Liposomal Tubacin: Strategies for the Formulation of a Highly Hydrophobic Anticancer Drug
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Physicochemical Properties of Liposomes
2.2.1. Plackett–Burmann Design (PBD)
2.2.2. Liposome Preparation
2.2.3. UHPLC Instrumentation and Chromatographic Conditions
2.2.4. Determination of Encapsulation Efficiency (EE%)
2.2.5. Batch-Mode Dynamic Light Scattering (DLS)
2.2.6. Asymmetrical Flow Field Flow Fractionation (AF4)
2.2.7. Freeze-Drying Process
2.2.8. Transmission Electron Microscopy (TEM)
2.3. Biological Characterization of Liposomes
2.3.1. Cell Culture
2.3.2. Mitochondrial Activity (WST-1)
2.3.3. Albumin Interaction
2.3.4. Immunofluorescence
2.3.5. Western Blot
2.3.6. Statistical Analysis
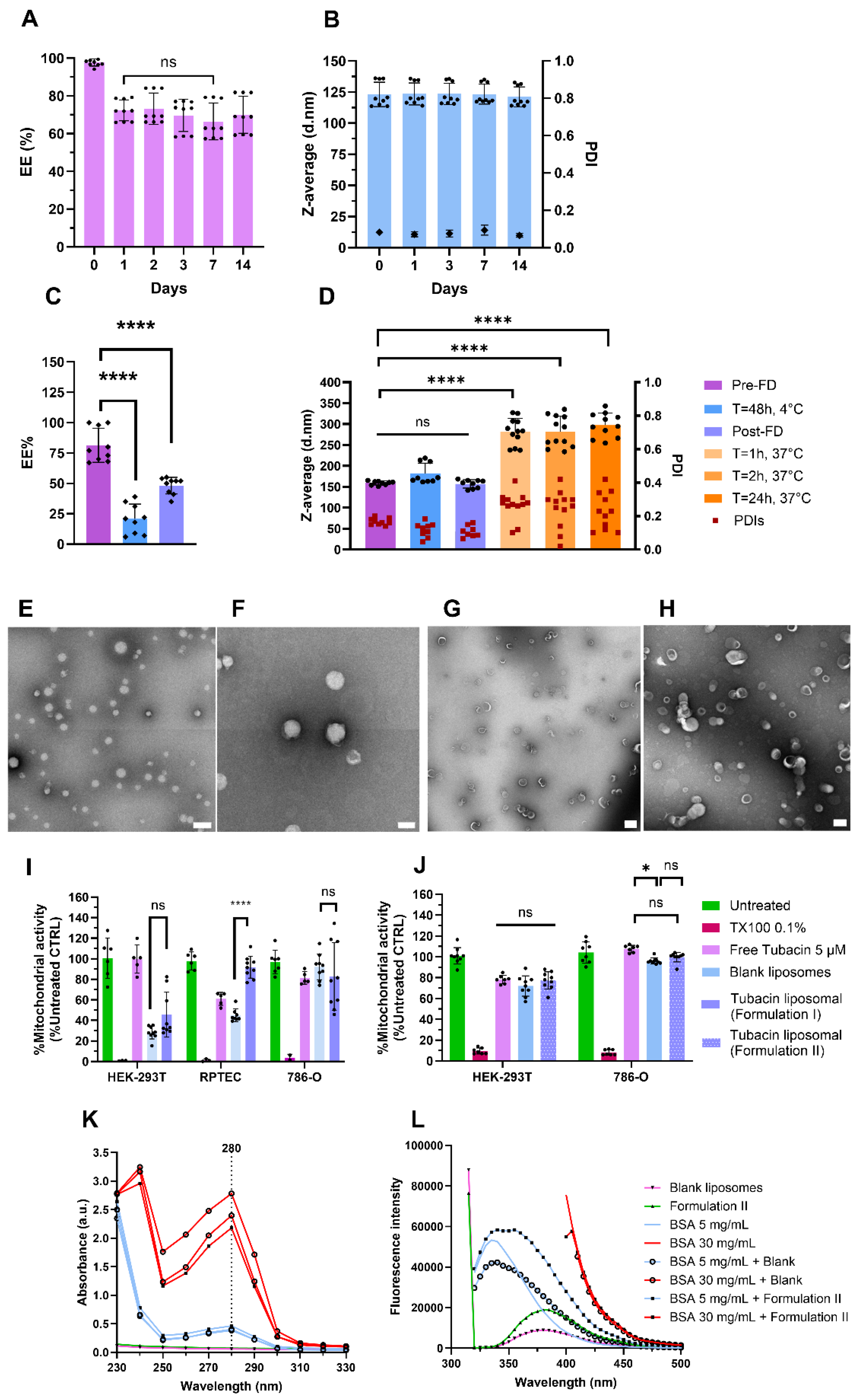
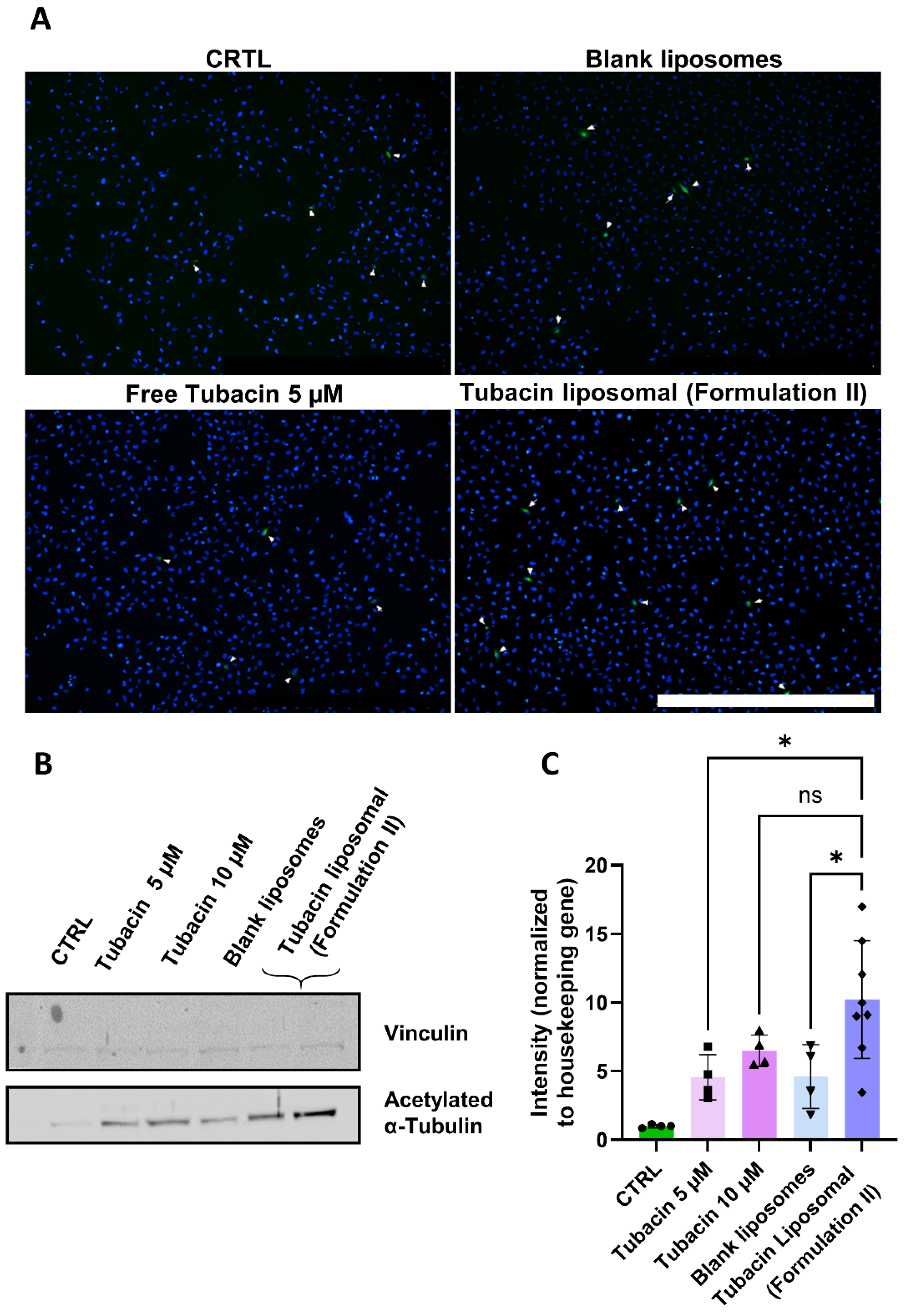
3. Results and Discussion
3.1. Plackett–Burmann Design
3.2. Formulation I Development: Selection of Optimal Tubacin-Loaded Liposome Formulation
3.3. Characterization of Formulation I and II
3.4. Formulation II Development
3.5. Tubacin Potency
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alchahin, A.M.; Mei, S.; Tsea, I.; Hirz, T.; Kfoury, Y.; Dahl, D.; Wu, C.L.; Subtelny, A.O.; Wu, S.; Scadden, D.T.; et al. A transcriptional metastatic signature predicts survival in clear cell renal cell carcinoma. Nat. Commun. 2022, 13, 5747–5762. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, K.; Yang, Z. Treatment strategies for clear cell renal cell carcinoma: Past, present and future. Front. Oncol. 2023, 13, 1133832. [Google Scholar] [CrossRef] [PubMed]
- Aweys, H.; Lewis, D.; Sheriff, M.; Rabbani, R.D.; Lapitan, P.; Sanchez, E.; Papadopoulos, V.; Ghose, A.; Boussios, S. Renal Cell Cancer—Insights in Drug Resistance Mechanisms. Anticancer. Res. 2023, 43, 4781–4792. [Google Scholar] [CrossRef]
- den Boogaard, W.M.C.; Komninos, D.S.J.; Vermeij, W.P. Chemotherapy Side-Effects: Not All DNA Damage Is Equal. Cancers 2022, 14, 627. [Google Scholar] [CrossRef]
- Schirrmacher, V. From chemotherapy to biological therapy: A review of novel concepts to reduce the side effects of systemic cancer treatment (Review). Int. J. Oncol. 2019, 54, 407–419. [Google Scholar] [CrossRef] [PubMed]
- Rausch, M.; Rutz, A.; Allard, P.-M.; Delucinge-Vivier, C.; Docquier, M.; Dormond, O.; Dyson, P.J.; Wolfender, J.-L.; Nowak-Sliwinska, P. Drug Repurposing to Identify a Synergistic High-Order Drug Combination to Treat Sunitinib-Resistant Renal Cell Carcinoma. Cancers 2021, 13, 3978. [Google Scholar] [CrossRef]
- Hany, D.; Zoetemelk, M.; Bhattacharya, K.; Nowak-Sliwinska, P.; Picard, D. Network-informed discovery of multidrug combinations for ERalpha+/HER2-/PI3Kalpha-mutant breast cancer. Cell Mol. Life Sci. 2023, 80, 80–105. [Google Scholar] [CrossRef]
- Ramzy, G.M.; Norkin, M.; Koessler, T.; Voirol, L.; Tihy, M.; Hany, D.; McKee, T.; Ris, F.; Buchs, N.; Docquier, M.; et al. Platform combining statistical modeling and patient-derived organoids to facilitate personalized treatment of colorectal carcinoma. J. Exp. Clin. Cancer Res. 2023, 42, 79–96. [Google Scholar] [CrossRef]
- Weiss, A.; Le Roux-Bourdieu, M.; Zoetemelk, M.; Ramzy, G.M.; Rausch, M.; Harry, D.; Miljkovic-Licina, M.; Falamaki, K.; Wehrle-Haller, B.; Meraldi, P.; et al. Identification of a Synergistic Multi-Drug Combination Active in Cancer Cells via the Prevention of Spindle Pole Clustering. Cancers 2019, 11, 1612. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Ku, S.; Ciamporcero, E.; Miles, K.M.; Attwood, K.; Chintala, S.; Shen, L.; Ellis, L.; Sotomayor, P.; Swetzig, W.; et al. HDAC 1 and 6 modulate cell invasion and migration in clear cell renal cell carcinoma. BMC Cancer 2016, 16, 617. [Google Scholar] [CrossRef]
- Pili, R.; Quinn, D.I.; Hammers, H.J.; Monk, P.; George, S.; Dorff, T.B.; Olencki, T.; Shen, L.; Orillion, A.; Lamonica, D.; et al. Immunomodulation by Entinostat in Renal Cell Carcinoma Patients Receiving High-Dose Interleukin 2: A Multicenter, Single-Arm, Phase I/II Trial (NCI-CTEP#7870). Clin. Cancer Res. 2017, 23, 7199–7208. [Google Scholar]
- Pili, R.; Liu, G.; Chintala, S.; Verheul, H.; Rehman, S.; Attwood, K.; Lodge, M.A.; Wahl, R.; Martin, J.I.; Miles, K.M.; et al. Combination of the histone deacetylase inhibitor vorinostat with bevacizumab in patients with clear-cell renal cell carcinoma: A multicentre, single-arm phase I/II clinical trial. Br. J. Cancer 2017, 116, 874–883. [Google Scholar]
- Ota, S.; Zhou, Z.Q.; Hurlin, P.J. Suppression of FGFR3- and MYC-dependent oncogenesis by tubacin: Association with HDAC6-dependent and independent activities. Oncotarget 2018, 9, 3172–3187. [Google Scholar] [PubMed]
- Mozzicato, A.M.; Bastrup, J.A.; Sanchez-Alonso, J.L.; van der Horst, J.; Gorelik, J.; Hagglund, P.; Jepps, T.A. Mesenteric artery smooth muscle cells from hypertensive rats have increased microtubule acetylation. Biochem. J. 2024, 481, 387–403. [Google Scholar] [PubMed]
- Aldana-Masangkay, G.I.; Rodriguez-Gonzalez, A.; Lin, T.; Ikeda, A.K.; Hsieh, Y.T.; Kim, Y.M.; Lomenick, B.; Okemoto, K.; Landaw, E.M.; Wang, D.; et al. Tubacin suppresses proliferation and induces apoptosis of acute lymphoblastic leukemia cells. Leuk. Lymphoma 2011, 52, 1544–1555. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yu, S.; Cai, X.; Wu, C.; Liu, Y.; Zhang, J.; Gong, X.; Wang, X.; Wu, X.; Zhu, T.; Mo, L.; et al. Targeting HSP90-HDAC6 Regulating Network Implicates Precision Treatment of Breast Cancer. Int. J. Biol. Sci. 2017, 13, 505–517. [Google Scholar]
- Shi, J.; Kantoff, P.W.; Wooster, R.; Farokhzad, O.C. Cancer nanomedicine: Progress, challenges and opportunities. Nat. Rev. Cancer 2017, 17, 20–37. [Google Scholar]
- Schelker, C.; Nowak-Sliwinska, P.; Borchard, G. HDACIs and TKIs combinations and their liposomal delivery for cancer treatment. J. Control Release 2023, 358, 59–77. [Google Scholar]
- Barenholz, Y. Doxil(R)—The first FDA-approved nano-drug: Lessons learned. J. Control Release 2012, 160, 117–134. [Google Scholar] [PubMed]
- Bulbake, U.; Doppalapudi, S.; Kommineni, N.; Khan, W. Liposomal Formulations in Clinical Use: An Updated Review. Pharmaceutics 2017, 9, 12–45. [Google Scholar] [CrossRef]
- Fulton, M.D.; Najahi-Missaoui, W. Liposomes in Cancer Therapy: How Did We Start and Where Are We Now. Int. J. Mol. Sci. 2023, 24, 6615. [Google Scholar] [CrossRef]
- Peng, T.; Xu, W.; Li, Q.; Ding, Y.; Huang, Y. Pharmaceutical liposomal delivery-specific considerations of innovation and challenges. Biomater. Sci. 2022, 11, 62–75. [Google Scholar] [CrossRef]
- Akbarzadeh, A.; Rezaei-Sadabady, R.; Davaran, S.; Joo, S.W.; Zarghami, N.; Hanifehpour, Y.; Samiei, M.; Kouhi, M.; Nejati-Koshki, K. Liposome: Classification, preparation, and applications. Nanoscale Res. Lett. 2013, 8, 102–111. [Google Scholar]
- Briuglia, M.L.; Rotella, C.; McFarlane, A.; Lamprou, D.A. Influence of cholesterol on liposome stability and on in vitro drug release. Drug Deliv. Transl. Res. 2015, 5, 231–242. [Google Scholar] [CrossRef]
- Lopez-Pinto, J.M.; Gonzalez-Rodriguez, M.L.; Rabasco, A.M. Effect of cholesterol and ethanol on dermal delivery from DPPC liposomes. Int. J. Pharm. 2005, 298, 1–12. [Google Scholar] [CrossRef]
- Jain, A.; Hurkat, P.; Jain, S.K. Development of liposomes using formulation by design: Basics to recent advances. Chem. Phys. Lipids 2019, 224, 104764–104780. [Google Scholar]
- Curic, A.; Reul, R.; Moschwitzer, J.; Fricker, G. Formulation optimization of itraconazole loaded PEGylated liposomes for parenteral administration by using design of experiments. Int. J. Pharm. 2013, 448, 189–197. [Google Scholar] [CrossRef]
- Sahu, A.K.; Jain, V. Screening of process variables using Plackett-Burman design in the fabrication of gedunin-loaded liposomes. Artif. Cells Nanomed. Biotechnol. 2017, 45, 1011–1022. [Google Scholar] [CrossRef]
- Muthu, M.S.; Kulkarni, S.A.; Xiong, J.; Feng, S.S. Vitamin E TPGS coated liposomes enhanced cellular uptake and cytotoxicity of docetaxel in brain cancer cells. Int. J. Pharm. 2011, 421, 332–340. [Google Scholar]
- Gouda, A.; Sakr, O.S.; Nasr, M.; Sammour, O. Ethanol injection technique for liposomes formulation: An insight into development, influencing factors, challenges and applications. J. Drug Del. Sci. Tech. 2021, 61, 102174–102184. [Google Scholar]
- Notman, R.; Noro, M.; O’Malley, B.; Anwar, J. Molecular Basis for Dimethylsulfoxide (DMSO) Action on Lipid Membranes. J. Am. Chem. Soc. 2006, 128, 13982–13983. [Google Scholar] [CrossRef]
- Cagdas, F.M.; Ertugral, N.; Bucak, S.; Atay, N.Z. Effect of preparation method and cholesterol on drug encapsulation studies by phospholipid liposomes. Pharm. Dev. Technol. 2011, 16, 408–414. [Google Scholar] [CrossRef]
- Kulkarni, A.A.; Vijaykumar, V.E.; Natarajan, S.K.; Sengupta, S.; Sabbisetti, V.S. Sustained inhibition of cMET-VEGFR2 signaling using liposome-mediated delivery increases efficacy and reduces toxicity in kidney cancer. Nanomedicine 2016, 12, 1853–1861. [Google Scholar]
- Lammers, T. Nanomedicine Tumor Targeting. Adv. Mater. 2024, 36, 2312169. [Google Scholar]
- Franco, M.S.; Oliveira, M.C. Liposomes Co-Encapsulating Anticancer Drugs in Synergistic Ratios as an Approach to Promote Increased Efficacy and Greater Safety. Anticancer. Agents Med. Chem. 2019, 19, 17–28. [Google Scholar] [PubMed]
- Kraft, J.C.; Freeling, J.P.; Wang, Z.; Ho, R.J.Y. Emerging Research and Clinical Development Trends of Liposome and Lipid Nanoparticle Drug Delivery Systems. J. Pharm. Sci. 2014, 103, 29–52. [Google Scholar] [PubMed]
- Kobayashi, H.; Watanabe, R.; Choyke, P.L. Improving conventional enhanced permeability and retention (EPR) effects; what is the appropriate target? Theranostics 2013, 4, 81–89. [Google Scholar]
- Notman, R.; den Otter, W.K.; Noro, M.G.; Briels, W.J.; Anwar, J. The permeability enhancing mechanism of DMSO in ceramide bilayers simulated by molecular dynamics. Biophys. J. 2007, 93, 2056–2068. [Google Scholar]
- Justo, O.R.; Moraes, Â.M. Analysis of process parameters on the characteristics of liposomes prepared by ethanol injection with a view to process scale-up: Effect of temperature and batch volume. Chem. Eng. Res. Des. 2011, 89, 785–792. [Google Scholar]
- Zhang, Z.; Tan, S.; Feng, S.S. Vitamin E TPGS as a molecular biomaterial for drug delivery. Biomaterials 2012, 33, 4889–4906. [Google Scholar]
- Jain, S.; Pandey, S.; Sola, P.; Pathan, H.; Patil, R.; Ray, D.; Aswal, V.K.; Bahadur, P.; Tiwari, S. Solubilization of Carbamazepine in TPGS Micelles: Effect of Temperature and Electrolyte Addition. AAPS PharmSciTech 2019, 20, 203–211. [Google Scholar] [PubMed]
- Ruozi, B.; Belletti, D.; Tombesi, A.; Tosi, G.; Bondioli, L.; Forni, F.; Vandelli, M.A. AFM, ESEM, TEM, and CLSM in liposomal characterization: A comparative study. Int. J. Nanomed. 2011, 6, 557–563. [Google Scholar]
- Liu, X.; Shan, K.; Shao, X.; Shi, X.; He, Y.; Liu, Z.; Jacob, J.A.; Deng, L. Nanotoxic Effects of Silver Nanoparticles on Normal HEK-293 Cells in Comparison to Cancerous HeLa Cell Line. Int. J. Nanomed. 2021, 16, 753–761. [Google Scholar]
- Brodaczewska, K.K.; Szczylik, C.; Fiedorowicz, M.; Porta, C.; Czarnecka, A.M. Choosing the right cell line for renal cell cancer research. Mol. Cancer 2016, 15, 83–98. [Google Scholar] [PubMed]
- Yu, S.H.; Possmayer, F. Lipid compositional analysis of pulmonary surfactant monolayers and monolayer-associated reservoirs. J. Lipid Res. 2003, 44, 621–629. [Google Scholar]
- Farooq, M.A.; Trevaskis, N.L. TPGS Decorated Liposomes as Multifunctional Nano-Delivery Systems. Pharm. Res. 2023, 40, 245–263. [Google Scholar]
- Tavares Luiz, M.; Delello Di Filippo, L.; Carolina Alves, R.; Sousa Araújo, V.H.; Lobato Duarte, J.; Maldonado Marchetti, J.; Chorilli, M. The use of TPGS in drug delivery systems to overcome biological barriers. Eur. Polymer J. 2021, 142, 110129. [Google Scholar]
- Pande, S. Liposomes for drug delivery: Review of vesicular composition, factors affecting drug release and drug loading in liposomes. Artif. Cells Nanomed. Biotechnol. 2023, 51, 428–440. [Google Scholar]
- Bergonzi, M.C.; Vasarri, M.; Marroncini, G.; Barletta, E.; Degl’Innocenti, D. Thymoquinone-Loaded Soluplus((R))-Solutol((R)) HS15 Mixed Micelles: Preparation, In Vitro Characterization, and Effect on the SH-SY5Y Cell Migration. Molecules 2020, 25, 4707–4724. [Google Scholar] [PubMed]
- Zhou, C.; Guo, C.; Li, W.; Zhao, J.; Yang, Q.; Tan, T.; Wan, Z.; Dong, J.; Song, X.; Gong, T. A novel honokiol liposome: Formulation, pharmacokinetics, and antitumor studies. Drug Dev. Ind. Pharm. 2018, 44, 2005–2012. [Google Scholar]
- Olusanya, T.O.B.; Haj Ahmad, R.R.; Ibegbu, D.M.; Smith, J.R.; Elkordy, A.A. Liposomal Drug Delivery Systems and Anticancer Drugs. Molecules 2018, 23, 907–924. [Google Scholar] [CrossRef]
- Caracciolo, G.; Pozzi, D.; Capriotti, A.L.; Cavaliere, C.; Piovesana, S.; La Barbera, G.; Amici, A.; Lagana, A. The liposome-protein corona in mice and humans and its implications for in vivo delivery. J. Mater. Chem. B 2014, 2, 7419–7428. [Google Scholar] [PubMed]
- Tretiakova, D.; Kobanenko, M.; Le-Deygen, I.; Boldyrev, I.; Kudryashova, E.; Onishchenko, N.; Vodovozova, E. Spectroscopy Study of Albumin Interaction with Negatively Charged Liposome Membranes: Mutual Structural Effects of the Protein and the Bilayers. Membranes 2022, 12, 1031. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yin, D.; Liao, J.; Wang, Y.; Gou, R.; Tang, C.; Li, W.; Liu, Y.; Fu, J.; Shi, S.; et al. Regulation of protein corona on liposomes using albumin-binding peptide for targeted tumor therapy. J. Control Release 2023, 355, 593–603. [Google Scholar] [PubMed]
- Mast, M.P.; Modh, H.; Champanhac, C.; Wang, J.W.; Storm, G.; Kramer, J.; Mailander, V.; Pastorin, G.; Wacker, M.G. Nanomedicine at the crossroads—A quick guide for IVIVC. Adv. Drug Deliv. Rev. 2021, 179, 113829. [Google Scholar]
- Verhoef, J.J.; Anchordoquy, T.J. Questioning the Use of PEGylation for Drug Delivery. Drug Deliv. Transl. Res. 2013, 3, 499–503. [Google Scholar]

| Formulation n° | Temp. (°C) | Rotation Speed (rpm) | Rotation Time (min) | Tubacin Input (µg/mL) | DMSO (µL) | DPPC (%) | Cholesterol (%) | TPGS (%) | EE (%) | Size (nm) | PDI |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 50 | 800 | 5 | 50 | 200 | 49.6 | 37.7 | 12.7 | 18 ± 8 | 236 ± 24 | 0.174 |
| 2 | 60 | 600 | 10 | 20 | 200 | 21.8 | 68.9 | 9.30 | 27 ± 2 | 135 ± 4 | 0.09 |
| 3 | 50 | 600 | 10 | 20 | 500 | 39.6 | 30.1 | 30.3 | 15 ± 6 | 120 ± 19 | 0.165 |
| 4 | 50 | 600 | 5 | 50 | 500 | 18.4 | 58.2 | 23.5 | 38 ± 13 | 136 ± 8 | 0.067 |
| 5 | 60 | 600 | 10 | 50 | 200 | 39.6 | 30.1 | 30.3 | 37 ± 18 | 163 ± 8 | 0.066 |
| 6 | 50 | 600 | 5 | 20 | 200 | 37.2 | 47.0 | 15.8 | 30 ± 4 | 148 ± 3 | 0.097 |
| 7 | 60 | 800 | 5 | 20 | 200 | 27.3 | 51.8 | 20.9 | 38 ± 5 | 149 ± 5 | 0.020 |
| 8 | 60 | 800 | 5 | 20 | 500 | 28.2 | 35.7 | 36.1 | 28 ± 5 | 130 ± 4 | 0.092 |
| 9 | 60 | 600 | 5 | 50 | 500 | 31.7 | 60.2 | 8.10 | 9.0 ± 3 | 141 ± 9 | 0.091 |
| EE% | Size | PDI | |
|---|---|---|---|
| Independent variables | p-value | ||
| Temperature | n.a. | 0.079 | 0.052 |
| Rotation speed | n.a. | 0.026/Positive | 0.067 |
| Tubacin concentration | 0.106 | 0.016/Positive | n.a. |
| DPPC | 0.029/Negative | n.a. | n.a. |
| Cholesterol | 0.119 | n.a. | n.a. |
| TPGS | 0.007/Positive | n.a. | 0.153 |
| Loading time | n.a. | n.a. | 0.103 |
| DMSO | 0.009/Negative | 0.027/Negative | 0.191 |
| Formulation | Temperature | Rotation (rpm) | Rotation Time (min) | Tubacin Input (mg/mL) | DPPC Input (mg) | Cholesterol Input (mg) | TPGS Input (mg) |
|---|---|---|---|---|---|---|---|
| I | 50 | 600 | 5 | 0.05 | 3 | 5 | 3 |
| Compound | Concentration (mg/mL) | Molar Ratio | %mol in the Bilayer | Total Lipids (mg/µmol) | Final Drug Content (µM/µmol) | Drug-to-Lipid Molar Ratio | |
|---|---|---|---|---|---|---|---|
| Formulation I | DPPC | 0.6 | 1.0 | 20 | 13/20.32 | 48.3/0.243 | 0.012 |
| Cholesterol | 1.0 | 3.1 | 64 | ||||
| TPGS | 1.0 | 1.6 | 16 | ||||
| Formulation II | DSPC | 0.3 | 1.0 | 48 | 3/4.06 | 34.5/0.173 | 0.043 |
| Cholesterol | 0.1 | 0.1 | 31 | ||||
| TPGS | 0.2 | 0.63 | 16 | ||||
| Kolliphor® HS-15 | 0.04 | 0.32 | 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schelker, C.; Revaclier, L.; Borchard, G.; Nowak-Sliwinska, P. Liposomal Tubacin: Strategies for the Formulation of a Highly Hydrophobic Anticancer Drug. Pharmaceutics 2025, 17, 491. https://doi.org/10.3390/pharmaceutics17040491
Schelker C, Revaclier L, Borchard G, Nowak-Sliwinska P. Liposomal Tubacin: Strategies for the Formulation of a Highly Hydrophobic Anticancer Drug. Pharmaceutics. 2025; 17(4):491. https://doi.org/10.3390/pharmaceutics17040491
Chicago/Turabian StyleSchelker, Cindy, Léa Revaclier, Gerrit Borchard, and Patrycja Nowak-Sliwinska. 2025. "Liposomal Tubacin: Strategies for the Formulation of a Highly Hydrophobic Anticancer Drug" Pharmaceutics 17, no. 4: 491. https://doi.org/10.3390/pharmaceutics17040491
APA StyleSchelker, C., Revaclier, L., Borchard, G., & Nowak-Sliwinska, P. (2025). Liposomal Tubacin: Strategies for the Formulation of a Highly Hydrophobic Anticancer Drug. Pharmaceutics, 17(4), 491. https://doi.org/10.3390/pharmaceutics17040491








