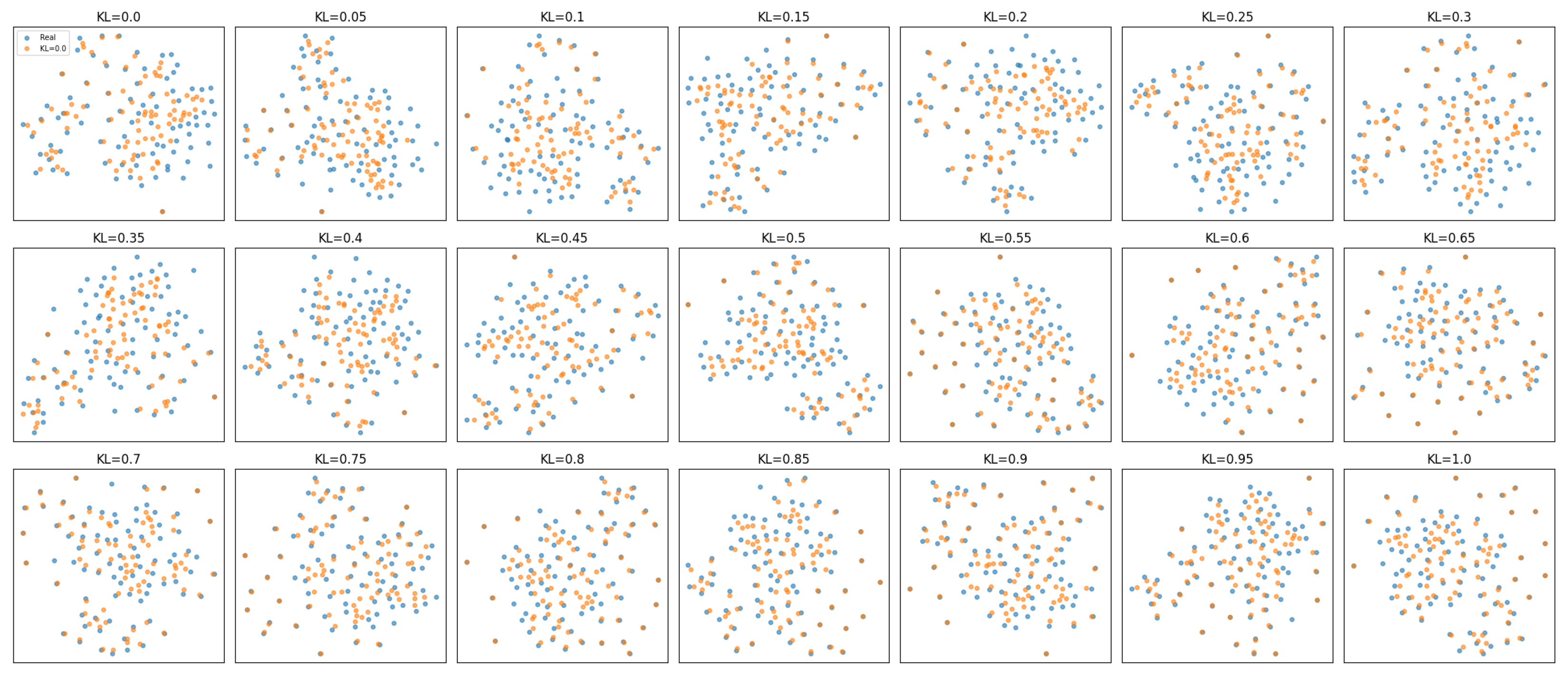
3.1. Resemblance-Dimension Evaluation
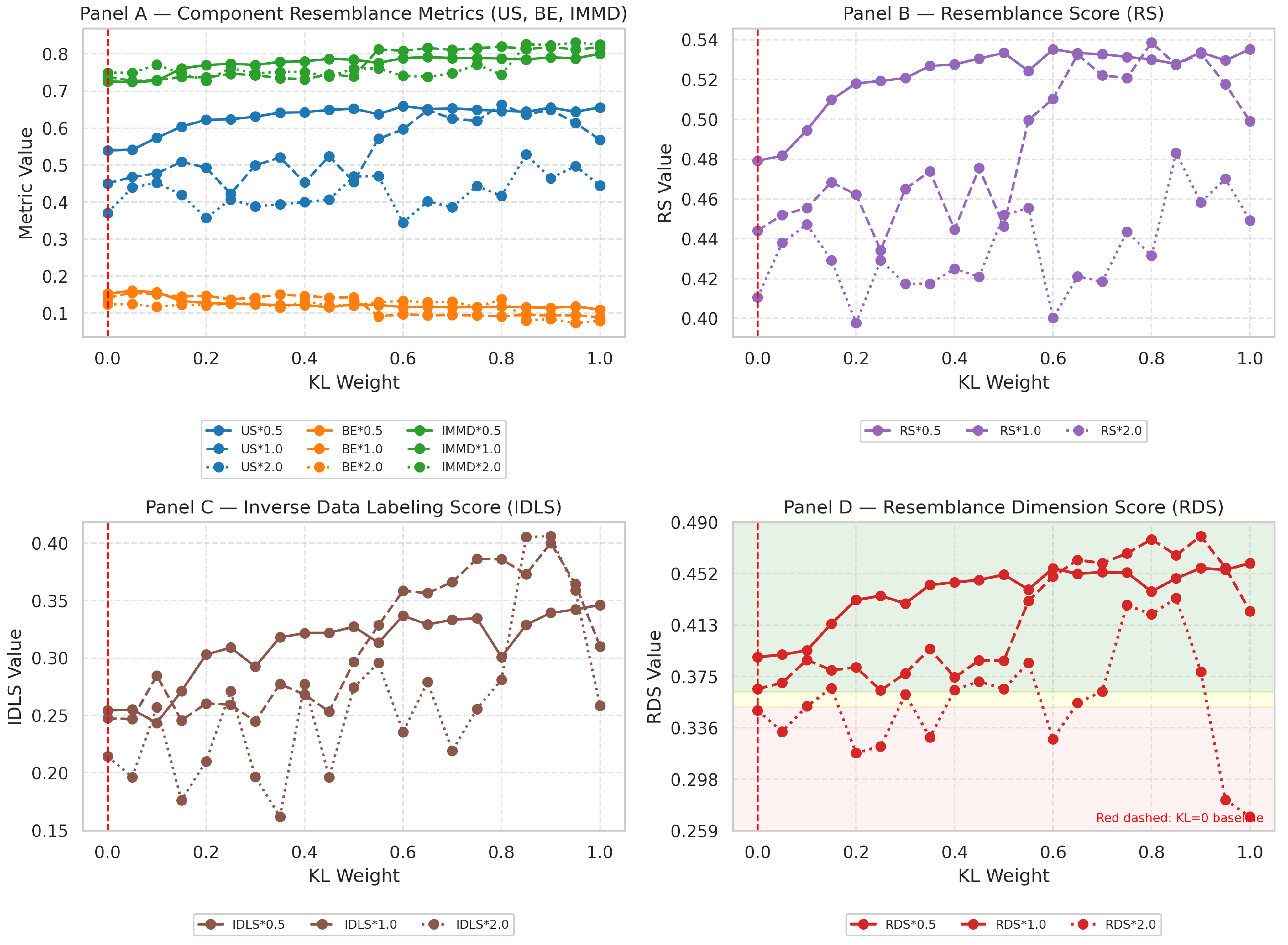
3.2. Panel A—Component-Level Metrics (US, BE, IMMD)
3.3. Panel B—Aggregate Resemblance Score (RS)
3.4. Panel C—Inverse Data-Labeling Score (IDLS)
3.5. Panel D—Resemblance-Dimension Score (RDS) with Quality Bands
Statistical Correlation Analysis
3.6. Computational Efficiency Analysis
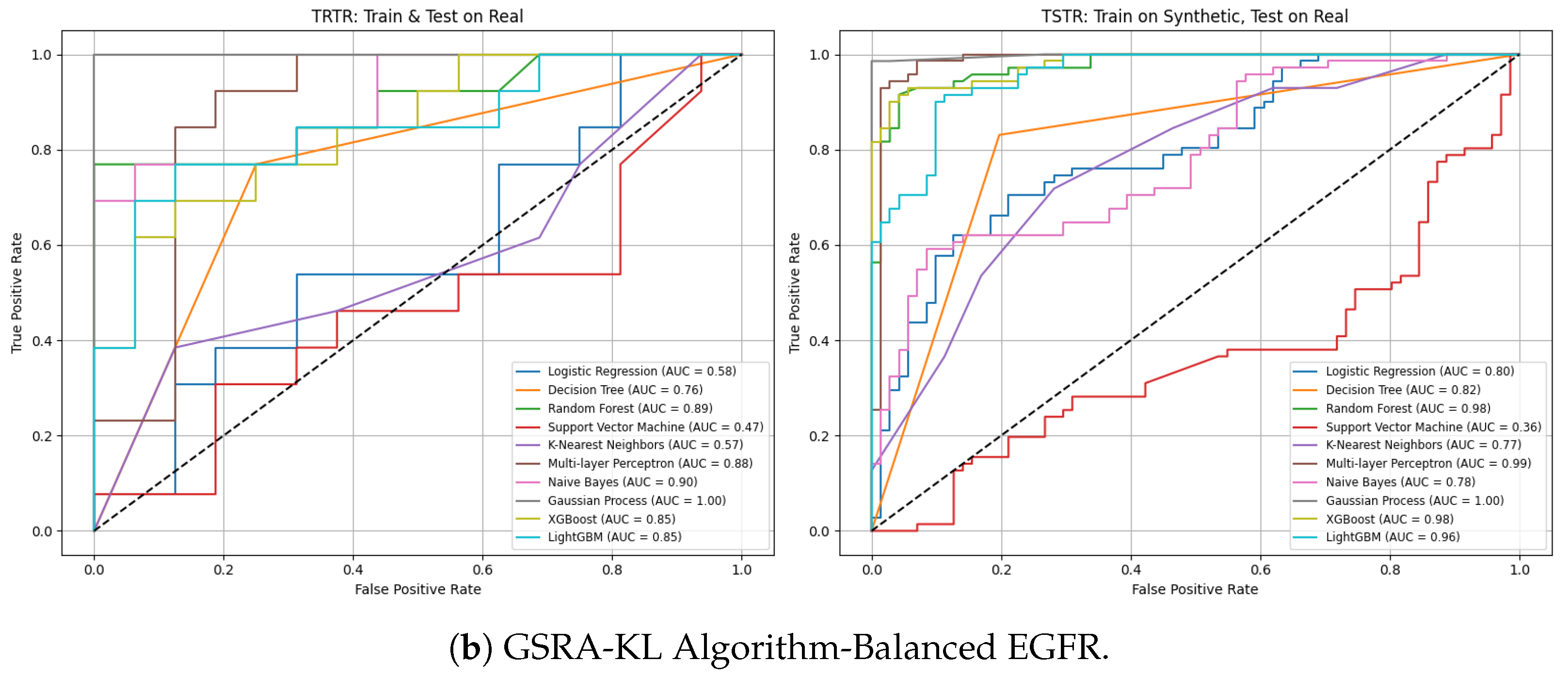
3.7. Utility-Dimension Evaluation
TRTR Versus TSTR Strategy
- Under an imbalanced class distribution, TRTR performance was notably poor, with a UDS of 0.2757.
- In contrast, balanced distribution significantly improved performance to an excellent UDS of 0.7090.
- Under imbalanced data, GSRA-KL resulted in a UDS of 0.5868 (poor), while TVAE achieved 0.7092 (excellent).
- Under balanced data, GSRA-KL and TVAE delivered excellent UDS values of 0.8138 and 0.8036, respectively.
4. Discussion
4.1. Statistical Analysis—Resemblance Dimension
4.2. Computational Cost Efficiency
4.3. Utility-Dimension Analysis
4.4. Observations and Future Research Direction
- Mitigation of Sample Scarcity and Class Imbalance: Although the dataset comprised only 83 patients with 12 EGFR-positive cases, the proposed GSRA–KL framework was specifically designed to address the challenges of sample scarcity and class imbalance. The scalable kl_weight hyperparameter was systematically varied over 21 values (). Each configuration was evaluated under three distinct hidden-layer sizes—equal to half of and double the input dimensionality—resulting in a total of 63 synthetic datasets derived from the limited CT radiomics cohort. This extensive experimentation enabled the framework to learn stable latent representations across diverse network complexities and varying strengths of KL-regularization. The generated datasets were comprehensively validated using resemblance metrics, the proposed utility-dimension score (UDS), and t-SNE visualizations to ensure distributional fidelity. These results demonstrate that GSRA–KL effectively mitigates small-sample constraints while maintaining statistical and predictive consistency. Nonetheless, validation on larger, multi-institutional datasets remains an important direction for confirming generalizability and clinical translation.
- Categorization of RDS Metrics: The RDS thresholds (excellent ; good –; poor ) were empirically determined based on our experiments and adaptations from previous studies [21,26]. While these ranges reflect the quality of synthetic datasets in the context of small, imbalanced EGFR mutation cohorts, they have not yet been established as universal clinical or technical benchmarks and require further investigation on multiple modalities and disease radiomics datasets.
- Baseline Comparison and Rationale: While this study primarily compares GSRA-KL with GSRA and TVAE, we acknowledge the relevance of other widely used generative and augmentation methods such as GANs, CTGAN, and SMOTE. The current selection was guided by methodological proximity, as both GSRA and TVAE can be used for radiomics feature synthesis and share architectural similarities with GSRA-KL. In contrast, GAN- and SMOTE-based models are generally optimized for lower-dimensional or class-imbalanced tabular datasets, and are less suited to the complex, high-dimensional correlations characteristic of radiomics features (263 in this study). Moreover, GAN-based models are known to exhibit instability during training and suffer from mode collapse, particularly when applied to limited or highly correlated feature spaces, leading to reduced diversity and fidelity in the generated data [30,31,32]. For these reasons, their full-scale inclusion was beyond the scope of this work. Nonetheless, both GAN and CTGAN (as representative tabular synthesis models) will be considered in future research to contrast further their performance and suitability for generating high-dimensional radiomics features.
- Training Instability: The proposed GSRA-KL framework establishes a pipeline for effective synthetic radiomics data generation, emphasizing that parameter and hyperparameter selection remain subjective and dataset-dependent. The current study proposes an evaluation framework for generating effective and high-quality synthetic dataset generation strategies, as well as for selecting viable techniques, as outlined in the current work. Although results show unstable performance at higher KL weights (especially for hidden size ), future work will explore stability enhancements through additional regularization (e.g., dropout tuning, early stopping) and cross-validation across multiple random seeds to improve the robustness and reproducibility of GSRA-KL training, as similar techniques have been shown to mitigate mode collapse and training variance in generative models [30,32]. Furthermore, future research will focus on formalizing this process by integrating adaptive and Bayesian optimization techniques [33,34] to automatically identify stable and optimal configurations for diverse radiomics datasets.
- Evaluation Metric Validation: While the proposed utility-dimension score (UDS) demonstrates promising results in evaluating the practical utility of synthetic data within this study, its validation remains confined to the datasets and models explored herein. To enhance its credibility and general applicability, future studies should extend UDS evaluation to diverse datasets and compare it against established measures such as FID, MMD, and discriminability indices. Such comparative analyses across various generative frameworks would provide deeper insight into the robustness and generalizability of UDS as a reliable metric for synthetic data assessment.
- Clinical Utility and Data Imbalance: In utility-dimension analysis, the UDS under imbalanced EGFR mutation data (UDS = 0.2757) appears poor. However, this result actually highlights the critical importance of balancing strategies, such as those implemented within the GSRA–KL framework. When the same data were balanced, UDS markedly improved to 0.7090 (excellent), demonstrating the framework’s capacity to enhance predictive performance once class imbalance is mitigated. This transition from poor to excellent UDS supports GSRA–KL’s intended role as a data-level augmentation approach rather than a classifier itself. Furthermore, future work will integrate GSRA–KL–generated balanced datasets into clinically relevant endpoints (e.g., treatment response and survival prediction), thereby extending its validation beyond TRTR and TSTR strategies and toward real-world clinical translation.
- Runtime Comparison and Optimization Environment: The reported 90% reduction in optimization runtime when employing Optuna compared to the PSO-based implementation was obtained under identical Google Colab computational settings using the same dataset, preprocessing pipeline, and GSRA-KL architecture. Both approaches optimized equivalent hyperparameters (e.g., penalty weight, sparsity weight, and sparsity target) with the same validation loss objective. The observed difference primarily reflects algorithmic design rather than hardware bias: the PSO implementation in pyswarm executes particle evaluations sequentially without adaptive stopping, whereas Optuna’s Tree-structured Parzen Estimator (TPE) sampler performs adaptive and asynchronous trial evaluation, enabling faster convergence within the same search space. While every effort was made to maintain consistent runtime conditions, minor variations in TPU/CPU scheduling inherent to the Colab environment may introduce marginal timing differences. Nonetheless, the comparative efficiency observed for Optuna is attributed to its inherently more scalable and sample-efficient search strategy. Future work will extend this analysis by performing fully normalized benchmarking across optimization algorithms and compute backends further to validate the fairness and reproducibility of runtime comparisons.
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Lung cancer | LC |
| Resemblance-dimension scores | RDS |
| Utility-dimension scores | UDS |
| Generic Sparse Regularized Autoencoders with Kullback-Leibler divergence | GSRA-KL |
| Kullback–Leibler divergence | KL |
| Tabular Variational autoencoder | TVAE |
| Particle Swarm Optimization | PSO |
| Non-small cell lung cancer | NSCLC |
| Epidermal growth factor receptor | EGFR |
| Tyrosine kinase inhibitors | TKI |
| Artificial intelligence | AI |
| Generic sparse regularized autoencoders | GSRA |
| Positron emission tomography | PET |
| Computed tomography | CT |
| Student’s t-test | ST |
| Mann–Whitney U-test | MW |
| Kolmogorov–Smirnov test | KS |
| Wasserstein Distance | WD |
| Inversely normalized WD (1-WD) | IWD |
| Univariate score | US |
| Bivariate Effectiveness | BE |
| Maximum Mean Discrepancy | MMD |
| Inversely normalized MMD (1-MMD) | IMMD |
| Resemblance Score | RS |
| Data-Labelling Analysis | DLA |
| Data-labelling score | DLS |
| Principal Component Analysis | PCA |
| Training on Real Data and Testing on Real Data | TRTR |
| Training on Synthetic Data and Testing on Real Data | TSTR |
| Standard Deviation | SD |
References
- Wéber, A.; Morgan, E.; Vignat, J.; Laversanne, M.; Pizzato, M.; Rumgay, H.; Singh, D.; Nagy, P.; Kenessey, I.; Soerjomataram, I.; et al. Lung cancer mortality in the wake of the changing smoking epidemic: A descriptive study of the global burden in 2020 and 2040. BMJ Open 2023, 13, e065303. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Rosell, R.; Moran, T.; Queralt, C.; Porta, R.; Cardenal, F.; Camps, C.; Majem, M.; Lopez-Vivanco, G.; Isla, D.; Provencio, M.; et al. Screening for epidermal growth factor receptor mutations in lung cancer. N. Engl. J. Med. 2009, 361, 958–967. [Google Scholar] [CrossRef]
- Maemondo, M.; Inoue, A.; Kobayashi, K.; Sugawara, S.; Oizumi, S.; Isobe, H.; Gemma, A.; Harada, M.; Yoshizawa, H.; Kinoshita, I.; et al. Gefitinib or chemotherapy for non–small-cell lung cancer with mutated EGFR. N. Engl. J. Med. 2010, 362, 2380–2388. [Google Scholar] [CrossRef]
- Ramalingam, S.S.; Vansteenkiste, J.; Planchard, D.; Cho, B.C.; Gray, J.E.; Ohe, Y.; Zhou, C.; Reungwetwattana, T.; Cheng, Y.; Chewaskulyong, B.; et al. Overall survival with osimertinib in untreated, EGFR-mutated advanced NSCLC. N. Engl. J. Med. 2020, 382, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The biology and management of non-small cell lung cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Tu, S.; Chen, Y.; Liu, T.; Lee, Y.; Yen, J.; Fang, H.; Chang, J. Mutation profile of non-small cell lung cancer revealed by next generation sequencing. Respir. Res. 2021, 22, 3. [Google Scholar] [CrossRef] [PubMed]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images are more than pictures, they are data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; De Jong, E.E.C.; Van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Parmar, C.; Grossmann, P.; Rietveld, D.; Rietbergen, M.M.; Lambin, P.; Aerts, H.J.W.L. Radiomic machine-learning classifiers for prognostic biomarkers of head and neck cancer. Front. Oncol. 2015, 5, 272. [Google Scholar] [CrossRef] [PubMed]
- Forghani, R.; Savadjiev, P.; Chatterjee, A.; Muthukrishnan, N.; Reinhold, C.; Forghani, B. Radiomics and artificial intelligence for biomarker and prediction model development in oncology. Comput. Struct. Biotechnol. J. 2019, 17, 995–1008. [Google Scholar] [CrossRef] [PubMed]
- Bortolotto, C.; Lancia, A.; Stelitano, C.; Montesano, M.; Merizzoli, E.; Agustoni, F.; Stella, G.; Preda, L.; Filippi, A.R. Radiomics features as predictive and prognostic biomarkers in NSCLC. Expert Rev. Anticancer Ther. 2021, 21, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Tu, W.; Sun, G.; Fan, L.; Wang, Y.; Xia, Y.; Guan, Y.; Li, Q.; Zhang, D.; Liu, S.; Li, Z. Radiomics signature: A potential and incremental predictor for EGFR mutation status in NSCLC patients, comparison with CT morphology. Lung Cancer 2019, 132, 28–35. [Google Scholar] [CrossRef]
- Li, H.; Gao, C.; Sun, Y.; Li, A.; Lei, W.; Yang, Y.; Guo, T.; Sun, X.; Wang, K.; Liu, M.; et al. Radiomics analysis to enhance precise identification of epidermal growth factor receptor mutation based on positron emission tomography images of lung cancer patients. J. Biomed. Nanotechnol. 2021, 17, 691–702. [Google Scholar] [CrossRef]
- Moreno, S.; Bonfante, M.; Zurek, E.; Cherezov, D.; Goldgof, D.; Hall, L.; Schabath, M. A radiogenomics ensemble to predict EGFR and KRAS mutations in NSCLC. Tomography 2021, 7, 154–168. [Google Scholar] [CrossRef]
- Zhao, W.; Chen, W.; Li, G.; Lei, D.; Yang, J.; Chen, Y.; Jiang, Y.; Wu, J.; Ni, B.; Sun, Y.; et al. GMILT: A novel transformer network that can noninvasively predict EGFR mutation status. IEEE Trans. Neural Netw. Learn. Syst. 2022, 35, 7324–7338. [Google Scholar] [CrossRef]
- Meng, Y.; Sun, J.; Qu, N.; Zhang, G.; Yu, T.; Piao, H. Application of radiomics for personalized treatment of cancer patients. Cancer Manag. Res. 2019, 11, 10851–10858. [Google Scholar] [CrossRef]
- Bidzińska, J.; Szurowska, E. See Lung Cancer with an AI. Cancers 2023, 15, 1321. [Google Scholar] [CrossRef]
- Wu, Y.; Wu, F.; Yang, S.; Tang, E.; Liang, C. Radiomics in early lung cancer diagnosis: From diagnosis to clinical decision support and education. Diagnostics 2022, 12, 1064. [Google Scholar] [CrossRef]
- Hernadez, M.; Epelde, G.; Alberdi, A.; Cilla, R.; Rankin, D. Synthetic tabular data evaluation in the health domain covering resemblance, utility, and privacy dimensions. Methods Inf. Med. 2023, 62, e19–e38. [Google Scholar] [CrossRef]
- Pezoulas, V.C.; Zaridis, D.I.; Mylona, E.; Androutsos, C.; Apostolidis, K.; Tachos, N.S.; Fotiadis, D.I. Synthetic data generation methods in healthcare: A review on open-source tools and methods. Comput. Struct. Biotechnol. J. 2024, 23, 2892–2910. [Google Scholar] [CrossRef]
- Wang, A.X.; Chukova, S.S.; Simpson, C.R.; Nguyen, B.P. Challenges and opportunities of generative models on tabular data. Appl. Soft Comput. 2024, 166, 112223. [Google Scholar] [CrossRef]
- Xu, L.; Skoularidou, M.; Cuesta-Infante, A.; Veeramachaneni, K. Modeling tabular data using conditional gan. Adv. Neural Inf. Process. Syst. 2019, 32, 7335–7345. [Google Scholar]
- Borisov, V.; Leemann, T.; Seßler, K.; Haug, J.; Pawelczyk, M.; Kasneci, G. Deep Neural Networks and Tabular Data: A Survey. IEEE Trans. Neural Netw. Learn. Syst. 2022, 35, 7499–7519. [Google Scholar] [CrossRef] [PubMed]
- Munir, M.A.; Shah, R.A.; Ali, M.; Laghari, A.A.; Almadhor, A.; Gadekallu, T.R. Enhancing Gene Mutation Prediction With Sparse Regularized Autoencoders in Lung Cancer Radiomics Analysis. IEEE Access 2024, 13, 7407–7425. [Google Scholar] [CrossRef]
- Akiba, T.; Sano, S.; Yanase, T.; Ohta, T.; Koyama, M. Optuna: A next-generation hyperparameter optimization framework. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Anchorage, AK, USA, 4–8 August 2019; pp. 2623–2631. [Google Scholar]
- Demšar, J. Statistical comparisons of classifiers over multiple data sets. J. Mach. Learn. Res. 2006, 7, 1–30. [Google Scholar]
- García, S.; Fernández, A.; Luengo, J.; Herrera, F. Advanced nonparametric tests for multiple comparisons in the design of experiments in computational intelligence and data mining: Experimental analysis of power. Inf. Sci. 2010, 180, 2044–2064. [Google Scholar] [CrossRef]
- Salimans, T.; Goodfellow, I.; Zaremba, W.; Cheung, V.; Radford, A.; Chen, X. Improved techniques for training GANs. Adv. Neural Inf. Process. Syst. 2016, 29, 2234–2242. [Google Scholar]
- Arjovsky, M.; Bottou, L. Towards principled methods for training generative adversarial networks. arXiv 2017, arXiv:1701.04862. [Google Scholar] [CrossRef]
- Gulrajani, I.; Ahmed, F.; Arjovsky, M.; Dumoulin, V.; Courville, A.C. Improved training of Wasserstein GANs. Adv. Neural Inf. Process. Syst. 2017, 30, 5769–5779. [Google Scholar]
- Snoek, J.; Larochelle, H.; Adams, R.P. Practical Bayesian optimization of machine learning algorithms. Adv. Neural Inf. Process. Syst. 2012, 25, 2951–2959. [Google Scholar]
- Bergstra, J.; Yamins, D.; Cox, D.D. Making a science of model search: Hyperparameter optimization in hundreds of dimensions for vision architectures. In Proceedings of the 30th International Conference on Machine Learning, Atlanta, GA, USA, 16–21 June 2013; Volume 28. [Google Scholar]


| Algorithm | Similar Features | Metrics | |||||||
|---|---|---|---|---|---|---|---|---|---|
| ST (↑) | MW (↑) | KS (↑) | ST (↑) | MW (↑) | KS (↑) | WD (↓) | IWD (↑) | US (↑) | |
| kl_weight = 0.0 | |||||||||
| GSRA-KL*0.5 | 263 | 225 | 177 | 0.7302 | 0.4235 | 0.2918 | 0.0399 | 0.9601 | 0.5395 |
| GSRA-KL*1 | 252 | 168 | 145 | 0.5157 | 0.2933 | 0.2422 | 0.0416 | 0.9584 | 0.4504 |
| GSRA-KL*2 | 215 | 124 | 114 | 0.3742 | 0.2003 | 0.2048 | 0.0478 | 0.9526 | 0.3708 |
| kl_weight = 1.0 | |||||||||
| GSRA-KL*0.5 | 263 | 251 | 229 | 0.7125 | 0.6144 | 0.4896 | 0.0294 | 0.9706 | 0.6553 |
| GSRA-KL*1 | 246 | 201 | 188 | 0.5775 | 0.4634 | 0.4146 | 0.0304 | 0.9696 | 0.5837 |
| GSRA-KL*2 | 232 | 171 | 143 | 0.4373 | 0.2739 | 0.2735 | 0.0356 | 0.9644 | 0.4445 |
| TVAE | 263 | 251 | 229 | 0.5942 | 0.5866 | 0.5248 | 0.0331 | 0.9699 | 0.6395 |
| Algorithm | kl_weight | BE (↑) | MMD (↓) | IMMD (↑) | RS (↑) |
|---|---|---|---|---|---|
| GSRA-KL*0.5 | 0.0 | 0.1525 | 0.2746 | 0.7254 | 0.4792 |
| GSRA-KL*1 | 0.0 | 0.1418 | 0.2622 | 0.7378 | 0.4440 |
| GSRA-KL*2 | 0.0 | 0.1245 | 0.2507 | 0.7493 | 0.4105 |
| GSRA-KL*0.5 | 1.0 | 0.1097 | 0.1995 | 0.8005 | 0.5352 |
| GSRA-KL*1 | 1.0 | 0.0891 | 0.1829 | 0.8171 | 0.4990 |
| GSRA-KL*2 | 1.0 | 0.0796 | 0.1750 | 0.8250 | 0.4492 |
| TVAE | - | 0.0730 | 0.1819 | 0.8181 | 0.5200 |
| Algorithm | Accuracy (↓) | AUC (↓) | Precision (↓) | Recall (↓) | F1-Score (↓) | DLS (↓) | IDLS (↑) | Comments |
|---|---|---|---|---|---|---|---|---|
| kl_weight = 0.0 | ||||||||
| GSRA-KL*0.5 | 0.7026 | 0.7923 | 0.6495 | 0.9762 | 0.7601 | 0.77614 | 0.22386 | Good |
| GSRA-KL*1 | 0.7320 | 0.8435 | 0.6915 | 0.9762 | 0.7859 | 0.80582 | 0.19418 | Poor |
| GSRA-KL*2 | 0.7288 | 0.8425 | 0.6869 | 1.0000 | 0.7796 | 0.8028 | 0.1972 | Poor |
| kl_weight = 1.0 | ||||||||
| GSRA-KL*0.5 | 0.6914 | 0.7679 | 0.6302 | 0.9500 | 0.7405 | 0.756 | 0.244 | Good |
| GSRA-KL*1 | 0.6592 | 0.7016 | 0.5998 | 0.8955 | 0.7048 | 0.71218 | 0.28782 | Good |
| GSRA-KL*2 | 0.6322 | 0.6900 | 0.5747 | 0.8336 | 0.6689 | 0.68008 | 0.31992 | Good |
| TVAE | 0.4235 | 0.5125 | 0.3758 | 0.6000 | 0.4604 | 0.47444 | 0.52556 | Excellent |
| Algorithm | KL Weight | RDS | Comments |
|---|---|---|---|
| GSRA-KL*0.5 | 0.0 | 0.3892 | Poor |
| 1.0 | 0.4596 | Excellent | |
| GSRA-KL*1 | 0.0 | 0.3654 | Poor |
| 1.0 | 0.4235 | Good | |
| GSRA-KL*2 | 0.0 | 0.3492 | Poor |
| 1.0 | 0.2695 | Poor | |
| TVAE | – | 0.5241 | Excellent |
| Metric | Layer Size | Correlation () | p-Value |
|---|---|---|---|
| US | 0.5* | 0.819 | 5.50 × 10−6 |
| 1.0* | 0.794 | 1.77 × 10−5 | |
| 2.0* | 0.431 | 0.051 | |
| IMMD | 0.5* | 0.899 | 3.14 × 10−8 |
| 1.0* | 0.855 | 8.16 × 10−7 | |
| 2.0* | 0.394 | 0.078 | |
| BE | 0.5* | –0.871 | 2.71 × 10−7 |
| 1.0* | –0.809 | 8.96 × 10−6 | |
| 2.0* | –0.270 | 0.236 | |
| RS | 0.5* | 0.823 | 4.54 × 10−6 |
| 1.0* | 0.794 | 1.77 × 10−5 | |
| 2.0* | 0.516 | 0.0167 | |
| IDLS | 0.5* | 0.865 | 4.21 × 10−7 |
| 1.0* | 0.831 | 3.06 × 10−6 | |
| 2.0* | 0.662 | 0.00107 | |
| RDS | 0.5* | 0.878 | 1.70 × 10−7 |
| 1.0* | 0.823 | 4.54 × 10−6 | |
| 2.0* | 0.219 | 0.339 |
| Algorithm: GSRA-KL | Best Hyperparameters | ||
|---|---|---|---|
|
Sparsity Regularization |
L2 Weight Regularization | Sparsity Proportion | |
| kl_weight = 0.0 | 2.46 × 10−4 | 2.16 × 10−4 | 1.31 × 10−1 |
| kl_weight = 1.0 | 1.28 × 10−3 | 1.01 × 10−4 | 1.05 × 10−1 |
| Metric | 0.5* | 1.0* | 2.0* | p-Value |
|---|---|---|---|---|
| mse | 0.007682 ± 0.000694 | 0.007452 ± 0.001735 | 0.007976 ± 0.001502 | 7.165 × 10−1 |
| optimization_time | 220.788208 ± 41.293179 | 237.399137 ± 58.739415 | 278.671245 ± 19.188771 | 4.115 × 10−2 |
| training_time | 11.237552 ± 3.404399 | 10.945072 ± 3.145071 | 12.941548 ± 1.542467 | 5.385 × 10−1 |
| total_execution_time | 232.026462 ± 43.408871 | 248.344791 ± 61.702421 | 291.613893 ± 20.362463 | 1.290 × 10−1 |
| ram_usage_gb | 3.251824 ± 1.181036 | 3.294094 ± 1.285505 | 4.207957 ± 1.640278 | 1.727 × 10−7 |
| disk_usage_gb | 29.107617 ± 0.003875 | 29.111608 ± 0.006725 | 29.121750 ± 0.012597 | 1.727 × 10−7 |
| Metric | Layer Size | Correlation () | p-Value |
|---|---|---|---|
| MSE | 0.5* | −0.784 | 2.56 × 10−5 |
| 1.0* | −0.743 | 1.15 × 10−4 | |
| 2.0* | −0.332 | 0.141 | |
| Optimization Time | 0.5* | 0.823 | 4.54 × 10−6 |
| 1.0* | 0.722 | 2.19 × 10−4 | |
| 2.0* | −0.705 | 3.56 × 10−4 | |
| Training Time | 0.5* | 0.745 | 1.05 × 10−4 |
| 1.0* | 0.649 | 1.45 × 10−3 | |
| 2.0* | −0.471 | 0.0310 | |
| Total Execution Time | 0.5* | 0.842 | 1.74 × 10−6 |
| 1.0* | 0.701 | 3.97 × 10−4 | |
| 2.0* | −0.690 | 5.43 × 10−4 | |
| RAM Usage (GB) | 0.5* | 0.500 | 0.0210 |
| 1.0* | 0.312 | 0.169 | |
| 2.0* | 0.421 | 0.0575 | |
| Disk Usage (GB) | 0.5* | 0.357 | 0.112 |
| 1.0* | 0.217 | 0.345 | |
| 2.0* | 0.392 | 0.0787 |
| Overall Means of Testing Metrics with Standard Deviation (SD) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dataset with Imbalanced Class Distribution | Dataset with Balanced Class Distribution | |||||||||
| Strategy | Accuracy | AUC | Precision | Recall | F1-Score | Accuracy | AUC | Precision | Recall | F1-Score |
| Real Radiomics Dataset | ||||||||||
| TRTR | 0.8000 | 0.4700 | 0.0250 | 0.0500 | 0.0333 | 0.6621 | 0.7750 | 0.6120 | 0.8077 | 0.6884 |
| SD | 0.1182 | 0.1638 | 0.0750 | 0.1500 | 0.1000 | 0.1270 | 0.1667 | 0.1171 | 0.0988 | 0.0873 |
| GSRA-KL based Synthetic Radiomics Dataset | ||||||||||
| TSTR | 0.8434 | 0.7437 | 0.4987 | 0.4250 | 0.4233 | 0.7958 | 0.8426 | 0.7934 | 0.8296 | 0.8075 |
| SD | 0.0733 | 0.1706 | 0.2918 | 0.2341 | 0.2119 | 0.1354 | 0.1833 | 0.1527 | 0.1024 | 0.1201 |
| TVAE based Synthetic Radiomics Dataset | ||||||||||
| TSTR | 0.7867 | 0.8863 | 0.4060 | 0.9083 | 0.5585 | 0.7789 | 0.8488 | 0.7524 | 0.8451 | 0.7928 |
| SD | 0.0598 | 0.0578 | 0.0734 | 0.0870 | 0.0818 | 0.0687 | 0.0707 | 0.0742 | 0.0854 | 0.0611 |
| Dataset with Imbalanced Class Distribution | Dataset with Balanced Class Distribution | |||||
|---|---|---|---|---|---|---|
| Algorithm | Strategy | UDS | Comments | Strategy | UDS | Comments |
| EGFR Mutation | ||||||
| - | TRTR | 0.2757 | Poor | TRTR | 0.7090 | Excellent |
| GSRA-KL | TSTR | 0.5868 | Poor | TSTR | 0.8138 | Excellent |
| TVAE | TSTR | 0.7092 | Excellent | TSTR | 0.8036 | Excellent |
| Metric | Scale 0.5* | Scale 1.0* | Scale 2.0* |
|---|---|---|---|
| IMMD | 0.90 | 0.85 | 0.39 |
| US | 0.82 | 0.79 | 0.43 |
| BE | –0.87 | –0.81 | –0.27 |
| RS | 0.82 | 0.79 | – |
| IDLS | 0.86 | 0.66 | – |
| RDS | 0.88 | 0.82 | – |
| Algorithm | PSO-Based Previous Study [26] | Optuna-Based Current Study | ||||
|---|---|---|---|---|---|---|
|
RAM (GB) |
Disk (GB) |
Time (s) |
RAM (GB) |
Disk (GB) |
Time (s) | |
| GSRA-KL () | 2.70 | 32.60 | 1729 | 1.45 | 29.10 | 168.59 |
| GSRA-KL () | 3.60 | 36.60 | 2731 | 2.83 | 29.11 | 278.14 |
| Retraining time (s) with best hyperparameters | ||||||
| GSRA-KL () | – | – | 15.63 | – | – | 7.59 |
| GSRA-KL () | – | – | 6.79 | – | – | 12.09 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Munir, M.A.; Shah, R.A.; Waheed, U.; Aslam, M.A.; Rashid, Z.; Aman, M.; Masud, M.I.; Arfeen, Z.A. Sparse Regularized Autoencoders-Based Radiomics Data Augmentation for Improved EGFR Mutation Prediction in NSCLC. Future Internet 2025, 17, 495. https://doi.org/10.3390/fi17110495
Munir MA, Shah RA, Waheed U, Aslam MA, Rashid Z, Aman M, Masud MI, Arfeen ZA. Sparse Regularized Autoencoders-Based Radiomics Data Augmentation for Improved EGFR Mutation Prediction in NSCLC. Future Internet. 2025; 17(11):495. https://doi.org/10.3390/fi17110495
Chicago/Turabian StyleMunir, Muhammad Asif, Reehan Ali Shah, Urooj Waheed, Muhammad Aqeel Aslam, Zeeshan Rashid, Mohammed Aman, Muhammad I. Masud, and Zeeshan Ahmad Arfeen. 2025. "Sparse Regularized Autoencoders-Based Radiomics Data Augmentation for Improved EGFR Mutation Prediction in NSCLC" Future Internet 17, no. 11: 495. https://doi.org/10.3390/fi17110495
APA StyleMunir, M. A., Shah, R. A., Waheed, U., Aslam, M. A., Rashid, Z., Aman, M., Masud, M. I., & Arfeen, Z. A. (2025). Sparse Regularized Autoencoders-Based Radiomics Data Augmentation for Improved EGFR Mutation Prediction in NSCLC. Future Internet, 17(11), 495. https://doi.org/10.3390/fi17110495








