1. Introduction
Advances in next-generation sequencing can be seen in the rapid decrease in sequencing costs attributed to genome sequencing, which dropped from USD 1 million in 2007 to USD 600 in 2021 [
1]. This drop has led to the exponential growth of sequencing data, which ranges from gigabytes to terabytes and even petabytes. The fact that the capacity of sequence data doubles within 12 months while the processor processing capacity doubles within 18 months results in a significant challenge. Consequently, it requires a more powerful sequence processing program to manage the data deluge [
2].
Sequence alignment is the most fundamental process in the analysis of genomic data. Across most variation detection and annotation processes, it is the starting point, leading to biologically significant consequences. It is a highly computing-intensive and resource-heavy process. In genomics, efficient sequence alignment certainly has practical implications, primarily for evaluating data correctly and aligning millions to billions of short-read sequences accurately. The greatest computational challenge, however, is handling the ever-growing data in genomic sequences. Among the operations that the alignment of sequences provides is variant analysis, which is crucial in the process of the interpretation of genetic information or the discovery of cryptic features in DNA, RNA, and protein sequences. It also enables researchers and scientists to analyze and interpret genomic data more compactly and precisely, advancing personalized medicine, genetic disease diagnosis, and drug discovery.
The FM index is renowned in the large bioinformatics community for its enormous ability to handle large-scale short-read sequence alignment projects [
3]. Its small size and fast query capabilities in the next-generation sequencing platform pose an optimal condition to accommodate the vast amounts of short-read data. Paolo Ferragina and Giovanni Manzini [
4] designed the succinct model of the FM index. This approach was adopted in 1994 when Michael Burrows and David Wheeler suggested compressing text using the Burrows–Wheeler Transform (BWT) before building the FM index. This allows characters in the text sequence to be permuted with the BWT, making the patterns of the transformed data similar to those of the original characters, generally allowing for the better compressibility of the modified data. In addition, the BWT was combined with the sequence alignment properties to form the FM index, further consolidating a wavelet tree data structure. This combination highly augmented efficiency during the pattern matching and searching performed within the compressed datasets, primarily for sequence data. Nowadays, the FM index is widely employed in many short-read sequence aligners to facilitate the efficient and accurate mapping of short reads to genome references [
5], including BOWTIE, BWA, and SOAP2.
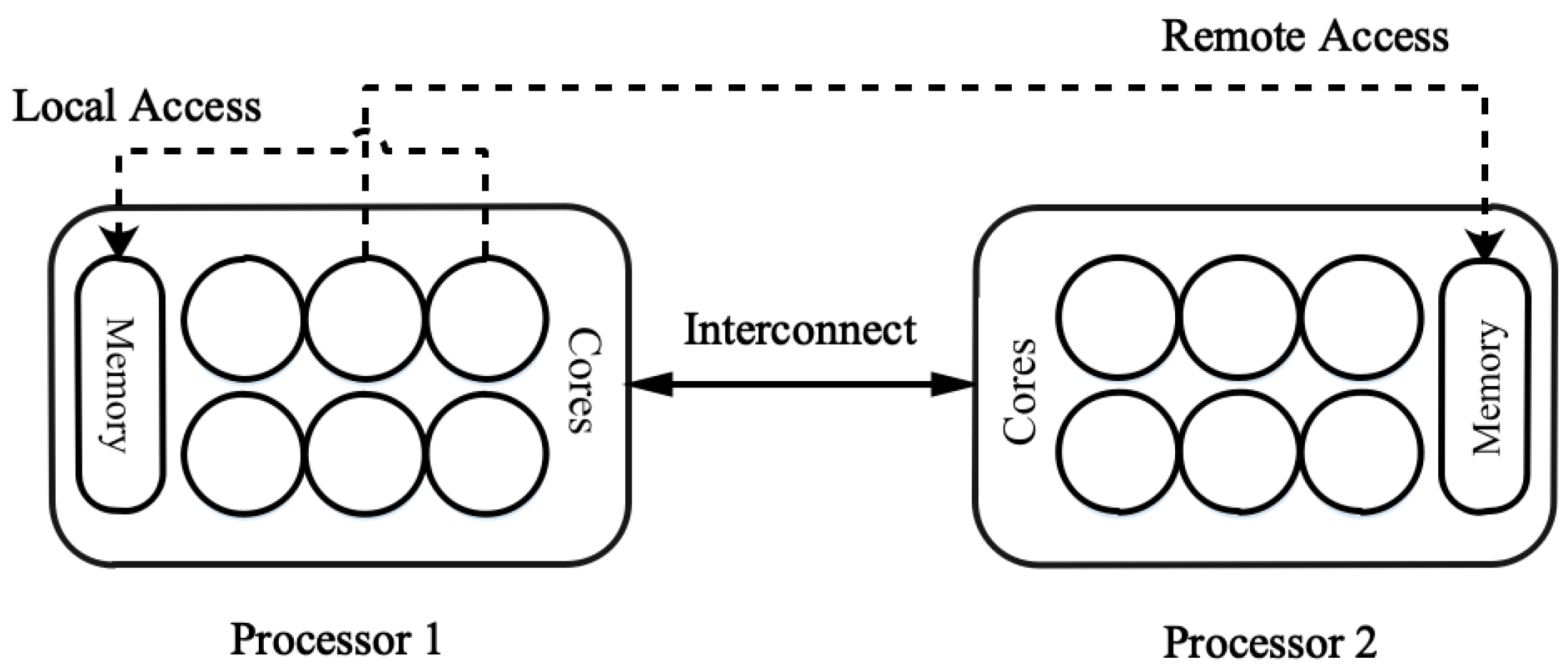
Heterogeneous architecture nodes could result in a reliable and efficient computational framework within an HPC system [
6]. The heterogeneous system utilizes different computing infrastructures that come with various processor architectures to support parallel processing and maximize the extent of computation that can be executed simultaneously [
7]. Thus, the heterogeneous system proves to be more reliable for sequence alignment and requires extensive computation. In addition, the heterogeneous system consists of NUMA architectures, which are prevalent in HPC systems. The effective utilization of NUMA can enhance the performance of crucial applications such as short-read sequence alignments. This NUMA architecture enables processing optimization, allowing processors to have much faster access to memory segments and significantly decreasing the bottleneck in data processing. Therefore, the enhancement in such heterogeneous systems can result from the implementation of NUMA-aware optimizations in the computational effectiveness of HPC environments [
8].
In this paper, it was observed that the common FM-based alignment algorithms showed limitations in terms of computational efficiency when dealing with longer and more complex datasets. We proposed a parallel approach for short-read sequence alignment using the longest-first alignment method combined with enhanced partitioning to improve the performance of FM aligners for the evaluation of large-scale genomic short-read sequence data. The proposed algorithm employs LF and EP to achieve efficient load balancing and parallelization on heterogeneous NUMA processors of the HPC system. We conducted numerous experiments using sequence datasets from the 1000 Genomes Project and various benchmark datasets to assess the alignment performance of various FM-based algorithms. The alignment performance of the LF-EP-based FM aligner was shown to be particularly outstanding in comparison to established well-known FM aligners, like SOAP2, BOWTIE2, BWA, and GEM3, according to the popular benchmark BALIBASE, OXBENCH, SABRE, and PREFAB. These results in the experiments suggest that the LF-EP-based FM aligner results in better speedup and more accurate alignment results in better quality than other state-of-the-art FM aligners, thus making it a reliable, precise approach for large-scale genomic short-read sequence data. Furthermore, the results also provide valuable insights into future study directions to improve the efficiency of FM aligners in heterogeneous architecture systems.
The following is a summary of the contributions of this investigation:
- (1)
We developed a convenient parallelism technique for NUMA systems that couples the longest-first distribution strategy with enhanced partitioning. This approach attains effective load balancing and optimal parallelization across heterogeneous NUMA processors on HPC systems. This contribution focuses primarily on the effective and rapid deployment of bioinformatics algorithms on modern heterogeneous architectures.
- (2)
We introduced the LF-EP-based FM aligner, which utilizes the FM index for short-read sequence alignment with a novel implementation tailored for popular heterogeneous computing platforms that constitute NUMA nodes. This proposed aligner shows significant performance improvements in scalability, speedup, and quality, which is convenient for genomic research where massive data volumes are processed rapidly.
- (3)
We conducted a comprehensive evaluation of the popular NUMA infrastructures. Our paper demonstrates improvements in parallelism through a thorough review of both main processor platforms. We tested our approach on a wide variety of NUMA architectures, including Intel Xeon and AMD Opteron.
- (4)
We compared it to prevailing FM aligners and sequence benchmarks. These FM aligners include BWA, SOAP2, BOWTIE2, and GEM and various sequence benchmarks—BALIBASE, OXBENCH, SABRE, and PREFAB. These comprehensive aligners and benchmarks not only highlight the practical advantages but also establish a baseline for future studies to be compared with.
The rest of the paper is organized as follows: a critical survey of related studies in the literature is performed in
Section 2. Basic concepts of sequence alignment, an in-depth review of the FM aligner, and details of heterogeneous architectures are presented in
Section 3. Our proposed algorithm, longest-first combined with enhanced partitioning, is provided in
Section 4. The experimental results and performance evaluation of the proposed algorithm are presented in
Section 5. Experimental findings are concluded in
Section 6, with prospective research directions detailed in brief.
2. Literature Study
The FM index finds a wide range of applications related to genomic studies, particularly in sequence alignment. It is a suitable approach for aligning short reads to large genome references with a small memory footprint [
9]. Most efficient sequence aligners, including BOWTIE [
10], BWA [
11], HISAT [
12], and SOAP2 [
13], exploit the FM index-based mechanism. The FM index has two types of indices: a global FM index for the entire genome and many small FM indices for genome fragments that span the whole genome together [
3].
Additionally, another area where the FM index is used in genomics studies is whole-genome alignment [
14]. Moreover, hardware-accelerated exact and inexact alignment using the FM index has been studied for FPGA-based alignment architectures [
15]. An evaluation study, in the context of RNA alignment, showed a significant reduction in memory consumption and runtime of FM index-based aligners compared to hash table-based aligners at nearly identical precision and recall rates [
16]. This test also showed the high tolerance of BOWTIE to indels and sequencing errors, so it was not the proper candidate for using high-tolerant but inexact hash-based aligners. While FM-based aligners have been widely adopted as the data structure for several years for the fast alignment of sequences, some performance limitations have been observed in memory-constrained systems when using FM-based aligners compared to hash-based aligners [
17].
Sequencing alignment is a collaborative research type in which heterogeneous architectures have been widely used. NUMA has emerged as one of the most widely utilized computing infrastructures [
18]. Research focusing on sequence alignments as a trade-off between data locality and data dispersion in the systems based on NUMA highlights an interesting research topic on NUMA-based heterogeneous architecture [
16]. Several studies [
19,
20,
21] have quantitatively investigated the performance of various memory allocation policies and data partitioning strategies in NUMA architectures. Many algorithms and tools have been developed, but they do not consider the impact of the computing structure on the overall performance of sequence alignment. For instance, BarraCUDA is one of the short-read sequence aligners that rapidly align sequences using GPUs [
22]. It uses a suffix-tree method to locate exact matches in the reference sequence. GSWABE provides all-to-all pairwise global, semi-global, and local alignment through a tile-based method that leverages the computational capability of CUDA-enabled GPUs [
23]. However, these new hardware techniques are not aimed at improving the efficiency of the utilization of the existing infrastructures.
Based on the above discussion, using NUMA-based heterogeneous architectures in HPC systems can generally improve sequence alignment algorithms [
24]. Processors with NUMA architectures and parallelization techniques enhance the speed of the alignment process. It is very important to note that the size of biological databases is continuously increasing, which results in data being more complex in the alignment process and also needing more time for computation [
25]. Additionally, there is a requirement for the development of alignment algorithms that can work with datasets where sequences have varying lengths, which is a must to achieve more precise results for applications requiring high precision [
26]. However, much more research is needed to be conducted regarding the investigation of the interplay between stages of alignment and the processor architecture to enhance the performance of the sequence alignment algorithms for NUMA-based heterogeneous architectures. The research aims to address these challenges and contribute to better and more effective alignment algorithms suitable for datasets exhibiting heterogeneity in computing architectures within heterogeneous processors of HPC systems.
4. The Proposed Algorithm
This research takes into consideration various NUMA computing architectures, which have been used in most of the latest developments in the HPC field. Heterogeneous clusters have become the first preference of most of the research community today because they can form a versatile architecture for computing systems [
32]. It is usually much easier to optimize load balance in homogeneous nodes as their similar designs allow for more coherent interaction. Therefore, our proposed scheme primarily focuses on addressing load balancing in heterogeneous systems comprising different types of NUMA architectures. Furthermore, we focus on homogeneous architecture to solve the potential parallelism within the processors. Our system comprises two phases, discussed as follows.
Heterogeneous phase: Load balancing across all the nodes of the cluster is the primary purpose behind our efforts in this phase. We use a longest-first (LF) approach to achieve load balancing among the heterogeneous architectures. This enables us to distribute the workload across many nodes
Homogeneous phase: The fundamental aim of this phase is to maximize the data parallelism on each homogeneous processor within the heterogeneous architectures. To achieve this objective, we have developed an enhanced partitioning (EP) technique by exploiting the available computational resources according to the maximum potential benefits on each node.
4.1. Longest-First Algorithm in the Heterogeneous Phase
The algorithm introduced in this research paper proposes a parallel methodology for partitioning a set of sequence reads, incorporating both the
k-mer distance and the maximum length of the sequence read datasets. The
k-mer distance is a measure of dissimilarity between genomic sequences with respect to the frequencies of subsequences of length
k. The realization of multiple jobs executed simultaneously with an optimal balance is intended to be carried out in a NUMA node-based heterogeneous computing environment.
| Algorithm 1. LF algorithm |
Input:- (1)
Sequence reads set . - (2)
k-mer distance set of is . - (3)
The longest length of a sequence read set is . - (4)
j-part partitioned sequence reads set .
Output:- (1)
d-part partitioned sequence reads set .
Steps:
- 1.
Calculate the k-mer distance for each element in parallel. - 2.
Rank sequence reads set in the decreasing order based on the values of k-mer distance set K. - 3.
Divide into subread sets , where . - 4.
Sort in decreasing order based on the longest length of subsequence reads set , make . - 5.
Set is the total sum of complementation time in . - 6.
For k = 1 to d. - 7.
Sort in increasing order , such that . - 8.
Set . - 9.
End for.
|
The following sequence depicts the parallel workflow of the longest-first algorithm in the heterogeneous phase.
Calculating the k-mer distance: For all the sequence reads in the input set, the first operation calculates the k-mer distance in parallel. The k-mer distance is calculated for any two sequence reads and measures the dissimilarity between them.
Ranking sequence reads: The sequence reads are ordered from the highest to the lowest based on k-mer distances. Therefore, the sequence reads with the highest k-mer distance are ranked at the top, the second highest k-mer distance follows, and so on.
Partitioning sequence reads: The ranked sequence reads are partitioned into several sets. There can optimally be different numbers of sets based on the value of j, where j is the number of partitioned sequence read sets. All the sets could be of almost equal size except for the last set, which can have any size based on the total number of sequence reads and the value of j.
Arranging subread sets: The subread sets are arranged in decreasing order from the longest size of the sequence reads in a subset. That means the subread with the longest sequence read is the one of the highest priority, and the subread with the second longest sequence read is the one of the second highest priority, and so on.
Merging partitions: The algorithm enters a loop that runs d times, where d is the final number of partitions. For each of those d iterations, the partitioned sequences are sorted in ascending order according to the sum of total complementation time. At each iteration, the sequence with the least complementation time is merged into the partitioned set, thereby updating the sum of complementation time for the partitioned set.
According to Algorithm 1, some crucial steps we conducted, including steps 3–8, are necessary to make an explanation. Step 3 describes how sequence reads are partitioned into subsets. It ensures that the sequence reads are divided as evenly as possible among the subsets.
There are components that exist in Step 3, including , the set of all sequence reads; , the set of partitioned subread sets; , the number of divided sequence reads; , the total number of sequence reads; and , the -th subread set.
One part of the formula describes the first subread sets. Each of these sets (for ) contains sequence reads. The floor function effectively balances the distribution by adding 1 to each subset. The other part of the formula describes the last subread set . The size of this last set is adjusted to ensure that the total number of sequence reads is accounted for. The size of is calculated as s minus the total number of reads allocated to the first sets, which is .
Step 3 has two main characteristics. One is balancing subsets. The formula ensures that the first subsets are filled with a nearly equal number of sequence reads. Each of these subsets receives 1 reads, where is the integer division of by . The other characteristic is adjusting the last subset. The last subset contains the remaining reads, which may not be divided evenly among the other subsets. This ensures that all reads are included in the partitioning without any being left out.
Step 4 involves sorting the partitioned sets of sequence reads based on the longest length of the subsequence reads within each set. The details are discussed in the following explanation. We sort
in decreasing order. The subsets
are sorted such that
based on the length of the longest sequence read in each subset. This means that the subset with the longest sequence read is first, the subset with the next longest sequence read is second, and so on. Let
denotes the length of the longest sequence read in subset
. The sorted subsets
should satisfy the following condition. The length of the longest sequence read in the first subset is
, in the second subset is
, and so on.
Steps 5–8 describe the final steps of the algorithm for partitioning and balancing the sequence reads based on their complementation times. These steps involve iterating through the final sets, sorting them, and merging them to ensure balanced load distribution. These steps first make an initialization. We ensure that each final set is not empty and is a subset of the sorted sets . This is followed by the iterative merging and sorting of the final sets. In the iteration process, we sort the final sets in increasing order based on the longest sequence read length . We ensure that the total complementation times are in increasing order: . Then, we merge sequence reads from into : . Finally, we update the total complementation time by . These steps ensure that the sequence reads are efficiently and effectively balanced across the final sets, minimizing the total complementation time and achieving an optimal distribution of the computational load. This approach maximizes the performance and efficiency of sequence alignment tasks in high-performance computing environments.
According to the discussions above, the proposed LF method is a parallel approach for sequence read partitioning based on the average k-mer distance and the longest length of an obtained sequence read set. This implementation works in heterogeneous computing environments where numerous tasks can be performed in parallel. Algorithm 1 starts by calculating the k-mer distance between sequences in parallel and then ranking the sequences in decreasing order. These are later partitioned into subread sets by considering the value of with the decreasingly ranked sequence reads. Finally, the subread sets are sorted in descending order by the sequence read length. The final partitioning loop is then run times to ensure that the tasks are equally distributed among the processing units. These configurations ensure the efficiency and synchronization of the implementation process.
4.2. Enhanced Partitioning Algorithm in Homogeneous Phase
As shown in
Figure 3, there are only two genome datasets involved in the sequence alignment procedure.
represents a short-read dataset and
represents a long genome reference.
is the individual dataset of short reads at node
.
corresponds to a particular block in short read
and
is the total amount of blocks of
.
is the responding genome reference at node
.
corresponds to a particular replica of the genome reference of
and
is the number of replicas of
. Therefore, short-read subsets in node
can be referred to as
and genome reference replicas in node
are referred to as
in
Figure 3. Within the sequence alignment procedure, three pivotal steps can be identified: establishing, mapping, and gathering. These steps are summarized as follows.
- (1)
Establishing stage: The aligner builds the index file for the genome reference ; the wall time of this stage is denoted as .
- (2)
Mapping stage: The aligner achieves an alignment sub-result by mapping short read against genome reference ; the wall time of this stage is denoted as .
- (3)
Gathering stage: The aligner collects sub-results from stage 2 and takes some necessary configurations to merge the final output ; the wall time of this stage is denoted as .
The majority of the total time in the sequence alignment is typically allocated to the establishing and mapping stages. It is noteworthy that the mapping stage necessitates a substantial allocation of computational resources and entails the most significant computing resource. The generation and dissemination of the index file can be performed in advance and shared among multiple instances during the experiments. Thus, the overall execution time of the three stages, denoted as , can be simplified to in general scenarios.
A range of s and g values is taken into consideration for a comprehensive view, which is detailed further in the following section.
- (1)
Case 1: s = g = 1. This case discusses the native implementation of the original structure of a sequence aligner built for mapping complete reads to a single genome reference.
- (2)
Case 2: s = 1, g > 1. This case is utilized for the purpose of conducting the alignment of complete reads that are shared, against a set of g replicates of the reference genome. The use of many replicas of the genetic reference inside a NUMA architecture system offers benefits in terms of system architecture, as this design incorporates a large number of memory blocks.
- (3)
Case 3: s = 1, 0 < g < 1. This case involves mapping the complete reads to a subsequence of the genome reference, specifically 1/g of the reference sequence. The genome reference is partitioned into 1/g segments, which are separately aligned with the whole set of read data.
- (4)
Case 4: s > 1, g = 1. This case is implemented by dividing the reads into s subsets, which are mapped against a complete replica of the genome reference. The dataset is partitioned into s segments, and a complete genome reference is distributed across the threads residing in the memory.
- (5)
Case 5: s > 1, g > 1. This case is implemented with s subsets of reads being mapped to g replicates of the genome reference. The short read and genome reference are divided or duplicated into s segments and g duplicates, respectively. A partitioned subread can be regarded as a genome reference that has been repeated, resulting in the equation s = g. Thus, this case is suitable for large memory systems.
- (6)
Case 6: s > 1, 0 < g < 1. This case is implemented by mapping s subsets of the reads to 1/g subsequence of the genome reference. Both the short read and the genome reference are divided into s and 1/g segments, respectively. A partitioned subread can be viewed as a genome reference that has been replicated, resulting in a value of s = 1/g. Therefore, this case is suitable for memory-sensitive systems.
Table 2 presents a taxonomy of the alignment approaches within the context of the EP algorithm, categorized by the number of read subsets and genome replicates involved. Additionally,
Table 2 displays operational descriptions and illustrative applications in genomics. The examples in cases 1–4 provide a comprehensive overview of the steps in the EP method evaluation and critically illustrate its effectiveness in performance. They also establish a baseline for a more detailed comparison and analysis in the experiments. Cases 5 and 6 are discussed to illustrate the best-case scenarios for the EP approach in terms of its implementation quality in a heterogeneous system with distinctive architectural features.
The pseudocode of the EP algorithm is provided in Algorithm 2. Such an algorithmic framework is typical in bioinformatics because it facilitates the efficient processing of large-scale sets of sequences. It is a scalable and parallelizable methodology applicable to the complex datasets commonly associated with genomic research.
| Algorithm 2. EP algorithm |
Input:- 1.
reads: The set of reads to be aligned. - 2.
genome_reference: The genome reference used for alignment. - 3.
num_read_partitions s: The number of subsets/partitions for reads. - 4.
num_genome_partitions g: The number of replicas/partitions for the genome reference.
Output:- 5.
alignment_results: The alignment results for the reads against the genome reference.
Steps:- 6.
Function partitionReads(read reads, int num_partitions) {return array of read partitions}. - 7.
Function replicateGenome(genome original_genome, int num_replicas) {return array of replicated genome partitions}. - 8.
Function partitionGenome(genome original_genome, float partition_ratio) {return array of genome partitions}. - 9.
Function alignPartition(readPartition reads, genomePartition genome) {return alignment results for that partition}. - 10.
Function enhancedAlignment(read reads, genome genome, int num_read_partitions, int num_genome_partitions) {
Steps:
- a.
Initialize an array alignment_results for storing results. - b.
If no partitioning is required (both num_read_partitions == 1 and num_genome_partitions are == 1). - c.
Directly align reads to the genome using a standard alignment function. - d.
Else: - e.
Partition reads using partitionReads(). - f.
Depending on the value of num_genome_partitions, either replicate the genome using replicateGenome() or partition it using partitionGenome(). - g.
If (num_genome_partitions == 1) {genome_partitions = genome;} - h.
Else if (num_genome_partitions > 1) { genome_partitions = replicateGenome(genome, num_genome_partitions);} - i.
Else {genome_partitions = partitionGenome(genome, 1/num_genome_partitions);} - j.
Loop through each combination of reads and genome_partitions: - k.
Align each read partition to each genome_partition using alignPartition(). - l.
Store the results in alignment_results. - m.
Outputs: alignment_results.
- 11.
}
|
5. Results and Evaluations
5.1. Experimental Environment
In general, the data utilized in our research are derived from the international 1000 Genomes Project, which commenced in January 2008. We specifically selected the human genome (ID: HS37D5) as our reference sequence, which can reach a size of up to three gigabytes. For one of our natural short-read datasets, we employed the SRR766060 paired-end genome, with each segment approximately measuring 8.7 GB in size. Furthermore, we incorporated four widely recognized benchmark datasets in their latest version for alignment quality evaluation purposes—BALIBASE, OXBENCH, SABRE, and PREFAB. Additionally, for speedup evaluation and sequence complexity experiments, we utilized specified sequence datasets obtained from several widely implemented read simulation tools in bioinformatics, such as randDNA and wgsim. The utilization of these compelling datasets has significantly contributed to obtaining reliable outcomes throughout the experiment we conducted.
The overview information on the experimental environment and configurations is shown in
Table 3. Several additional widely used FM algorithms—SOAP2, BOWTIE2, BWA, and GEM3—were used in this research to comprehensively compare several significant issues. The experiments were also conducted on two different NUMA nodes, Intel Xeon and AMD Opteron, which are also commonly used in heterogeneous systems. These NUMA nodes have the capacity to support four processors with 32 cores, 64 threads, and 128 GB of memory. Although each processor has four NUMA nodes, they are interconnected differently among the cores. AMD Opteron 6376 utilizes a core network with full ties, while Intel Xeon E5 4620 employs a ring structure with circular connections among its cores, as shown in
Figure 1.
5.2. Quality Assessment of the Alignment Results
In this section, we compare the LF-EP FM aligner’s performance with the original FM aligner and other state-of-the-art FM algorithms such as SOAP2, BOWTIE2, BWA, and GEM3. We use BALIBASE, OXBENCH, SABRE, and PREFAB benchmarks to evaluate alignment quality. These benchmarks are highly respected in sequence alignment because they effectively evaluate aligner performance with respect to different features of alignment accuracy. This aims to assess aligner effectiveness across multiple benchmarks under various scenarios and sequence attributes. The benchmarks have been converted to the universally accepted FASTA format (as short-read sequences), which facilitates the following analytical processes.
In our evaluation, we use two accuracy metrics: Quality Score (QS) and Total Column Score (TCS). The QS measures the fraction of correct residue pairs aligned relative to the total number of residue pairs in the alignment. On the other hand, the TCS measures the correct alignment of columns relative to the total number of columns in the alignment. The TCS shows that the aligners are capable of reflecting the structural features of the sequences. The selection of the algorithms and the benchmarks is made in such a way that they represent a broad spectrum of methodologies for sequence alignment, allowing for the creation of a global assessment of the alignment performances of the aligners. The same FM-based algorithms are used in aligners tested in the benchmarks. Therefore, it might be expected that these various aligners would show slight differences in result quality. The absolute quantities QS and TCS are dimensionless; only their relative ratio is of interest.
Table 4 illustrates the QS and TCS values computed for different benchmarks employed in the present study. Since the QS and TCS values are dataset-dependent, we assess their relative values to provide a comparative assessment of the effectiveness of different sequence aligners. From the analysis in
Table 4, the proposed LF-EP-based FM aligner surpassed the rest, with the highest scores in both QS and TCS. Evaluation results reveal that the LF-EP-based FM aligner is among the most highly reliable and accurate FM aligners compared with previous aligners, thus signifying the ability to integrate the LF-EP algorithm into the framework of FM aligner.
Furthermore, SOAP2 was observed to perform at almost the same level as the LF-EP FM aligner, albeit a little lower. It can be observed that QS and TCS scores of SOAP2 are the second highest across most benchmarks, indicating that this tool is quite promising and could improve its alignment performance with further development. Additionally, BOWTIE2 shows a small decrease in performance quality compared to SOAP2, suggesting it might be suboptimal for strict quality standards. BWA also shows a similar drop in performance to BOWTIE2 but maintains impressive QS and TCS scores consistently during the evaluations. This finding positions BWA as a practical choice based on particular task needs and circumstances. Conversely, GEM3 displays the least impressive performance among the specified FM aligners. Although adequate for some tasks, its effectiveness is restricted in situations where top-tier accuracy is essential.
Therefore, the LF-EP-based FM aligner introduced in this research outperforms the other popular FM aligners across the four benchmarks, demonstrating its excellent reliability and precision. This finding substantiates the efficacy of implementing the LF-EP algorithm. Although SOAP2 demonstrates robustness and dependability, there is still room for improvement. Furthermore, the evaluation indicates that the performance quality of BOWTIE2, BWA, and GEM3 decreases progressively. Therefore, their suitability might fluctuate with the changing needs of specific tasks, ranging from high to lower levels.
5.3. Sequence Complexity Analysis
In biological sequences, which include DNA, RNA, and protein sequences, etc., average pairwise distance refers to the average genetic variation among all possible pairs within a particular set of sequences. It is used to show similarities or differences among a group of sequences. This complexity is examined by testing the datasets with 1000 sequences generated using the randDNA program. The average pairwise distance between sequences is varied from 50 to 500. The experiments are implemented on the selected datasets using the proposed algorithm along with other popular FM aligners such as BWA, SOAP2, BOWTIE2, and GEM3. The speedup factor is determined by measuring the performance efficiency of the BWA aligner in processing sequences with an average pairwise distance of 50.
The runtimes of different FM aligners on sequence datasets in the heterogeneous system of Intel Xeon and Intel Xeon Phi are depicted in
Figure 4. As the sequence distance increases, the speedup factor decreases for all aligners. This implies that the acceleration effect weakens in proportion to the sequence distance increase. The LF-EP-based FM aligner exhibits the largest speedup factor, reaching 4.4× at a sequence distance of 50, which demonstrates that the LF-EP-based FM aligner has a substantial accelerating effect on short-distance sequences. Additionally, the LF-EP-based FM aligner performs proficiently even at longer distances such as 300, 400, and 500. The LF-EP-based FM aligner consistently displays the highest speedup factor across all sequence distances, hence confirming its suitability for managing high-complexity data. Overall, GEM3 and BOWTIE2 demonstrate lower speedup factors than the LF-EP-based FM aligner. Most of their speedup factors are below 3, indicating less significant speed improvements. Therefore, GEM3 and BOWTIE2 may require further optimization to improve their speedup factors on high-complexity data. At shorter sequence distances (50 and 100), SOAP2 and BWA have speedup factors below 1, indicating that their speed is comparable to the original speed. Nonetheless, their speedup factors decrease as the sequence distance increases. Thus, SOAP2 and BWA are appropriate for shorter sequence distances, but their performance may have speedup limitations as the distance increases.
In conclusion, the research findings indicate that different aligners respond differently to variations in sequence distance, which affects their performance capabilities. The LF-EP-based FM aligner has been proven to be more efficient than SOAP2 in aligning sequences of both short and long distances. SOAP2, however, consistently demonstrates the least effective performance across all sequence distances. The BOWTIE2 algorithm exhibits a pronounced decline in its speedup as the sequence distance increases. The LF-EP-based FM aligner is the optimal tool for conducting complex sequence alignments on the Xeon–Xeon Phi heterogeneous system. It consistently demonstrates superior acceleration across various read lengths and is particularly well suited for large-scale datasets encompassing a mix of short and long sequences within a heterogeneous computing environment. The LF-EP-based FM aligner is an optimal solution for the management of a diverse range of sequence data due to its remarkable scalability.
5.4. Speedup Evaluation of FM Aligners
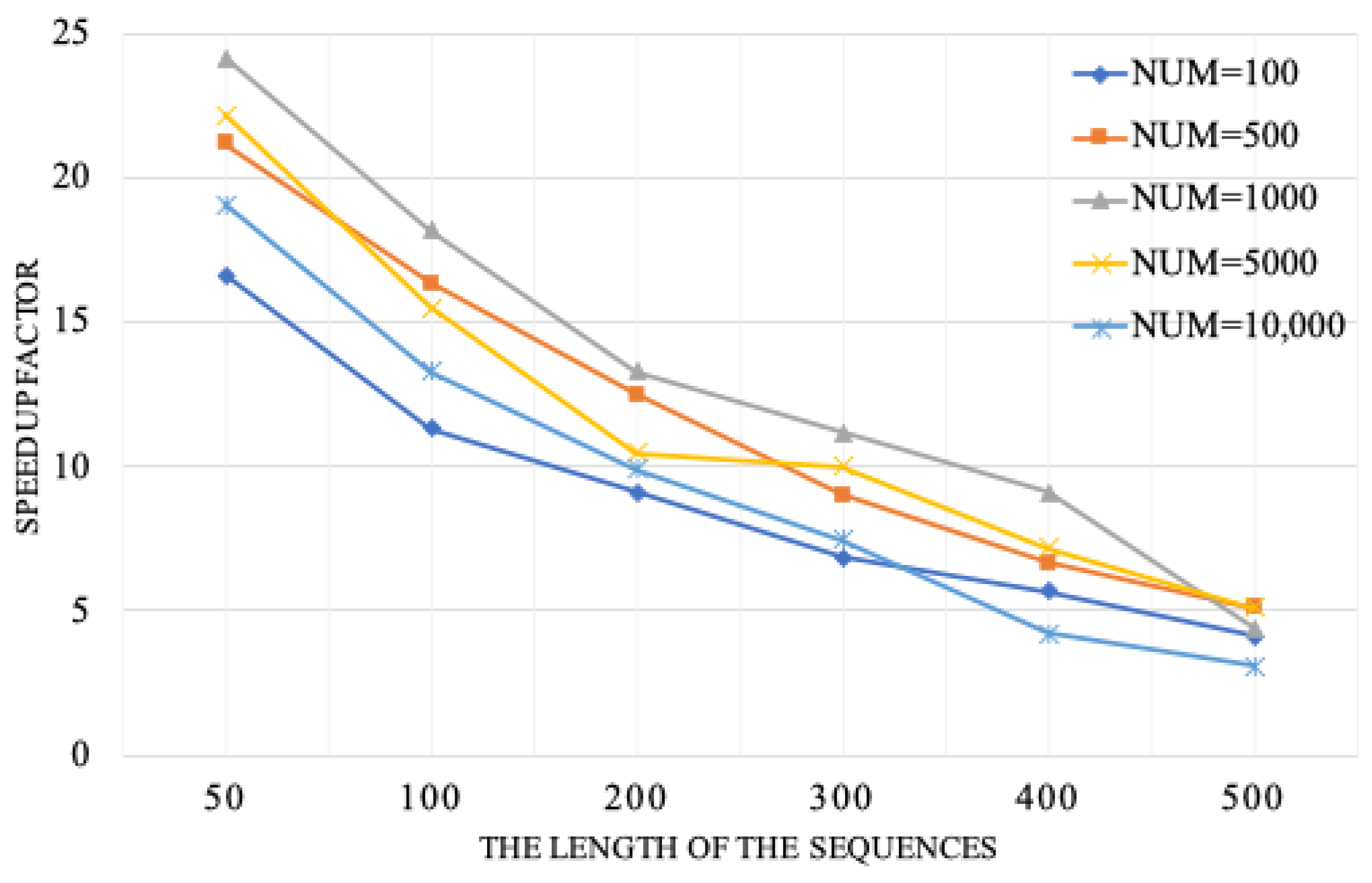
To accurately assess the speedup provided by the LF-EP-based FM aligner, we performed the following task of analyzing several sequence datasets. Some sequence mimic techniques, such as randDNA and wgsim, were used to establish these artificial datasets. In order to provide a complete picture of the speedup evaluation in these aligners, we included sequences whose average lengths ranged from 50 to 500, as well as a varied number of sequences ranging from 100 to 10,000.
In
Figure 5, we provide a detailed comparison of the speedup achieved with the LF-EP-based FM aligner versus the original FM aligner, which was implemented in the native mode. We calculate the speedup based on the original FM aligner divided by the LF-EP-based FM aligner in each instance. Our performance analysis of the proposed algorithm shows an excellent speedup of up to 24.2×. It is seen that the speedup decreases as the average sequence length increases. This is because the computational complexity of the FM aligner is sequence length-based. As the analyzed sequences become more extensive, the demand for physical memory and computing power becomes high, resulting in a performance bottleneck for FM aligner implementation in such a heterogeneous architecture.
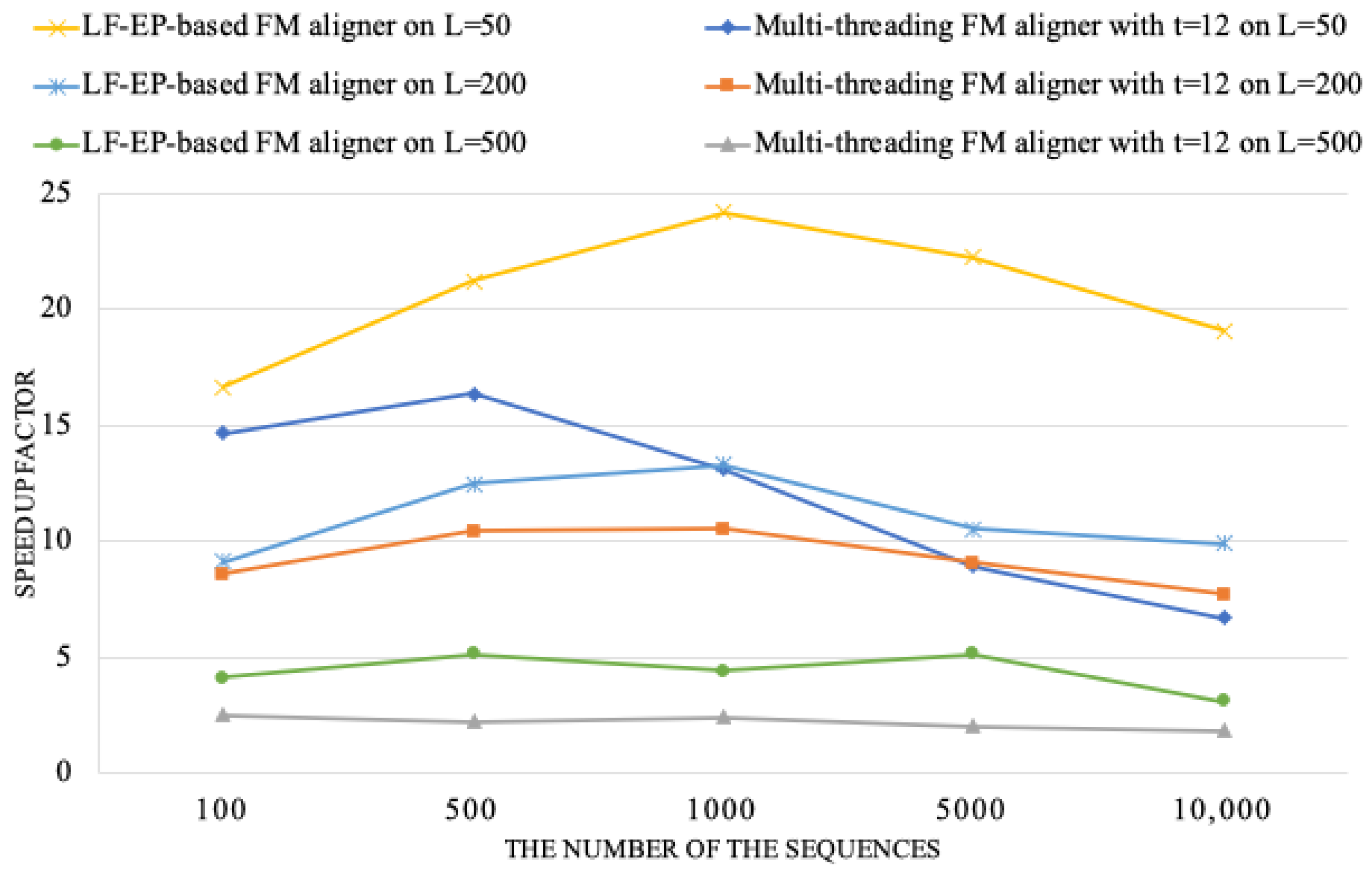
Furthermore, our study investigates those performance differences that really exist between the LF-EP-based FM aligner and the multi-threading FM aligner. This further analysis enables us to understand the relative strengths and limitations of each approach.
Figure 6 gives a valuable overview of the speedup achieved by the LF-EP-based FM aligner concerning the variant of the multi-threading FM aligner. Speedup is obtained concerning the performance of the original FM aligner.
Three sequence datasets were generated using mimic sequence tools in the experiments, each varying in average sequence length and number of sequences. The first dataset had an average length of 50, the second an average length of 200, and the third an average length of 500. Each dataset contained sequences ranging from 100 to 10,000. The results shown in
Figure 6 indicate that both the LF-EP-based FM aligner and the multi-threading FM aligner demonstrate remarkable speedup for the dataset with an average sequence length of 50. Additionally, a 24.2× speedup is obtained for the LF-EP-based aligner in a heterogeneous system, while this speedup is 16.3× for the multi-threading FM aligner. These results indicate substantial benefits of the LF-EP-based aligner, leading to a largely reduced execution time compared to the multi-threading alignment.
Thus, performance is significantly more different when analyzing other datasets whose average sequence lengths are 200 and 500. In these cases, although still impressively showing the speedup, the FM aligner based on the LF-EP method attains a lower value of 8.9×. The drop in speedup may be attributed to various factors, particularly the complexities of longer sequences and the corresponding increase in computational demands.
The efficacy of our parallel sequence alignment approach is apparent in its enhanced performance on sequences of moderate length, such as 50 or 200 lengths. However, it unveils intrinsic constraints when examining lengthier and more intricate data. The approach under consideration demonstrates significant improvements in computational efficiency when tested on artificially generated datasets, with average sequence lengths varying from 50 to 200 and average sequence quantities ranging from 100 to 1000. However, it is noticed that when the size of the datasets became greater than 200 in length or greater than 1000 in number, the expected improvement in computational performance became less significant. It also reveals some potential when larger datasets were used with our approach. In summary, our parallel approach demonstrates significant potential under specific circumstances but requires more refinement to be applicable to a wide range of sequence lengths and varied amounts of sequences.
6. Conclusions and Future Works
In this research, we develop and assess an optimized approach to the FM-based approach of short-read alignment on heterogeneous NUMA architectures. The novel integration of the longest-first algorithm and enhanced partitioning technique significantly enhances load balancing and data parallelism, addressing the computational challenges exhibited due to the exponential growth in sequencing data.
The proposed LF-EP-based FM aligner was benchmarked against popular tools such as BALIBASE, OXBENCH, SABRE, and PREFAB. These experiments demonstrate that our method, in terms of alignment quality and computational efficiency, outperforms some popular existing FM-based aligners. Our process has shown remarkable speedup and reduction in execution time, particularly in the handling of complex and large-scale genomic datasets within HPC systems. This approach we conducted is scalable and highly efficient, and our alignment management is robust in handling data deluge, thus assuring huge potential in personalized medicine for genetic disease diagnostics and drug discovery. Furthermore, our comprehensive quantification and correlation of sequence complexity underline the robustness of the LF-EP-based FM aligner in the management of high-complexity data from artificial data and the 1000 Genome Project.
Therefore, the LF-EP-based FM aligner is an impressive improvement to the sequence alignment technique. It is a very reliable and effective tool for genomics research. This research contributes to and opens new projects on the optimization of computational frameworks in heterogeneous environments within the field of bioinformatics, thereby enhancing the processing and interpretation ability of vast amounts of genomic data.
In the future, it is worth exploring and further developing the FM-based mechanism in areas of genomic studies [
33]. Consequently, it is highly recommended that integration with other robust algorithms and data structures be conducted so that it becomes more enhanced, improved, and effective towards sequence alignment in the heterogeneous NUMA architectures across various HPC platforms such as CPU plus GPU, FPGA, etc. [
34]. Furthermore, evaluations are recommended to be conducted on other reference sequences from different types of organisms. Therefore, this could ensure we validate our algorithm in detail concerning its efficiency and robustness.