1. Introduction
Volunteer computing (VC) uses consumer devices, such as desktop and laptop computers, tablets, smartphones, appliances and cars, to do high-throughput scientific computing. People can participate in VC by installing a program that downloads and executes jobs from servers operated by science projects. About 900,000 devices are currently participating in VC. These devices have 5 million CPU cores and 560,000 GPUs, and provide an average throughput of over 100 PetaFLOPS.
There are currently about 30 VC projects in many scientific areas and based at many institutions. The research enabled by VC has resulted in numerous papers in Nature, Science, PNAS, Physical Review, Proteins, PloS Biology, Bioinformatics, J. of Mol. Biol., J. Chem. Phys, and other top journals [
1].
Most VC projects use BOINC, an open-source middleware system [
2] (one major project, Folding@home, does not use BOINC [
3]). BOINC is a client/server system. Each project operates a server, which distributes jobs. Volunteers run the client program on their computing devices. A client can be “attached” to any set of projects. It periodically communicates with the project servers to report completed jobs and request new ones.
Originally, BOINC volunteers had to browse the set of projects and decide which ones to attach their clients to. More recently, BOINC has moved to a “coordinated model” in which volunteers select science areas rather than projects. A central coordinator dynamically assigns devices to projects, based on volunteer preferences and other factors. This model has several advantages. Most notably, projects no longer must recruit volunteers, and new projects can be assured a certain level of computing power. This reduces the barriers to entry for new projects.
The coordinated model is implemented in a system called Science United [
4]. Science United is designed to act as a “global scheduler” in a literal sense: dividing the world’s computing resources among the world’s computational scientists. The way it does this—its “scheduling policy”—must satisfy certain constraints: for example, jobs with particular RAM or GPU requirements can only be run on devices with those resources. Furthermore, it must try to balance several possibly conflicting goals: to maximize computational throughput, to honor volunteer preferences, and to allocate computing power to projects in proportion to per-project “shares”.
This paper describes global scheduling in Science United.
Section 2 outlines the architecture of BOINC.
Section 3 describes volunteer preferences.
Section 4 describes the scheduling policy.
Section 5 discusses the implementation and results, and
Section 6 proposes future work.
2. The Architecture of BOINC
2.1. BOINC Projects
Each science project using BOINC operates its own server, using the BOINC server software. The server dispatches jobs and provides a web interface for project administrators and volunteers.
The BOINC computing model assumes a highly heterogeneous resource pool [
5]. Jobs are submitted not for specific executables but for abstract “meta-apps” that can include any number of concrete “app versions”. Each app version has:
A “platform”: Windows/Intel, Mac OS on Intel or Apple Silicon, Linux on Intel or ARM, Android/ARM, and so on.
A set of additional hardware requirements, such as a GPU of a particular vendor and model, or a particular CPU feature such as SSE3.
A set of additional software requirements, such as a minimum GPU driver version, or the availability of a VM hypervisor such as VirtualBox.
A description of the processor usage, such as the number of CPUs or the usage (possibly fractional) of a GPU.
For example, a project might have an Autodock meta-app, comprising several versions of Autodock for different platforms and GPUs. Many projects have meta-apps containing dozens of app versions. Some projects (like GPUGrid.net [
6]) have only GPU applications; others (like LHC@home [
7]) have only VM applications. Thus, a particular volunteer device may be able to run jobs for some projects and not others.
When the BOINC scheduler dispatches a job to a client, it selects an app version to run the job with. It tries to use available resources, and it tries to choose the fastest version based on data from previous jobs.
Jobs have storage and RAM requirements and are dispatched only to qualifying devices. They also have a FLOPs estimate and a deadline. The BOINC scheduler estimates the runtime on a device, and dispatches jobs only to devices that are likely to complete the job by the deadline.
Because of these factors, it’s possible that, at a particular time, a project has many queued jobs but none that can be dispatched to a particular device.
Job submissions at a project may change over time. Some projects have an unbounded stream of jobs. Others may be sporadic: they might have large bursts of work, followed by weeks or months of inactivity.
2.2. The BOINC Client
The BOINC client runs on volunteer devices. Versions of the BOINC client are available for all major computing platforms.
The volunteer can specify a set of “computing preferences” that control when computing and file transfers can be carried out, how many CPUs can be used, how much RAM and storage can be used, and so on.
Each device is modeled as a set of “processing resources”, each possibly with multiple instances. The types of processing resources currently recognized by BOINC are CPUs, NVIDIA GPUS, AMD GPUs, and Intel GPUs. The client maintains a queue of jobs, each of which uses a subset of the processing resources. The client runs jobs in a way that (a) tries to fully utilize the available processing resources; (b) tries to meet the deadlines of jobs; and (c) respects limits on RAM usage based on the working-set size of jobs. It runs jobs at low process priority to minimize impact on system performance.
A client can be “attached” to one or more projects. It maintains estimates of the duration of the remaining jobs for each processing resources. When this falls below a threshold, it selects one of the attached projects and issues a “scheduler RPC” to its server, requesting jobs for one or more resources. The reply to this RPC includes, for each job, URLs of the app version’s files and the job’s input files. The client fetches these files and adds the jobs to the queue. The scheduler RPC and file transfers all use HTTP so that the client can function behind firewalls.
When the client is attached to a project, it caches the app version files so that these in general are downloaded only once. There is also a provision for caching job input files that are used by multiple jobs. Thus, each project has a “disk footprint” on the device; this may be many GB.
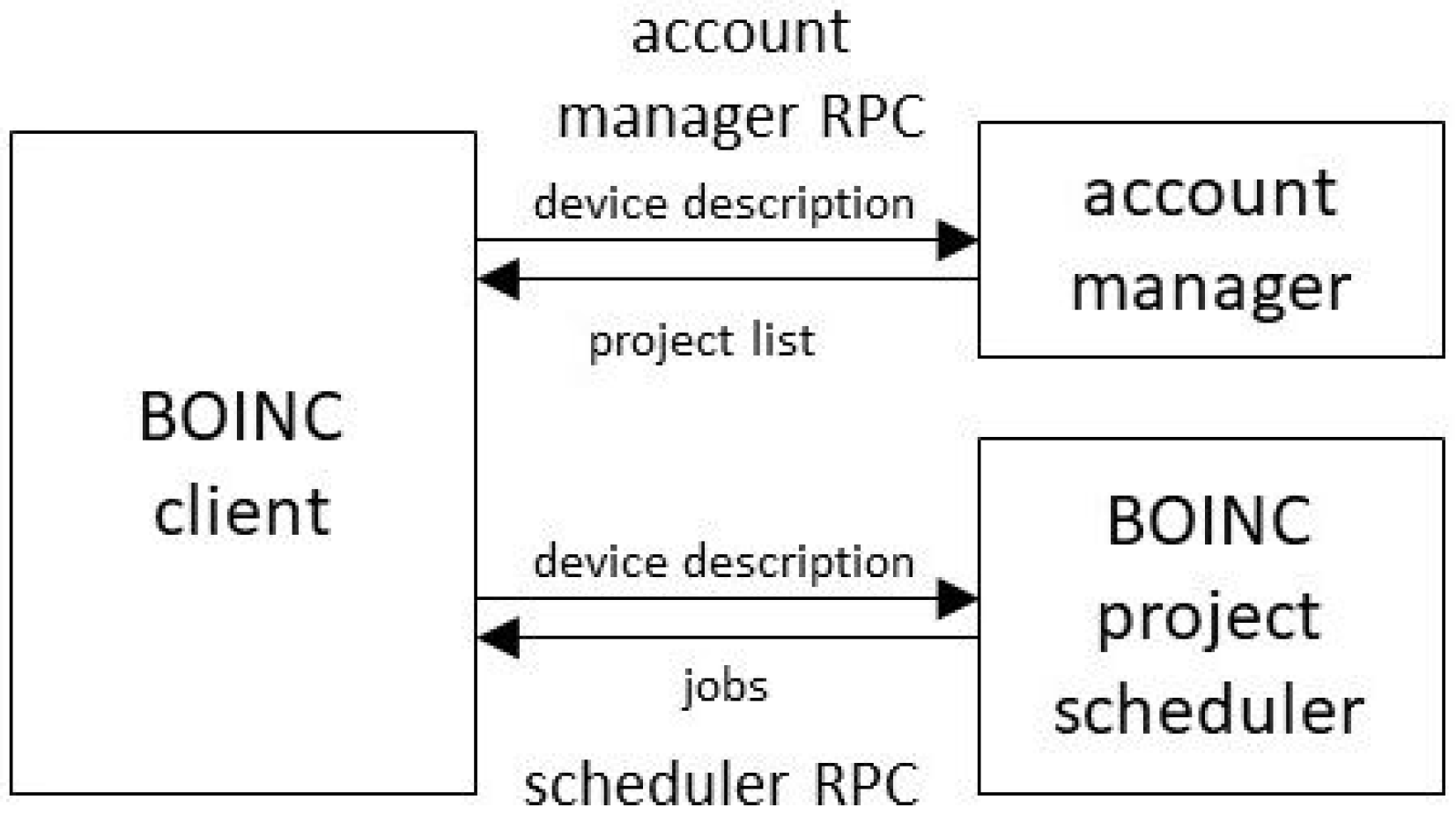
2.3. Account Managers
Instead of attaching a client directly to projects, a volunteer can attach it to an intermediate called an “account manager” (AM). This allows a level of indirection between clients and projects. The BOINC client periodically (typically once per day) issues an “account manager RPC” to the AM. The RPC reply contains a list of projects to which the client should attach. The client then communicates with those projects to get and report jobs; see
Figure 1.
The AM architecture was originally introduced to support web sites that let users browse and select projects: Gridrepublic, BAM!, and GRCPool [
8,
9,
10]. We extended the architecture to support Science United, in which users do not explicitly select projects.
The account manager RPC request message includes a detailed description of the device: its processing resources, software versions, and so on. It also includes a list of the projects to which the client is attached, and for each project, the amount of recent computing carried out and whether recent scheduler RPCs failed to return work; see
Section 4.5.
The RPC reply message includes a list of projects to which the client should attach. Each project descriptor can include a flag telling the client to finish any queued jobs for that project and then detach from it. The descriptor can also include flags for each processing resource, indicating whether the client should request jobs for that resource from the project. Thus, an account manager can control precisely how the device’s processing resources are allocated to projects.
3. Science United
In BOINC’s original model, a project recruits volunteers by publicizing itself and creating web pages describing its research. Volunteers survey available projects, and attach their clients to one or more.
The intention of this model was to create a “free market” in which scientists compete for computing power by promoting themselves and their research, and volunteers decide, based on their personal values and interests, which projects to support. The goal was that VC would divide computing power among scientists based on the aggregated knowledge and values of the public. This was inspired by the Iowa Political Stock Market, which used an analogous approach to predicting election results, with the viewpoint that “Markets allocate scarce resources to their most valued use” [
11].
The free market model did not work as intended. Most volunteers “locked in” to a few projects and did not seek out new ones [
12]. Additionally, in spite of the prospect of cheap computing power, relatively few scientists created BOINC projects. In retrospect this is understandable. In the free-market model, creating a project is risky: there’s a substantial investment [
13], with no guarantee of any return, since no one may volunteer. The model requires that projects publicize themselves, demanding resources and skills (media relations, web design, outreach) that are not readily available to most scientists. Volunteers demand a steady supply of jobs, and the computing needs of many research groups are sporadic.
To address these problems, we developed a “coordinated model” for BOINC-based volunteer computing. The client/server technology remains the same, but the volunteer interface is new. Instead selecting projects, volunteers now select the science areas they want to support. This aligns with the motivations of most volunteers: support of science goals has been shown to be the major motivation for participation in VC [
12].
A central “coordinator” then assigns volunteer devices to projects; this assignment can change over time. The coordinator is called “Science United” [
4]. New volunteers go to the Science United web site. As part of the signup process, they register their science-area preferences. They can also express preference for research locations—countries or institutions.
The volunteer downloads the BOINC client from Science United and installs it. Science United is an account manager. When the client first runs, it connects to Science United and obtains a list of projects to attach to, based on the volunteer’s preferences and other factors. This assignment can change over time; a volunteer may compute for a project that did not exist when they first registered.
Science United maintains a set of “vetted” BOINC projects, and it knows the science area and geographical attributes of each project. New BOINC projects can apply to be included in this list.
Science United’s coordinated model has several advantages over the free-market model:
Volunteers do not need to browse projects; in fact, they need not even be aware of the existence of projects.
Projects do not need to publicize themselves, or to operate a web site.
Scientists can apply to Science United to have prospective BOINC projects pre-vetted. At that point they can be guaranteed a certain amount of computing throughput, depending on their science area, their location, and what types of computing devices their applications can use. Thus the risk in creating a project is reduced.
Science United acts as a unified brand for VC. Publicity campaigns (mass media, social media, co-promotions, etc.) can refer to this brand, rather than the brands of individual projects, thus allowing more effective promotion.
Projects with sporadic computing needs do not risk losing volunteers.
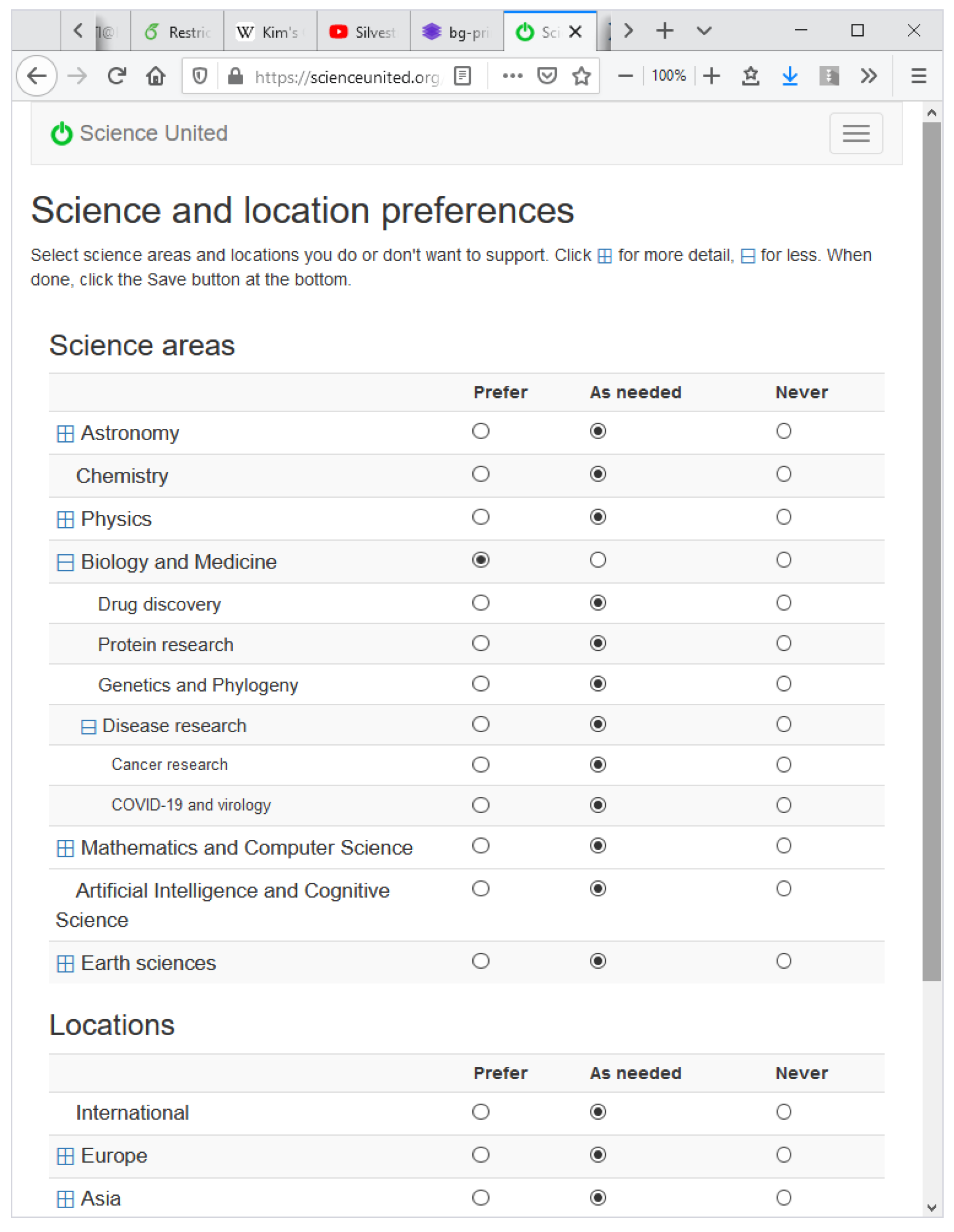
3.1. Science-Area and Location Preferences
As a basis for Science United volunteer preferences, we have defined a system of “keywords” for describing the nature and origin of jobs. There are two keyword categories: “science area” and “location” (the geographical and institutional location of the job submitter). Keywords form a hierarchy: each level N + 1 keyword is a child of a single level N keyword.
The set of keywords and the hierarchy can change over time. When a new keyword is added, the initial setting is “maybe” for all volunteers. Volunteers are notified of the new keyword.
Examples of BOINC projects and their associated keywords are shown in
Table 1.
3.2. Keyword Preferences
When a volunteer registers with Science United, they specify preferences for science areas and locations. A set of preferences maps keywords to {yes, no, maybe}. “No” means do not run jobs with that keyword. “Yes” means preferentially run jobs with that keyword. The user interface shows top-level keywords and lets the user drill down to lower levels; see
Figure 2.
Active Science United volunteers selected on average 4.8 “yes” keywords and 0.83 “no” keywords out of 25 science keywords and 41 location keywords. A total of 87% of the keyword preferences are for science areas; the remainder are for location. The most common “yes” science area keywords are Astronomy (68.8% of volunteers) and Biology and Medicine (67.1%). The most common common “no” keyword is Mathematics and Computer Science (8.0%). Among location keywords the most “yes” keywords are America (7.12%) and Europe (7.0%), and the most common “no” keyword is China (4.3%).
3.3. Project and Job Keywords
Each project has an associated set of keywords describing its science areas and location. The set of project keywords can change over time, reflecting changes in the project’s workload.
Science United enforces volunteer preferences at the project level. If a project has a keyword for which a volunteer has specified “no”, the volunteer’s devices are not attached to that project. In the future we plan to support projects whose jobs have differing keywords; see
Section 6.4.
4. The global Volunteer Computing Scheduling Problem
The central function of Science United is to divide computing power among projects. It does this by assigning volunteer devices to projects. These assignments can change each time the device issues an AM RPC (typically once per day). This section discusses the factors in the assignment policy, and describes the current policy.
4.1. Platforms and Processing Resources
Recall from
Section 2.1 that each device supports one or more “platforms” (Windows/x64, Mac/x64, Linux/ARM, etc.) and has a set of “processing resources”, including a CPU and possibly one or more GPUs of various vendors (NVIDIA, AMD, Intel). In addition, a device may have virtualization software (VirtualBox) installed. This information is conveyed to Science United in account manager RPC requests.
Recall also that each BOINC project has a set of “app versions”, each of which runs on a particular platform, uses a specific set of processing resources, and may require VirtualBox. Projects export this information through a Web RPC. Science United periodically imports this from all projects.
4.2. Project Shares
Science United allows projects to be allocated different amounts of computing resources. For example, we currently allocate more computing to COVID-related projects. This mechanism works as follows.
Let M(P) denote the maximum possible rate of computing (measured in FLOPS) for a project P, given Science United’s current resource pool. M(P) is determined by P’s keywords and applications. P can use a device D only if P’s keywords are compatible with the preferences of D’s owner, and it can use D’s processing resources (CPU and GPUs) only if it has appropriate applications. Thus M(P) can vary widely between projects.
In Science United, each project
P has a “share”
S(
P). Shares are assigned administratively (see
Section 6.3), and may change over time. Roughly speaking,
S(
P) determines how much computing is available to
P compared to other projects with similar
M(
P), over a time scale on the order of one week.
4.3. Resource Usage Accounting
Science United does accounting of processing resource usage. This serves several purposes: it provides a basis for enforcing project shares, it gives an estimate of the system-wide throughput, and it provides basis for volunteer incentive such as graphs of work carried out recently, work milestones, and so on.
BOINC has a system for estimating the FLOPs performed by completed jobs. It is “cheat-resistant”: it is difficult to get credit for computation not actually performed. However, the system is based in part on job replication, and credit for a job may not be granted until a replica of the job is completed, which could take weeks. This makes it unsuitable for Science United’s purposes.
Instead, Science United uses a quantity called “estimated credit” (EC), which is maintained by the BOINC client on a per-job and per-project basis, based on the runtime of jobs and the peak FLOPS of the processors they use. EC is a cruder estimate than credit, and it is not cheat-resistant. However, it accumulates continuously, with no need to wait for job completion or validation.
The AM RPC request message includes a list of currently attached projects and their CPU and GPU EC totals; these are used to update accounting records.
Science United maintains a database of historical accounting data. It maintains daily records (CPU and GPU EC and processing time, and number of jobs succeeded and failed) for each user and project, and in total. Once per day it adds these to the total for each entity, and creates a new daily record.
4.4. Share-Based Prioritization
Science United enforces project shares by prioritizing projects that have used less than their share of resources recently. It maintains, for each project
P, its average rate of computing over the last week, denoted
A(
P). This is measured in FLOPS, based on the estimated credit described above (in other words, the time each CPU and GPU was used, times the peak FLOPS of the processor). We let
Afrac(
P) is the fraction of total computing carried out by
P. Similarly, let
Sfrac(
P) is
P’s fraction of the total share. Then, let
E(P) represents the excess computing that P has received, relative to its share, over the last week. It is used to prioritize projects in the assignment algorithm (see below). At any point, computing resources are preferentially assigned to projects P for which E(P) is least.
This model handles both continuous and sporadic workloads well. For a project P with sporadic workload, E(P) will usually be near zero. When P generates a burst of work, it will have priority over the continuous-workload projects, and the work will get carried out quickly.
When a computer is assigned to a project, there is a delay of up to a day (the client polling period) until its computation is reported to Science United. Hence, the same project (the one for which E(P) is least) would be assigned to all hosts during that period. To avoid this, when we assign a computer to P we increment A(P) by an estimate of the computing (measured in EC) the computer will do in one day. Once per day A(P) is reset based on the accounting history.
4.5. Preventing Device Starvation
Can a project P supply jobs that use a processing resource R on a device D? This is central to the global scheduling problem, but it is not a simple question. We define:
Usable(P, D, R): this predicate is true if P has an app version for D’s platform that uses R, and D has virtualization software if needed by the app version. This may change over time as P adds and removes app versions.
If Usable(P, D, R) holds, it means that P can potentially supply jobs to D that use R. However, it does not mean that P can supply such jobs right now. P’s server may be temporarily down, or it may not have jobs that use R, or its jobs that use R may have keywords disallowed by the user’s preferences, or its jobs may require a different GPU driver version than the one on D, and so on.
If these conditions hold, and P is the only attached project, R is “starved”, i.e., the client has no jobs that use it. To maximize throughput, we want to avoid this situation.
Science United does not know about these factors, so it cannot directly know whether P can currently supply the needed jobs. Instead, it learns this from the BOINC client. The client keeps track of (a) projects for which the last scheduler RPC failed; and (b) (project, resource) pairs for which the last scheduler RPC requested work for the given resource, but none was returned. This information is included in the AM RPC request.
We then define:
Usable_now(P, D, R): this predicate is true if (a) Usable(P, D, R) is true, and (b) the most recent scheduler RPC from D to P succeeded and returned jobs using R.
Science United tries to attach D to projects that can use all its processing resources immediately, i.e., for which Usable_now(P, D, R) holds. However, it also must attach projects for which Usable(P, D, R) holds but Usable_now(P, D, R) does not, because the conditions that caused R to be unusable may be temporary. P’s server may come back up, or new jobs may be submitted. The only way for Science United to detect this is to attach D to P and let it request work for R. This will typically happen at least once per day: when a scheduler RPC fails, or fails to return jobs, the client uses exponential backoff with a maximum delay of one day.
4.6. Goals of the Scheduling Policy
The Science United scheduling policy has several goals:
To honor volunteer keyword preferences by preferentially assigning projects with the volunteer’s “yes” keywords.
To allow projects to be allocated different shares of the resource pool (see above).
To maximize total throughput. For example, if a host has a GPU, it should be assigned at least one project that can supply jobs that use the GPU.
To limit the number of attachments per device. Each attachment has a disk-space overhead; the client caches applications files for the project, which may include large VM image files.
These goals may conflict. For example, suppose a project P1 has a large share may and keywords with few “yes” preferences, while project P2 has a small share and keywords with lots of “yes” preferences. If we give P1 lots of computing, volunteers will notice that they’re not computing for the science areas they requested. The policy must balance these conflicting goals.
4.7. The Project Assignment Algorithm
BOINC clients using Science United periodically (once per day) issue an AM RPC. The request message includes a list of currently attached projects, with account and scheduler RPC failure information for each one as described in
Section 4.5.
The reply message includes a list of projects. Some of these are currently attached projects flagged as “outgoing”: this instructs the client to finish existing jobs for the project, then detach from it. For each of the other projects, the reply includes a list R(P) of processing resources that the client should use for P.
With these factors in mind, here is a sketch of the project assignment algorithm currently used by Science United:
First, we discard projects P that cannot be used because either (a) P has a keyword for which the volunteer has a “no” preference; or (b) Usable(P, D, R) is false for all processing resources, i.e., P does not have app versions that can use the device.
For each remaining project we compute a “score”
S(
P) which is the weighted sum of several components:
where
K is the number of project keywords for which the volunteer has a “yes” preference. Projects with such keywords will be preferred over projects with only “maybe” preferences.
E(
P) is the project’s allocation balance see
Section 4.4.
V is one or zero depending on whether the device has VirtualBox installed and the project has a VirtualBox app version that can run on the device. This preferentially assigns devices with virtualization capability (which are relatively scarce) to projects that require it.
C1, C2 and C3 weight these components, reflecting a balance between conflicting goals. They have been chosen empirically; currently C2 is 01 and the others are one.
We then compute a set A of projects to include in the reply This is carried out as follows. For each processing resource R, we find the highest-scoring project P for which Usable_now(P, D, R) is true. We add P to A, and include R in the resource list for P. If there are projects that have a higher score than P, and for which Usable(P, D, R) is true, we similarly add them to A.
The ability to specify the processing resources used by a project is important in some cases. For example, suppose the highest-scoring project P can only use the CPU, and the device also has a GPU. The scheduler can assign a lower-scoring project and have it use only the GPU, giving P full use of the CPU.
If the client is currently attached to a project not in A, we add it to A and set the “outgoing” flag, telling the client to finish existing jobs and detach from the project.
5. Implementation and Status
Science United was launched in 2017. Currently (August 2021) it has about 4100 active volunteers and 5800 computers, of which 4100 have usable GPUs. Averaged over the last month, these computers process 160,000 jobs per day and have a throughput of 400 TeraFLOPS.
The scheduling policy described in
Section 4 has evolved over time and has been in its current form for two years. It seems to satisfy the goals listed in
Section 4.6: project throughputs are stable and reflect shares, device starvation is rare, and no volunteers have complained about preferences not being respected.
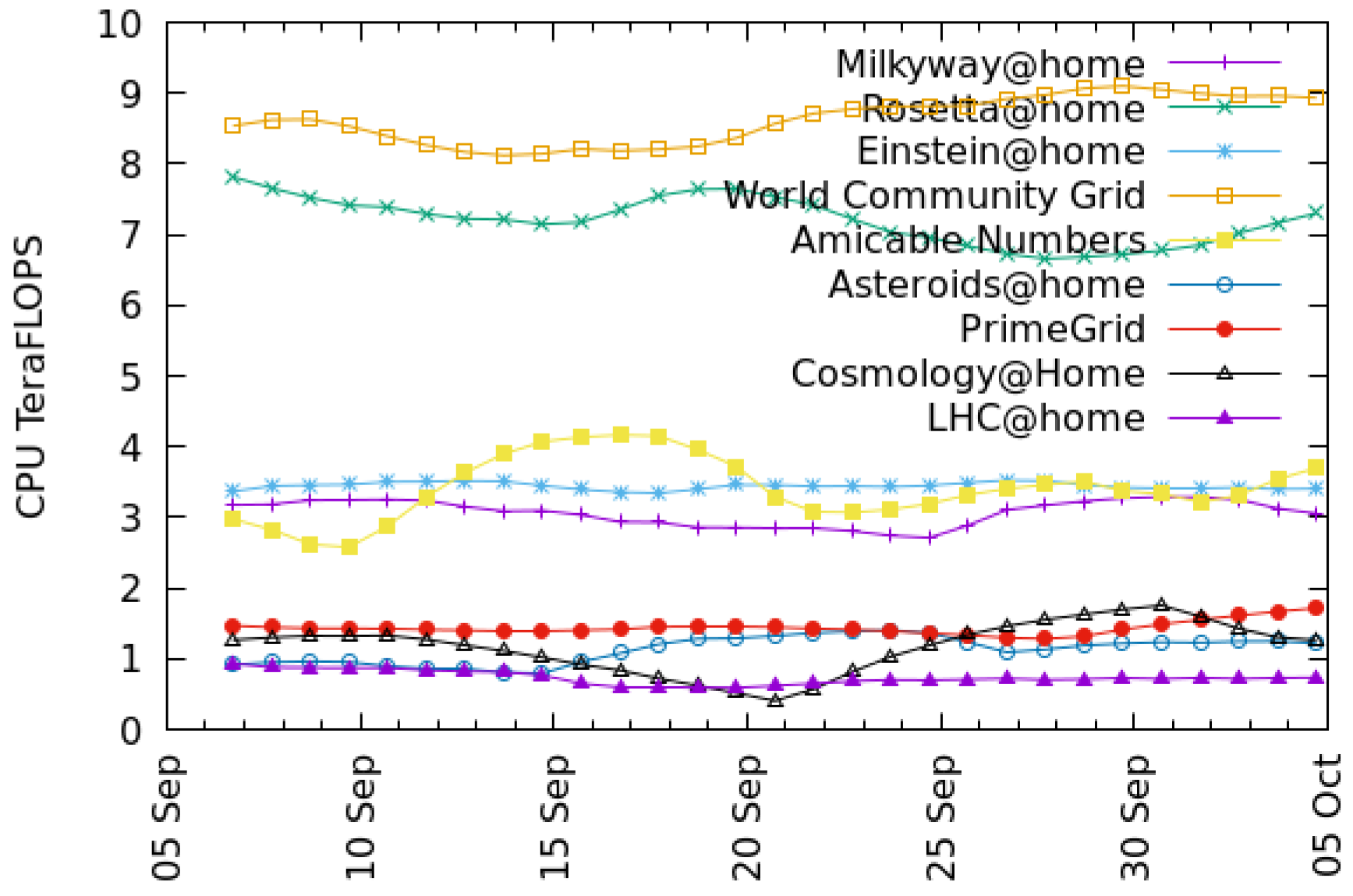
Figure 3 and
Figure 4 show recent throughput histories for CPU and GPU, respectively. Projects such as Rosetta@home appear only in the CPU graph because they have no GPU app versions.
Science United is implemented in PHP. It uses a MySQL database to store volunteer and project information, accounting data, and so on. The Science United source code is distributed under the LGPL v3 license and is available on Github.
SU required some modifications to the BOINC client, such as the starvation monitoring described in
Section 4.5. These changes were introduced in the 7.16 client release.
6. Future Work
6.1. Throughput Guarantees
In some HTC systems, a job submitter can be guaranteed a minimum throughput over a given period of time with high probability. Can we offer analogous guarantees with VC resources?
The performance of a pool of volunteer computers varies over time, in terms of both throughput and job latency. However, with a large resource pool, these quantities change slowly, and we can establish the statistics of this change. For example, given the total throughput T at a given time, we could find a T0 < T such that total throughput will remain above T0 for a week with a given confidence level.
Similarly, given the Science United resource pool and a particular set of projects and shares, the throughput of a project should remain fairly constant over time. These throughputs can be manipulated, within limits, by changing shares.
How can we predict, given a particular set of project shares, how much throughput each project will get? This depends on many factors: app versions, keywords, the project assignment algorithm, and so on. It’s unlikely that it can be determined analytically. Instead, we plan to implement an emulator that does a trace-based simulation of the entire Science United system and predicts the throughput of each project. Using this emulator we will be able to compute a mapping from project shares to project throughput, and to find project shares for which a particular project achieves a given throughput. This will provide a basis for guaranteeing throughput to projects over fixed periods.
Such guarantees would be project-level. Can we provide performance guarantees to a particular job submitter within a project that serves multiple job submitters? This is more complex but it may be possible. The BOINC server software allocates resources among computing job submitters within the project [
2]. It’s possible that the combination of a project-level allocation and a submitter-level allocation can provide performance guarantee to individual job submitters.
6.2. Minimizing Project Churn
Each time is attached to a client, the client must download a possibly large set of files. If the a project is detached and attached repeatedly, this may imposed a large network load at both server and client. It may be desirable to modify the scheduling algorithm to minimize this “project churn”. If a project has a large disk footprint on a client, we might want the client to remain attached, with a zero resource share, so that files do not need to be downloaded again the next time the project is attached.
6.3. Project Vetting and Allocation
Science United must make two administrative decisions: which projects to vet (i.e., to include in Science United), and what shares to assign to projects. Our current policy is to vet projects for which:
The project is non-commercial.
The project’s computing is directed toward a scientific or technical goal (broadly interpreted to include things such as mathematics and cryptography).
The project follows various security practices, such as application code-signing on secure offline machines.
Project shares might be influenced by the extent to which the project’s leadership is scientifically qualified (as demonstrated, e.g., by publications). In practice, we generally give all projects equal shares; in the COVID era we have given larger shares to projects doing COVID-related research.
In the future, we may establish a committee, including representatives of scientific and volunteer communities, to make these decisions.
6.4. Diverse Projects
Some projects have applications in multiple science areas or run jobs on behalf of multiple institutions; we call these “diverse” projects. The current Science United design does not handle such situations well, because keywords are at the project level. If a project P has 1% of jobs with keyword K, and a volunteer has a “no” preference for K, they will not be able to run the other 99% of P’s jobs.
With a few changes, Science United can handle diverse projects correctly. First, diverse projects must associate keywords with individual jobs, as part of the submission process. For example, if a job is submitted by a cancer researcher at UC Berkeley, its keywords would include “cancer research” and “UC Berkeley”. The project would maintain the fraction of recent jobs with each keyword, and export this data to Science United.
Then, if a volunteer has a “no” preference for keyword K, Science United would exclude a project only if its job fraction for K is one. Volunteer preferences are then enforced by the project’s BOINC job dispatcher. For each of the job’s keywords, if the volunteer specified “yes”, the job is preferentially sent; if “no”, the job is not sent.
6.5. Supporting a VC “Test Drive”
By eliminating the need for projects to recruit volunteers, Science United reduces the barrier to entry for scientists wanting to use VC. However, the effort to set up a BOINC server and get vetted by Science United is still significant. We would like to provide a mechanism whereby a scientist can immediately—in a few minutes—begin processing a significant number of jobs using volunteered computers. We have designed a mechanism for providing “test drives” of this sort.
This is based on a “BOINC app library”: a repository of versions of trusted, widely-used applications (such as Autodock [
14]) for various platform/coprocessor combinations. The BOINC client periodically downloads information about this set of app versions. An attachment to a project can be flagged as “untrusted”, in which case the client only allows the project to use app versions from the BOINC app library.
Science United volunteers can agree to be “test-drive resources”, meaning that they’re willing to run jobs for unvetted job submitters, but only using apps from the library.
A scientist can “test-drive” BOINC by creating a server and registering it with Science United (but not going through the vetting process). The Science United scheduler will attach test-drive devices to such projects in untrusted mode, providing the scientist with a limited amount of computing, for a limited time, and with a limited set of applications.