Abstract
Noncoding RNAs (ncRNAs) are a heterogeneous group of RNA molecules whose classification is mainly based on arbitrary criteria such as the molecule length, secondary structures, and cellular functions. A large fraction of these ncRNAs play a regulatory role regarding messenger RNAs (mRNAs) or other ncRNAs, creating an intracellular network of cross-interactions that allow the fine and complex regulation of gene expression. Altering the balance between these interactions may be sufficient to cause a transition from health to disease and vice versa. This leads to the possibility of intervening in these mechanisms to re-establish health in patients. The regulatory role of ncRNAs is associated with all cancer hallmarks, such as proliferation, apoptosis, invasion, metastasis, and genomic instability. Based on the function performed in carcinogenesis, ncRNAs may behave either as oncogenes or tumor suppressors. However, this distinction is not rigid; some ncRNAs can fall into both classes depending on the tissue considered or the target molecule. Furthermore, some of them are also involved in regulating the response to traditional cancer-therapeutic approaches. In general, the regulation of molecular mechanisms by ncRNAs is very complex and still largely unclear, but it has enormous potential both for the development of new therapies, especially in cases where traditional methods fail, and for their use as novel and more efficient biomarkers. Overall, this review will provide a brief overview of ncRNAs in human cancer biology, with a specific focus on describing the most recent ongoing clinical trials (CT) in which ncRNAs have been tested for their potential as therapeutic agents or evaluated as biomarkers.
1. Introduction
Cancer has a significant impact, not only on the physical and mental health of the person affected but also on society, considering the significant economic impact due to the high costs of the development and use of effective therapies. Furthermore, according to the World Health Organization (WHO) (see Table 1 for this and other abbreviations used throughout the text), cancer resulted in almost 10 million deaths in 2020, making it one of the leading causes of death worldwide [1].

Table 1.
This table lists all abbreviations used in this review. The first and second columns contain the abbreviations and corresponding definitions of the general terms mentioned in the text. The third and fourth columns show the abbreviations and corresponding definitions of the different types of cancer mentioned in the text.
Over time, the definition of cancer has changed several times in relation to new discoveries, especially in the field of molecular biology and genetics, and more generally with technical–scientific advancements. Many works use definitions that are largely consistent with that provided by the NCI: “Cancer is a disease in which some of the body’s cells grow uncontrollably and spread to other parts of the body” [2]. However, this definition does not consider the genetic and molecular aspects that lead to the malignant transformation of the cell or how these aspects vary over time. Consequently, this definition deeply simplifies the scenario that leads to carcinogenesis, the understanding of which is of fundamental importance for its diagnosis, prognosis, and therapy.
Each of the approximately 36 trillion cells that constitute the adult male body, and approximately 28 trillion in the female one [3], can potentially develop a tumor, which may have a solid or liquid structure. Solid tumors form an abnormal mass (or lumps) in specific organs or tissues, whereas liquid tumors do not form a solid mass, and cancer cells can circulate through either the bloodstream or lymphatic system [4,5,6].
The NCI lists more than 150 different types of tumors organized by organ location [7], but this number significantly increases considering that many tumors can be further divided into distinct subtypes based on their different “mutational signatures”. The incidence of these tumors is highly variable. In fact, while some tumors are particularly widespread in the population, such as BrC or PrC (1–3%), others, such as stomach cancer and LarC, occur in less than 15/100,000 people/year and are considered rare by the NCI [8].
Cancer-causing mutations are irreversible cellular mutations that affect DNA, both nDNA and mtDNA, and occur in regions that are necessary for proper cellular function and health. Cancer-causing mutations are known as driver mutations, and the affected DNA regions can be PC or nPC regions of the genome [9,10,11,12]. Initially, cancer-causing mutations induce changes in one or a few cells through a multistep process. These changes accumulate over time, arise from independent events, and undergo selection. These mutations allow a healthy cell to acquire new functional capabilities, collectively described as hallmarks of cancer, leading to its neoplastic transformation [13].
Furthermore, mutations continue to accumulate during subsequent cell divisions, producing heterogeneous progeny hosting different genetic profiles. This aspect implies that the tumor is composed of multiple subclones that share a common ancestor. These subclones can further diverge and expand simultaneously over time, acquiring different characteristics, such as increased fitness or intratumor variability, i.e., the simultaneous coexistence of genetically, molecularly, and phenotypically distinct cell populations [14]. Comprehensive reviews regarding this topic are available in the literature [15,16,17].
PC regions are sequences contained in genes that are translated into proteins. In particular, PCs represent only about 1–2% of the human genome, the remainder of which, about 98%, is composed of nPC regions [18]. This aspect is particularly important because an analysis of the SNVs identified in GWAS shows that more than 88% of disease- and trait-associated variants fall within nPC regions of the genome [19,20], showing that mutations causing protein alteration and dysfunction represent only a minority of the possible causes of tumorigenesis.
NPC regions are highly heterogeneous DNA sequences, both in terms of function and genomic localization, and participate in every biological process. Some nPC sequences have structural functions, such as the telomere and the centromere, which are both composed mainly of satellite DNA (high frequency of repetitive DNA sequences) and are considered essential both for the stability of the genome and for the correct carrying out of cell division [21,22]. Other nPC regions have a regulatory function and can be interspersed in the genome as transposons, enhancers, silencers, and insulators or form part of the canonical structure of a gene, such as the gene promoter and mRNA UTR. Finally, there are some types of nPCs that can be transcribed into RNA molecules, which in turn divide into constitutive or regulatory RNAs.
The distinction between the various types of nPC is unclear, and, often, the same region can be involved in multiple pathways. For example, TE are repeated and interspersed sequences capable of moving from one position to another in the genome of the same cell. Transposons, through their movement, produce genetic diversity mainly by a phenomenon called exon shuffling. Indeed, when the excision is not perfect, a TE can carry with it genomic sequences, and, if the juxtaposition of two previously unrelated exons occurs, potentially new gene products can be created. TE insertion can also cause damage if it occurs within a sequence that becomes nonfunctional or abnormally regulated with respect to cellular needs [23,24]. TEs are involved in the biogenesis of some ncRNAs with regulatory functions, such as miRNAs or piRNAs [25,26]. The processing of some constitutive RNAs can also generate regulatory ncRNAs [27,28,29,30,31,32].
The two main types of nPC regions with regulatory functions localized within the structure of a gene are the promoter, which plays a role in regulating the rate of transcription initiation of the gene, and the UTR, with sequences present at both the 3′ and 5′ ends of the gene and playing crucial roles in the post-transcriptional regulation of gene expression. The gene may also contain another type of nPC represented by introns, which are nucleotide sequences that separate two contiguous exons and from which, in some cases, the ncRNA could originate. Some mutations in the constituent elements of genes (promoters, UTRs, introns, and exons) could alter the levels of gene expression or lead to the synthesis of a nonfunctional protein product. If these mutations contribute to promoting carcinogenesis, they are called driver mutations, and the affected genes are called driver genes [33]. Cancer driver genes are broadly divided into two functional classes: oncogenes and tumor suppressor genes [34]. Oncogenes are usually involved in controlling cell proliferation and division, and, through gain-of-function mutations, they increase their activity compared to normal conditions. Meanwhile, tumor suppressor genes usually inhibit cell growth and division, promote DNA repair, and activate cell cycle checkpoints. The inactivation of tumor suppressors by loss-of-function mutations eliminates regulatory control over their targets, thus promoting the development of carcinogenesis.
Constitutive and Regulatory ncRNAs
Finally, other types of nPC sequences are transcribed into RNA molecules. Interestingly, approximately 75–80% of the human genome is transcribed into RNA, indicating that it transcribes tens of thousands of RNA molecules. These RNA molecules are collectively called ncRNAs [35,36,37] and can be divided into constitutive and regulatory ncRNAs (Figure 1) on the basis of their main function, with the former being abundantly and ubiquitously expressed in all cell types and providing essential functions to the organism, such as transcription or translation, and the latter being involved in the regulation of target gene expression.
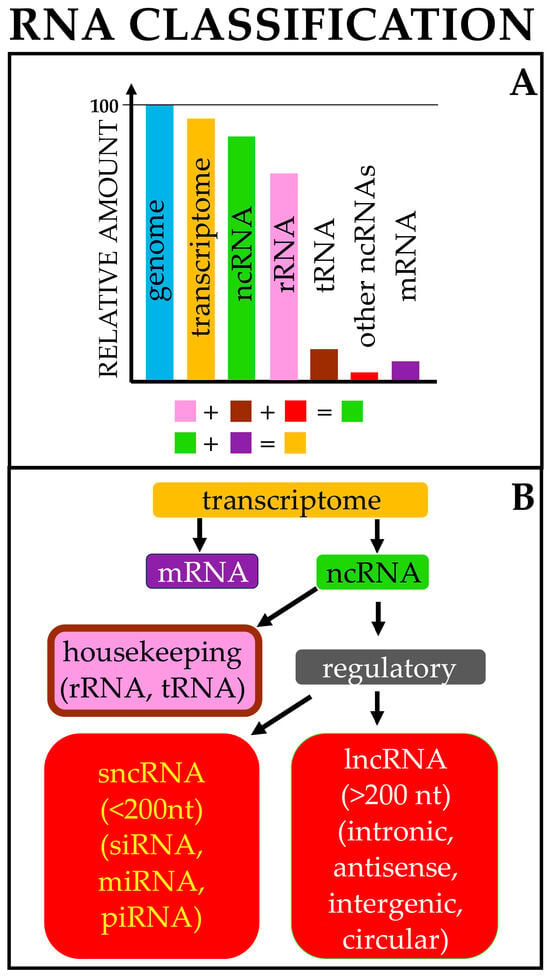
Figure 1.
RNA origin and classification. (A) Approximately 95% of the genome is transcribed (transcriptome). Of this, around 90–95% is composed of noncoding RNAs (ncRNAs), mostly rRNAs and tRNAs; the remainder consists of mRNAs. (B) Broad classification of cellular RNAs; for each class, only representative RNAs are reported. The list is not intended to be comprehensive. Color codes are the same in both figure parts.
Constitutive RNAs include tRNAs, rRNAs, snRNAs, snoRNAs, and TERCs. The function of tRNAs is to carry a specific amino acid, which, during translation, is added to the nascent protein chain [38]. Ribosomal RNAs fold to form secondary structures and play a structural and functional role within ribosomes, thus contributing to the enzymatic activity of the ribosome complex that is required for protein synthesis. There are four rRNA molecules in the ribosome that differ in sequence length and sedimentation coefficient: 5.8 S (156 nt), 18 S (1869 nt), 28 S (5070 nt), and 5 S (121 nt) [39,40]. snRNAs are involved in the formation of the spliceosome, a complex that allows the correct excision of introns (splicing) from the pre-mRNA sequence. There are many types of spliceosomes, distinguished by the combination of proteins and snRNAs used for the activity of the complex. There are five snRNAs and they have an approximate length of 100–200 nt: U1, U2, U4, U5, and U6 [41]. snoRNAs have a sequence length between 60 and 300 nt, are widely present in the nucleoli of eukaryotic cells, and are mainly encoded in the intron region of the gene transcribed by RNA polymerase II [42]. snoRNAs guide chemical modifications of other RNAs, such as rRNAs, tRNAs, snRNAs, and some types of mRNAs [43,44]. The modifications imposed by snoRNAs affect the stability and folding of RNAs, as in the case of rRNAs and tRNAs. In addition, several studies suggest that some snoRNAs can play miRNA-like roles because they are involved in the regulation of gene expression through the regulation of alternative splicing or the inhibition of the mRNAs of target genes [45,46].
Regulatory ncRNAs, which include miRNAs, siRNAs, piRNAs, lncRNAs, and circRNAs, will be discussed in detail in the following paragraphs.
The described genomic elements that constitute both the protein-coding and non-protein-coding regions are dependent on each other, implying that mutations affecting one region can affect the other and vice versa. For example, DNA methylation is an epigenetic mechanism that involves the transfer of a methyl group by specific enzymes, namely DNMTs, and accessory proteins on CpG islands. A CpG island refers to an area in the genome with a higher frequency of short stretches of palindromic DNA, in which a cytosine nucleotide is followed by a guanine nucleotide (with “p” indicating the phosphate bond between them). The methylation of CpG islands, usually found in promoters, allows them to regulate gene expression by recruiting proteins involved in gene repression or by inhibiting the binding of transcription factors to DNA. However, DNA methylation can also influence the expression of regulatory RNAs, including lncRNAs [47] and miRNAs [48,49]. In particular, miRNAs are small, ss ncRNAs that, through various mechanisms (described in Section 4.2), are involved in the regulation of gene expression. Aberrant DNA methylation can lead to either the upregulation or downregulation of miRNA expression, which can be associated with tumorigenesis [50,51]. Conversely, miRNAs can regulate DNA methylation in two ways: by modulating DNMTs’ activity [52,53] or by modulating the functions of accessory proteins that play a role in DNA methylation [54,55].
The existing link between PC and nPC regions suggests a plethora of very complex mechanisms by which carcinogenesis originates, which are very difficult to elucidate due to changes over time. Therefore, understanding how tumor evolution influences disease progression and how these processes are influenced by environmental factors and therapeutic treatments remains fundamental to develop not only new diagnostic and prognostic markers but also new therapeutic approaches.
For many decades, cancer patients have relied on three main different therapeutic approaches that can be used individually or in combination with each other: the surgical removal of the tumor mass, radiotherapy, and chemotherapy. These approaches have had an overall positive effect on morbidity and mortality in many types of cancer, despite the observation that the response to treatment can greatly vary from patient to patient, as it is influenced by parameters such as the cancer type and stage.
Over the years, a better understanding of cancer pathogenesis has enabled the development of new therapeutic approaches, including targeted therapy, immunotherapy, stem cell transplantation, and hormone therapy [56,57].
These new approaches have provided alternative solutions to the side effects related to conventional treatments. Recently, an innovative approach has been represented by therapies based on the use of regulatory RNAs. This has been possible thanks to improved next-generation sequencing and recent advances in high-throughput sequencing technologies, bioinformatics analysis tools, and computational platforms, which have enabled researchers to study in greater depth the genomic and transcriptomic profiles of many human diseases, including cancer. These technologies have made it possible to identify and classify thousands of regulatory RNA molecules that have both oncogenic and tumor-suppressive roles in cancer. Overall, the deregulation of regulatory RNAs influences cancer development and progression through the modification of cellular processes such as cell proliferation, apoptosis, invasion, and metastasis [58].
The discovery of such a diverse number of regulatory ncRNA species has changed the way that researchers think about the physiology and development of diseases, which, for decades, has been focused on the study of protein-coding genes. Furthermore, due to the involvement of regulatory ncRNAs in every cellular process, their large numbers compared to proteins, their higher sensitivity and specificity compared to traditional tumor markers, and their easy detection in many body fluids, they can be considered a reservoir with incalculable potential, not only for the development of future therapeutic applications for cancer treatment and precision medicine but also in providing more effective tools for early cancer diagnosis or drug response prediction. For these reasons, cancer-focused clinical trials involving regulatory ncRNAs as novel biomarkers or therapies are increasing every year.
This review examines different classes of regulatory ncRNAs (miRNAs, siRNAs, piRNAs, lncRNAs, and cirRNAs), describing their biogenesis, functions, and clinical applications. The aim of this review is to provide not only the most complete overview possible of regulatory ncRNAs, in relation to the tumor cell biology, reporting both the positive aspects and the challenges to overcome for their use in clinical practice, but also to provide an indication of how research is evolving by describing clinical studies evaluating the use of regulatory ncRNAs in the oncology field.
2. Regulatory ncRNAs: General Overview
ncRNAs are a class of RNA molecules that are transcribed but not translated into proteins. Regulatory RNAs are a subclass of ncRNAs that are implicated in the regulation of gene expression. However, recent discoveries have rendered the classical definition of ncRNAs ambiguous, because many studies have shown that a certain number of ncRNAs harbor small ORFs that can encode micropeptides (less than 100 amino acids) [59], as in the case of “bifunctional RNAs”, which are so called because they can both function as ncRNAs and be translated into peptides [60,61,62]. These unconventional peptides play functional roles in normal and pathological processes, including cancer [63].
Regulatory RNAs represent a very large group of polynucleotides whose cataloging can be based on different and mostly arbitrary criteria, such as the structure of the ncRNA (linear, circular), their endogenous functions, and their lengths. The length is conventionally used to distinguish the two main subcategories of regulatory RNAs: small noncoding RNAs (sncRNAs), composed of RNA molecules with less than 200 nts, and lncRNAs, composed of transcripts longer than 200 nt [64,65,66] (Figure 1).
sncRNAs include, among others, miRNAs, siRNAs, and piRNAs; these represent the three pathways of RNAi. In general, RNAi is an evolutionarily conserved and sequence-specific mechanism that is triggered by dsRNAs. RNAi not only provides a defense mechanism against invading viruses and TEs but also plays a role in regulating gene expression at either the transcriptional or the post-transcriptional level. The conservation of this mechanism and the possibility and ease of designing dsRNAs have allowed RNAi to be exploited for gene therapy and clinical application in numerous diseases, including cancer.
Meanwhile, lncRNAs include lincRNAs, circRNAs, antisense RNAs, and pseudogenes; their functions are highly heterogeneous, as they may serve as sncRNA sponges, structural elements, decoys, antisense molecules, etc., on a case-by-case basis.
Functional analyses have revealed that regulatory RNAs are mainly involved in the control of gene expression, adopting different mechanisms, and they participate in virtually all cellular processes [67,68,69]. Considering their heterogeneous characteristics, it is plausible to suggest that, in perspective, the classification of ncRNAs might change because of the general improvement in investigation techniques and the better understanding of the mechanisms in which they participate. This will also help to reconcile the definition of “noncoding RNA” with the evidence that some ncRNAs can be translated into small peptides.
3. mtDNA and Cancer Implications
Human mtDNA is 16,659 bp long, has no histone support, and is composed of two strands, called the heavy (H) and light (L) strands, organized in a circular molecule. The mitochondrial genome is exclusively maternally inherited, and sperm mtDNA is not transmitted to the next generation [70,71].
mtDNA is present in the cell in multiple copies, with generally between 1 and 10 copies per mitochondrion [72]. However, the number of mitochondria per cell varies widely, as does the number of mtDNA copies, and this depends on the cell type and its energy needs [73,74,75,76,77].
Usually, the copies of mtDNA are all identical, a condition called homoplasmy. In contrast, heteroplasmy is the condition in which there is more than one mtDNA variant in a cell. For example, if mutations occur in one or more copies of mtDNA, a mixed population of mutant and wild-type genomes will coexist in the cell. Healthy copies of mtDNA can functionally complement the damaged ones up to a critical threshold depending on the type of mutation. Once this threshold is exceeded, the defect associated with the mtDNA mutation becomes manifest.
The mitochondrial genome harbors 37 intronless genes, including 2 rRNAs, 22 tRNAs, and 11 mRNAs (two of which are bicistronic) [78,79]. In addition, a portion of the mitochondrial 16s rRNA contains a small ORF that encodes a small peptide known as humanin, which has neuroprotective activity and is also implicated in carcinogenesis [80,81,82,83].
3.1. mtDNA Deregulation in Carcinogenesis
Many mitochondrial dysfunctions associated with carcinogenesis are attributable to mutations in nDNA. However, several studies have found mtDNA mutations in over 50% of the tumors analyzed [84,85,86,87]. Compared to nDNA, mtDNA is more prone to damage. In fact, recent studies showed that mtDNA has a 10- to 100-fold higher rate of de novo germline mutation than nDNA [88,89]. Accumulated damage to mtDNA causes mitochondrial dysfunction, often associated with the impaired functioning of respiratory chain complexes and intracellular signaling pathways, which drives the pathogenesis of a variety of human diseases, especially neurodegenerative disorders and cancer. The causes are multiple, including age, a higher mtDNA replication rate, and less effective mtDNA damage repair mechanisms. For example, as healthy cells age, they accumulate nDNA and mtDNA damage due to environmental exposure and cellular processes, yet the mechanisms that regulate this damage’s induction are still unclear [90,91].
3.1.1. Replication and Repair Mechanisms Cause High mtDNA Mutation Rates
DNA polymerase gamma (PolG) is responsible for mtDNA replication and repair, and, until recently, it was considered the only polymerase present in the mitochondria. However, recent data suggest that several polymerases display activity in the mitochondria, such as polymerase theta (PolQ) [92]. Among the various DNA polymerases, PolG is the most reliable. Nonetheless, although mutations associated with its activity are rare, mtDNA replicates much more frequently than nDNA, which increases the likelihood of PolG-induced mutational events. In addition, PolG’s activity is influenced by over 300 point mutations that have been mapped in its coding gene and are associated with many inherited mitochondrial disorders [93].
PolQ is another polymerase that works in the mitochondrion and belongs to the family A DNA polymerases, like PolG. Fidelity measurements of PolQ revealed that it generates single-base-pair substitutions at a 10- to 100-fold higher rate than other characterized family members, making it one of the least reliable members of the family A DNA polymerases [94,95].
Damage to mtDNA, similarly to that to nDNA, can trigger the action of several mechanisms to ensure genome stability and guarantee the normal function of the mitochondria. In fact, in the mitochondrion, the mechanisms of excision repair, direct reversal, mismatch repair, and possibly ds break repair seem to be active. Instead, NER has not been confirmed as a system for mtDNA damage repair. However, these mechanisms seem less effective than those in the nucleus, and the key components of these pathways have not been characterized as well as those in the nuclear system [96].
For these reasons, the mtDNA sequence may contain SNPs, which, in some cases, have been correlated with carcinogenesis. For example, several mtDNA mutations are correlated with HCC progression, namely G3842A, which creates a premature stop codon in the mtND1 gene; A11708G, which results in amino acid substitutions in the mtND4 gene; and 12418insA, which result in frame shift mutations in the mtND5 gene [97]. In CRC, the mutations T4216C, T3394C, and C3497T cause an amino acid substitution, and the mutation 3565_3566insC causes a frame shift; these are associated with carcinogenesis and CRC progression [98,99,100,101,102,103]. mt-ND2 mutation G4776A enhanced the cell growth of HNC cells via the induction of HIF1α [104]. Yuan and coworkers analyzed the mutations of the mitND6 gene by sequencing the mtDNA of tumor tissue from 87 patients with primary LAC. The analysis identified eight missense mutations in the mtND6 gene, which resulted in amino acid changes, and three nonsense mutations in the same gene, which resulted in premature translation termination; these were significantly correlated with the pathological stage of the tumor, lymph node metastasis, and a shorter survival rate in LAC patients [105]. Beadnell et al. suggest that SNPs T3394C and C3497T in the MT-ND1 gene are correlated with distant metastasis [106].
Among mtDNA mutations, point mutations are the most frequent; however, mtDNA can be affected by different types of alterations that are linked to carcinogenesis. For example, the deletion from position 8470 to 13,447 in mtDNA, also known as the common deletion or mtDNA4977, is the most frequently observed deletion in human mtDNA. The common deletion is an important factor in the carcinogenesis of several tumors, including HCC [107,108], CRC [109,110], and brain tumors [111].
3.1.2. Influence of mtDNA Copy Number in Tumors
An increased or decreased number of mtDNA copies, or CNV, is a condition often observed in tumors and correlated with cancer progression and severity. For example, Mennuni and Al-Awadhi suggest that high mtDNA levels accelerate the progression of LAC [112] and CC [113]. Alwehaidah et al. demonstrate that high mtDNA copy numbers play a significant role during the initiation of ThC [114]. In TNBC, cell proliferation and resistance to doxorubicin (a commonly used chemotherapy agent) are correlated with high CNV values [115]. Meanwhile, in BrC [116,117], brain tumors [72], bone cancer [118], cancers of the oral tract [119], and HCC [84,120], cancer progression and severity has been associated with a reduction in the mtDNA copy number. Finally, tumors have been described whose progression and severity can be related to either a decrease or an increase in CNV. For example, CRC tumors isolated from different patients and analyzed for CNV showed both conditions [121,122]. In general, the results obtained from the analysis of different tumors show very heterogeneous behavior in relation to CNV, and the regulatory mechanisms are partly unknown. Thus, understanding CNV’s contribution to carcinogenesis requires further efforts.
3.1.3. Mitochondria and Numtogenesis Process
In some cases, a process called numtogenesis occurs, in which small fragments of mtDNA or the entire mitogenome are transferred into nDNA [123]. In this case, the mtDNA does not undergo mutations but instead becomes the cause of nDNA mutation. Indeed, the insertion into the nDNA leads to genetic instability through the destruction of regulatory sites or PC sequences. Numtogenesis can promote the onset of several pathologies, including cancer [22,124].
4. miRNAs
4.1. Biogenesis of miRNAs
The genomic sources from which miRNAs originate are multiple and differently organized. Indeed, miRNAs can be found not only within protein-coding genes—particularly in introns [125], at exon–intron junctions [126,127], or, more rarely, in exons [128,129,130]—but also in repetitive elements like TE [25,131,132] or in lncRNAs [129]. However, the origins of many miRNA molecules remain unknown [133]. Additionally, miRNAs can be organized as either single independent transcriptional units (annotated as miRNA host genes) or as multiple miRNAs embedded inside the same transcribed locus (Figure 2).
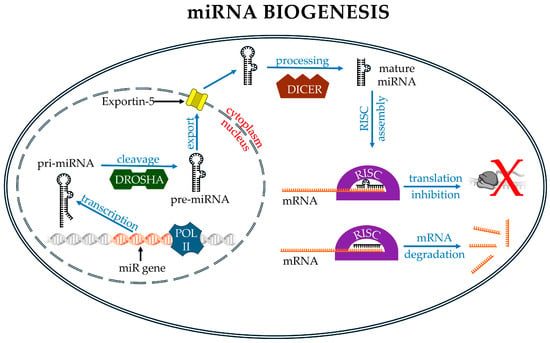
Figure 2.
Main events in miRNA biology. In the nucleus, the gene containing the miR sequence is transcribed by RNA polymerase II, which produces a pri-miRNA. This molecule is then cleaved by Drosha to form a pre-miRNA, which is exported into the cytoplasm by Exportin-5. In the cytoplasm, the pre-miRNA is further processed by the DICER complex to produce a mature double-stranded miRNA. Upon loading into the RISC, one of the two RNA strand binds to its target mRNA, promoting either its translational repression (partial match, red X) or degradation (perfect match). Image partially built using freely available resources at NIH BioArt (https://bioart.niaid.nih.gov/).
In the latter case, miRNAs form clusters whose transcription produces longer polycistronic primary miRNAs (pri-miRNAs), ranging in length from 1 to 10 kb. The pri-miRNAs contain the 5′ cap and the 3′ polyA tail and bear one or more hairpins, in which the mature miRNA sequence is located. The pri-miRNA is processed into a pre-miRNA in the nucleus by means of a complex known as the microprocessor. The microprocessor is a multiprotein complex in which Drosha and DiGeorge syndrome critical region 8 (DGCR8) constitute a minimal functional core. Drosha is a dsRNA-specific endoribonuclease III that binds at the stem–flank junction of the hairpin structure and mediates the cleavage of pri-miRNAs. DGCR8 forms a dimer that binds the terminal loop of the hairpin and interacts with Drosha, ensuring the accurate cleavage of pri-miRNAs [134,135,136]. The interaction between Drosha and DGCR8 induces a cleavage that generates a single pre-miRNA molecule with a length of approximately 55–70 nt. Pre-miRNAs are transported into the cytosol by the Exportin-5 (XPO5)/RanGTP complex. In the cytosol, pre-miRNAs are recognized by the Dicer/TRBP complex. Dicer, like Drosha, is a type III endoribonuclease. Dicer removes the loop structure from the pre-miRNA and forms mature miRNAs; these are ds segments whose lengths vary, according to different authors, in the range of 17–25 nts. The mature miRNA is loaded onto Argonaute (AGO) family proteins (AGO1–4), forming the pre-RNA-induced silencing complex (RISC). Within this complex, one of the two strands of the miRNA, the guide strand (antisense strand), is selected and retained, while the other, the passenger strand (sense strand), dissociates from the complex. The selection of the correct guide strand from the RNA duplex occurs through multiple mechanisms. For example, structural studies support a model that describes the thermodynamically asymmetric nature of duplex RNAs. Consequently, the strand with the most accessible 5′ end in the AGO binding pocket is the one that forms the thermodynamically most favorable bond and will function as the guide strand [137,138]. In addition, the identity of the 5′ nucleotide that specifically binds AGO can also influence the choice of the leader strand [139]. The RISC assembled with the guide strand becomes functional and is directed towards its targets by means of complementarity recognition between the guide strand and the target RNA. Additional non-canonical miRNA pathways exist and can be mainly distinguished as Drosha-independent or Dicer-independent pathways. For example, splicing and intron-debranching mechanisms can produce pre-miRNA-like structures that do not need to be processed by the microprocessor complex. These pre-miRNAs are then exported to the cytoplasm, where processing continues in the canonical pathway [140].
miRNA biogenesis using the non-canonical, Dicer-independent pathway is very rare. An example is the biogenesis of miR-451. Initially, pri-miR451 follows the canonical pathway in the nucleus: it is processed by Drosha and exported to the cytoplasm. However, the Drosha-mediated cleavage of pri-miR-451 produces a pre-miR-451 that is too short to be recognized by Dicer. Therefore, the cleavage of pre-miR-451 is mediated by AGO2. AGO2 has RNase-H-like endonuclease activity that can cleave specific miRNA precursors. Subsequently, the activity of a poly(A)-specific ribonuclease (PARN) is required to cleave the pre-miR-451 3′ end and produce mature miR-451. The functionality of miR-451 is dependent on its association with AGO2 [141,142].
4.2. Functional Role of miRNAs
The total number of miRNAs in the human genome is estimated to be several thousand. However, the most well-known miRNA databases, such as miRBasev.22.1 [143] and miRGeneDB v2.1 [144], report approximately 2000 miRNA molecules annotated for Homo sapiens. This discrepancy reveals the limited knowledge of miRNAs and their functions but, at the same time, suggests enormous potential that has not yet been explored. Interestingly, miRNAs can perform their activity either in the cell nucleus or in the cytoplasm.
In recent years, several studies have shown that miRNAs may perform functions within the nucleus, playing a role in transcriptional regulation. There, miRNAs can directly regulate miRNA biogenesis [145] or associate with target gene promoters [146] or enhancers [147,148].
Promoter-binding miRNAs and enhancers represent a subclass of miRNAs called NamiRNAs. NamiRNAs are transcribed by polymerase II from miRNA-coding genes with enhancer features. NamiRNAs help to assemble the complex containing polymerase II to allow the transcription of an activated enhancer. The resulting products are called eRNAs. NamiRNAs and eRNAs work together to activate gene expression [149,150].
In the cytoplasm, miRNAs are involved in gene regulation at the post-transcriptional level (Figure 2). Using different mechanisms, miRNAs can (I) promote the degradation of mRNAs using mechanisms such as deadenylation, decapping, and exonucleolytic decay [151]; (II) induce translational repression through the formation of the miRNA–RISC complex, which prevents the binding of the ribosome to the mRNA [152,153]; (III) bind other ncRNAs, thus indirectly controlling mRNA function. In particular, miRNAs may be part of a network consisting of various competing endogenous RNAs (ceRNAs), also called endogenous miRNA sponges. ceRNAs include various types of ncRNAs, such as lncRNAs or circRNAs. These ncRNAs likely prevent miRNAs from binding to miRNA-binding sites’ targets (miRNA recognition elements, or MREs) present in the mRNA [154,155,156].
miRNAs recognize and bind by sequence complementarity (Watson–Crick base pairing) to MREs present on different types of RNA molecules, including pseudogenes, lncRNAs, and circRNAs [157,158], and in the 3′ UTRs—or, more rarely, in the 5′ UTRs—of mRNAs [159,160,161,162]. The region of the miRNA complementary to the MRE is known as the seed sequence. The seed sequence represents a small part of the entire miRNA sequence and is usually included between nucleotides 2 and 7/8 from the 5′ end. The complementarity between the miRNA seed region and the MRE is responsible for the recognition of the correct miRNA target. Complementarity can be complete if all nucleotides of the seed sequence are involved in the interaction or partial if only some are involved. In partial interactions, bulges (structures formed when bases in one strand have no pairing partner in the opposite strand), wobble base pairs (a pairing between two nucleotides from two different strands that does not follow the Watson–Crick base pairing rule), or nucleotide mismatches (incorrectly paired nucleotides) may be present [163,164,165]. Moreover, a miRNA can bind its targets using additional mechanisms—for example, “pairing with the 3′ region” or using “centered sites”. Pairing with the 3′ region is a mechanism through which the region of the miRNA used for interaction with the mRNA is not limited to the seed sequence. In fact, this mechanism involves additional nucleotides located towards the 3′ end of the miRNA. This mechanism is used by miRNAs that use both full and partial pairing of the seed sequence [166,167]. Centered sites are non-canonical sites in miRNA targeting, consisting of contiguous base pairings of 11–12 nucleotides that occur starting from the third or fourth nucleotide and extending into the central region of the miRNA [168].
In general, the interaction between miRNAs and target RNAs is partial, since only a small part of the miRNA sequence is used. Furthermore, the involvement of few nucleotides increases the chance of a miRNA annealing on multiple RNA targets. This implies that a single miRNA may recognize many targets and, at the same time, allows a single target to be recognized by multiple miRNAs [58,169].
In addition to the well-known translational repression action, emerging evidence has revealed that some miRNAs can increase mRNAs’ stability and/or translation rate, resulting in target mRNAs’ upregulation [170,171,172]. The mechanism by which a miRNA induces gene upregulation is still partly unknown, but it seems favored under specific conditions. For example, some miRNAs, including let-7, activate translation during cell cycle arrest, but, in proliferating cells, they show the opposite effect [173,174]. Moreover, in quiescent cells, such as oocytes [175,176], or during amino acid starvation [177], it has been shown that miRNAs can upregulate gene expression.
The action of miRNAs may extend beyond the cell of origin thanks to their incorporation into exosomes. Exosomes are vesicles that are secreted by cells and are enclosed by a lipid membrane bilayer with a variable diameter of 30–150 nm. Exosomes transport many types of molecules, including proteins, lipids, DNA fragments, and different RNA species, including miRNAs. Thus, exosomes may connect two neighboring or distant cells by transporting messenger molecules. For example, it has been shown that exosomal miRNAs (exo-miRNAs) participate in various processes of tumorigenesis, including (but not limited to) tumor invasion and metastasis [178,179,180], cell proliferation [181], angiogenesis [182], and EMT [183,184]. Additionally, exosomes derived from tumor cells act as messengers and control tumor cells’ behavior within the tumor microenvironment [185]. Exo-miRNAs can influence the cancer treatment response as well [186,187,188]. Exo-miRNAs are primarily studied for their potential use as non-invasive biomarkers but represent an exciting frontier in cancer therapeutics research. However, to date, their translation into clinical practice poses significant challenges and requires further investigation.
Finally, a particularly interesting aspect concerns exogenous miRNAs or xeno-mirs. Cells, in addition to endogenous miRNAs and those received through exosomes, can contain miRNAs that are derived from other organisms and are mainly taken in through the diet. Xeno-mirs can influence cellular functions and play a role in maintaining the health of the organism [189,190,191]. In fact, some xeno-mirs act on the functions of the gut microbiota, and this, in turn, can play a role in the development of pathologies including coronary artery disease, neural degenerative diseases, and cancer [190,192]. The role of miRNAs in cancer was identified for the first time in 2002 by Callin and collaborators, who demonstrated that miR-15 and mi-R16 map at chromosome 13q14, a region frequently deleted in CLL. This deletion causes the absence or downregulation of both miRs in the majority of CLL cases [193].
In every type of cancer analyzed so far, the anomalous expression of several miRNAs has been observed, and, depending on their targets, miRNAs can act as either oncogenes or tumor suppressors [194,195].
4.3. Therapeutic Applications of miRNAs
Among ncRNAs, miRNAs are the most investigated in cancer, and their broad role in tumorigenesis makes them excellent candidates for the development of new and personalized therapeutic strategies. Nonetheless, several drawbacks limit their use in clinical practice. The lack of an effective delivery system capable of protecting RNA molecules from degradation by nucleases is one of the main issues. In addition, a molecular transport system that guarantees their release specifically in tumor cells, without inducing adverse effects such as an excessive immune response, is still a problem in the design and delivery of these molecules.
At present, miRNA-based therapeutics involve either miRNA inhibition or miRNA replacement [196]. In the first approach, chemically modified ASOs, such as LNAs or Antagomirs, are used, while, in the second approach, miRNA mimics are used. An important consideration is that miRNAs act as inhibitors of gene expression; therefore, strategies that aim at their inhibition have the effect of activating the expression of the target gene of the miRNA [197].
ASOs represent a large and heterogeneous group of ss DNA or RNA molecules, of approximately 15–21 chemically modified nucleotides [198], that bind to complementary miRNA sequences, modulating their functions (Figure 3).
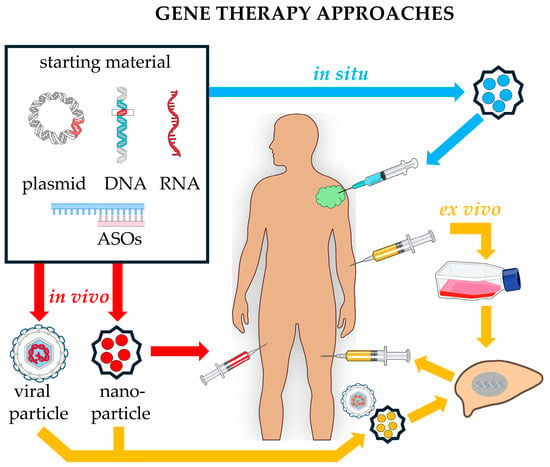
Figure 3.
Comparing different approaches in gene therapy. In the in vivo approach (red arrows), the starting material is incorporated into a vector (a viral or nanoparticle) and then injected into the patient. In the in situ approach (blue arrows), using appropriate vectors, the starting material is directly injected into the site of interest (e.g., a tumoral mass), where it exerts its effects. In the ex vivo procedure (yellow arrows), cells are explanted from the patient and cultured in vitro. Upon growth and selection, some cells are transformed using appropriate DNA vectors, such as a virus, to insert the sequence of interest into recipient cells, which are then transplanted back to the same donor. Image built using freely available resources at NIH BioArt (https://bioart.niaid.nih.gov/).
Chemical modifications endow ASOs with characteristics such as stability and cellular availability, target affinity, and cellular uptake, making them superior to sequences consisting of only canonical oligonucleotides. In fact, canonical oligonucleotides are easily degraded by both serum exonucleases and intracellular endonucleases, a condition that strongly impairs their therapeutic efficacy. Depending on their chemical modifications, ASOs have evolved through three generations. The first generation consists of ASOs with modifications of the phosphodiester bond. In these, one of the free oxygens of the phosphate group is replaced by a specific chemical group (sulfur, methyl, or amine group), generating the PSs, methylphosphonates and phosphoramidates. PSs represent the most widely used group of first-generation ASOs since this chemical modification confers an improvement in the stability of the structure, with consequently increased resistance to nuclease degradation and the elongation of its half-life. Members of the first generation can activate an RNAse H response; this is a ubiquitous enzyme that cleaves the RNA strand in a DNA–RNA duplex. In the case of ASO treatment, the activation of RNAse H allows the degradation of the target RNA within the ASO/RNA complex. Despite these positive aspects, first-generation ASOs are toxic and poorly specific [199]; consequently, researchers have explored other types of chemical modifications, which has led to the emergence of second-generation ASOs.
The second generation of ASOs consists of alkyl modifications at the 2′-position of the ribose, which leads to the formation of 2′-O-methyl and 2′-O-methoxyethyl nucleotides [200,201]. These modifications, on the one hand, improve their specificity and decrease their toxicity, but, on the other hand, they inhibit the ability to activate RNAse H. Therefore, second-generation ASOs are useful in cases where transient inhibition but not RNA degradation is required. The third generation is, in contrast, very heterogeneous in terms of the chemical modifications tested. This allow us to improve, depending on the chemical modification used, different characteristics, such as the binding affinity, nuclease resistance, pharmacokinetics, and thermal stability of ASOs. Example of these molecules are reported below.
4.3.1. LNAs in miRNA Inhibition-Based Therapy
One of the most widely used molecules belonging to third-generation ASOs is the LNA. LNAs are DNA or RNA sequences formed from a ribose sugar moiety modification in which the 2′-oxygen is connected to the 4′-carbon through a methylene bridge [202,203]. The structure of an LNA possesses characteristics that are useful for its use as a therapeutic agent, such as stability under nuclease-mediated degradation, excellent sequence specificity, good solubility in the aqueous phase, low toxicity, and high stability both in vivo and in vitro [204]. In relation to the structure of the molecule, synthetic LNAs are divided into two main groups: mixmers and gapmers. Mixmers are oligonucleotide sequences formed by LNA and DNA nucleotides that are randomly placed next to each other. Gapmers are also oligonucleotide sequences but, in this case, the DNA nucleotides are located in the center of the sequence, while the LNA nucleotides are found on either side [202]. Mixmers and gapmers can bind both DNA and RNA, and this makes their use particularly versatile. In general, LNAs work using different mechanisms of action: (I) they can induce the destruction of the target (activating RNase H or the RISC), (II) they can cause splicing alterations, or (III) they can induce a steric blockage in the target RNA.
The activation of RNase H is obtained using gapmers. Indeed, the DNA sequence contained within the LNA gapmer forms a DNA/RNA hybrid. This hybrid activates RNase H, with the consequent destruction of the target RNA [205,206]. Helmen and collaborators described the use of molecules consisting of a combination of mixmers and siRNAs, called siLNAs. siLNAs are not only compatible with the intracellular machinery of siRNAs but also mediate the activation of the RISC and the destruction of their target RNAs. Notably, siLNAs show a longer serum half-life than unmodified siRNAs [207].
LNAs can be used to induce alterations in the splicing process. In fact, their binding to the splice site at an intron/exon boundary can block the splicing of a specific intron and thus direct the splicing process towards the production of a specific product [208]. This use is particularly interesting because the relative abundance of alternatively spliced mRNA variants in tumor cells is different from that in healthy cells, suggesting an important contribution of mRNA alternative splicing in carcinogenesis [209].
A further mechanism of action promoted by LNAs is steric blockage, i.e., the reversible attack of the LNA molecule to the complementary miRNA. Obad and collaborators describe a method in which short LNA sequences (8-mer LNA oligonucleotides) can simultaneously inhibit multiple miRNAs that share the same seed sequence, with the concomitant upregulation of direct targets [210].
4.3.2. Antagomirs in miRNA Inhibition-Based Therapy
Antagomirs are a group of ASO-derived anti-miRNAs, also known as anti-miRNA ASOs or blockmirs, that function by binding complementary miRNAs, thus inhibiting their action on target genes. Antagomirs are characterized by a sequence consisting of ssRNA analogs conjugated to cholesterol. In vivo laboratory tests on murine models showed that the use of cholesterol in the antagomir improves its cellular delivery compared to other ASOs. In addition, antagomirs possess some good pharmacological characteristics, such as specificity, efficiency, and prolonged effects [211].
An example of their therapeutic use is described in the work of Wang J. and collaborators [212], who used miR-BART1-5p-antagomirs to inhibit a specific miRNA produced by EBV. EBV produces different miRNAs that can be secreted via exosomes from infected cells and affect the tumor microenvironment. Among these miRNAs, miR-BART has been associated with growth and invasion in several types of tumors, such as Hodgkin lymphoma and GC. In nasopharyngeal carcinoma EBV-miR-BARTs are associated with the mechanisms of VM, i.e., the process that leads to the formation of microvascular channels composed of tumor cells, and angiogenesis. The authors generated a therapeutic targeting exosome system with miR-BART1-5p-antagomirs and observed an inhibitory effect on both VM and angiogenesis. This effect is probably due to the increase in the expression levels of proteins such as Ras, c-Raf, MAPK, VEGF, PI3K, Akt, mTOR, and HIF1-α, which are important effectors of the signaling pathways that regulate angiogenesis and VM [212].
4.3.3. miRNA Replacement Therapy
miRNA replacement therapy is based on the use of synthetic miRNAs (also known as miRNA mimics), whose function is to re-establish the normal levels of expression and function of a specific endogenous miRNA that is underexpressed in tumor cells [213]. There are several types of mimics, which differ in their structure and the type of nucleotide chemical modifications used in the synthesis of the miRNA molecule [214,215]. However, both the initial steps required for the development of miRNA mimics and the chemical modifications of the nucleotides used are not well defined and are often covered by intellectual property rights [214].
The mechanism of action used by miRNA mimics involves their loading onto RISC and silencing their target mRNAs through the normal miRNA signaling pathway. The miRNA mimics used to activate RISC can be either ss or ds. In the case of ss (miRNA precursors), the mimic contains a sequence identical to the guide strand of the mature miRNA. The miRNA precursors can be both pri-miRNAs and pre-miRNAs. The pri-miRNA is transfected into cells and enters the nucleus, where it undergoes the first processing step. It is then translocated into the cytoplasm, where it is cleaved by Dicer and transformed into a mature miRNA. Instead, the pre-miRNA, after entering the cell, is directly cleaved by Dicer [214,216]. If ds, the miRNA mimic contains both the guide strand and the passenger strand. A comparison of ss miRNAs with ds miRNAs shows that ds miRNAs are 100 to 1000 times more effective than ss miRNAs [196,217]. The difference in efficacy observed between ss and ds miRNAs is due to the ds structure, which can facilitate the correct loading of the RNA molecule into the RISC, thus enhancing the gene silencing effect. Therefore, the design of mimetic miRNAs with a duplex structure, such as agomirs (see below), has become a major direction in therapeutic development.
Agomir miRNAs are short ds artificial RNAs in which the antisense strand has the same chemical modifications described for antagomirs. The applied chemical modifications allow not only higher affinity for the cell membrane, consequently increasing the transfection efficiency, but also greater intracellular enrichment compared to other types of miRNA mimetics, due to better resistance to degradation and greater stability in cells [218].
Overall, the use of miRNA mimics as potential therapeutic agents in cancer is still in the early stages of clinical development, but their potential as drugs is clear.
4.4. miRNA-Based Therapies in CTs
CTs evaluating ASOs and their derivatives as a possible therapeutic strategy in the oncology field are increasing. Their use is mainly based on their ability to selectively target mRNAs and consequently silence cancer-associated proteins [219,220,221]. In this way, pathogenic processes associated with the protein function are interrupted at the molecular level. However, their use towards ncRNAs, such as miRNAs, is still very limited. In fact, a search on the ClinicalTrials.gov website, which is a database of clinical research studies conducted around the world, including their results, for therapies that use miRNAs as a therapeutic target produces only eight results to date (Table 2). These studies mainly use LNA and miRNA mimetics as therapeutic tools.

Table 2.
List of main oncology CTs, described in detail in the text, which use ncRNAs as therapeutic targets. Data retrieved from ClinicalTrials.gov website on 22 December 2024.
NCT04811898 was a trial completed in 2021, in which a 13-mer LNA inhibitor of miR-221 (LNA-i-miR-221) with a full PS-modified backbone was analyzed for its safety and tolerability in patients affected by refractory multiple myeloma and advanced solid tumors. This LNA downregulates miR-221 and upregulates its targets, i.e., CDKN1B/p27 and PTEN. The study demonstrated that LNA-i-miR-221 has an excellent safety profile and anti-tumor activity, thus representing the first clinical evidence of the use of an LNA for the treatment of tumors [222].
The drug MRG-106 (cobomarsen) is an LNA inhibitor of miR-155 that stops cell proliferation and induces cell apoptosis in MF-CTCL cell lines and HTLV-1+ CTCL cells. MF-CTCL is a type of non-Hodgkin lymphoma localized in the skin; it is the most common form of cutaneous T-cell lymphoma. HTLV-1 is a virus that infects T cells and can cause leukemia and lymphoma. HTLV-1+ CTCL cells are CTCL cells infected by the HTLV-1 virus. Cobomarsen has been used in three clinical studies: NCT02580552, NCT03713320, and NCT03837457. NCT02580552 was a phase 1 CT completed in 2020. In this trial, cobomarsen was tested in MF-CTCL, CLL, DLBCL, and ATLL [223,228]. Given the successful results of the phase 1 CT in terms of clinical safety, efficacy, and pharmacokinetics, an additional CT (NCT03837457—phase 2) using the same drug was carried out. The primary aim of this second CT was to investigate the efficacy and safety of cobomarsen for the treatment of MF-CTCL in subjects who had confirmed disease progression following treatment with Vorinostat in the SOLAR clinical study (MRG106-11-201). The trial was terminated in 2020 with the following justification: “study no longer needed because eligible subjects may receive treatment with cobomarsen in a crossover arm of the SOLAR CT (NCT03713320)” [229]. The phase 2 CT NCT03713320 was the third trial involving the use of cobomarsen. The primary objective of the trial was to study the efficacy and safety of this molecule for the treatment of MF-CTCL. The study aimed to compare the effects of cobomarsen and Vorinostat, a previously approved drug used for the treatment of CTCL. Unfortunately, the trial was terminated early for business reasons and not due to concerns regarding cobomarsen’s safety or efficacy [230].
CT NCT01829971 was the first-in-human, phase 1 study of a miRNA-based cancer therapy. The purpose of this trial was to evaluate the safety of the drug MRX34, which is a synthetic ds miR-34a mimic encapsulated in a liposomal nanoparticle. Patients participating in the trial received at least one dose of MRX34 intravenously. Patients had solid tumors, including HCC, Me non-cutaneous excluding uveal, SCLC, TNBC, Sa, BlC, RC, and OC. The trial was terminated early in 2017 because five immune-related serious adverse events occurred among the 85 patients studied, resulting in four patient deaths [224,231,232]. The MRX34 drug was also evaluated in a phase 1B CT, NCT02862145. This trial involved the use of MRX34 combined with dexamethasone in Me cancer patients. The trial aimed to investigate the biomarkers, pharmacodynamics, and pharmacokinetics of MRX34. The participants were Me patients with easily accessible lesions who were monitored through serial biopsies and serial blood sample collection. Consequent to the adverse events observed during the first trial, the NCT02862145 trial was withdrawn in 2017 before participants were enrolled. However, despite the side effects, it was observed that treatment with MRX34 decreased the expression of miR-34 target genes, oncogenes, and immune escape genes in cancer patients. Therefore, miR-34a is still a promising target for miRNA-based cancer therapy.
The phase 1 CT NCT02369198 was the first human trial of the drug Targomir. The trial aimed to evaluate the safety and activity of Targomir in MPM and advanced NSCLC. Targomir is a new technology in the context of miRNA mimic-based therapies [225,233]. It consists of three parts: (a) a miRNA mimic based on miR-16, as several different forms of cancer have been linked to the miR-16 family’s role as tumor suppressors [234]; (b) a drug delivery system called EnGeneIC Dream Vector (EDV), where EDVs are non-living bacterial mini-cells (nanoparticles) that enable the efficient packaging of drugs, proteins, or nucleic acids inside them; (c) as a targeting moiety, an anti-epidermal growth factor receptor (EGFR) antibody that directs the EDVs to cancer cells expressing EGFR [225,233]. Indeed, it is known that EGFR is consistently deregulated in both LC and mesothelioma; it can therefore be used to target Targomir specifically to tumor cells [235,236]. NCT02369198 was completed in 2017 and involved 27 patients, of whom 26 received at least one dose of Targomir (one patient died before starting the treatment). During the trial, 21 deaths occurred, of which 20 were related to tumor progression and one was due to bowel perforation (caused by a second primary tumor). The experimentation allowed the researchers to establish not only the maximum tolerated dose but also the early signs of anti-tumor activity in patients with MPM [233]. Overall, this study offered new hope for mesothelioma patients, of whom less than 10% currently survive for more than 5 years [237]. Furthermore, the positive results obtained support a future phase 2 CT to evaluate the efficacy of Targomir therapy alone or in combination with conventional chemotherapy.
The drug INT-1B3 is an LNP-formulated miR-193a-3p mimic that was evaluated in the phase 1/1B CT NCT04675996. This trial was a first-in-human clinical study aiming to evaluate the safety, pharmacokinetics, pharmacodynamics, and preliminary efficacy of INT-1B3 in the treatment of patients with advanced solid tumors. In previous preclinical work, the function of synthetic miR-193a-3p mimic 1B3 was tested in cell lines derived from several cancers, such as TNBC, NSCLC, Me, CRC, and HCC. Treatment with 1B3 resulted in the upregulation of the tumor-suppressive PTEN pathway and the downregulation of many oncogenic pathways in cancer-derived cells. In addition, despite the different genetic backgrounds of these cancer cell lines, 1B3 showed consistent effects in suppressing cell proliferation, cell cycle progression, and cell migration and inducing apoptosis, cell senescence, and DNA damage. These results suggest the potential of IB3 in a broad range of cancers. The NCT04675996 trial started in 2020, and the last study records uploaded to the ClinicalTrials.gov website were provided in February 2024; the trial is currently described as “terminated due to insufficient funding” [226,238].
4.5. Recent CTs Evaluating miRNAs as Biomarkers
To date, the most numerous clinical trials based on miRNAs are those evaluating their possible use as tumor markers, as reviewed by Kim and Croce [239]. Therefore, this review aims to provide up-to-date information on the most recent clinical trials, registered on the ClinicalTrials.gov website, that have used miRNAs as potential diagnostic or prognostic biomarkers in various types of cancer (Table 3).

Table 3.
Summary of oncology CTs evaluating sncRNAs as biomarkers. Data retrieved from ClinicalTrials.gov website on 22 December 2024.
NCT06738225 is a CT that will start in 2025 and evaluate miR-15b and miR-21 as diagnostic biomarkers of CRC [240] by comparing their expression levels in CRC patients and healthy individuals.
NCT06610851 and NCT06203496 are CTs that seek to improve the knowledge of GB recurrence. NCT06610851 is a study initiated in 2024 that aims to identify miRNAs that can be used to monitor patients undergoing surgery for grade 2 and 3 GB, thus allowing the early diagnosis of recurrence. GB represents tumors of the central nervous system and is divided into four histological grades of malignancy. The treatment of GB is based on tumor removal and radiotherapy/chemotherapy treatments, depending on the grade of the glioma and the quality of the excision. In the case of recurrence, to be detected as early as possible, the patient can receive second-line chemotherapy. However, for grade 2 and 3 GB, monitoring is imperfect because it is not possible to detect tumor recurrence at an early stage. For these reasons, the use of early biomarkers, such as miRNAs, to monitor patients with grade 2 and 3 GB allows the timely diagnosis of recurrence. NCT06203496 focuses on changes over time in the plasma levels of pro-oncogenic miRNAs, after the surgical removal of a grade 4 GB, to assess whether they can be used to identify false-positive recurrences on magnetic resonance imaging.
In CT NCT06730035, EV circulating in plasma and the miRNAs that they contain are analyzed. The aim is to observe whether changes in the content of EV during neoadjuvant radiotherapy for locally advanced rectal tumors could provide early indications of the tumor’s response to treatment. Neoadjuvant radiotherapy is generally performed before surgery to reduce the size of the tumor to be removed. Therefore, obtaining early information on the response to neoadjuvant radiotherapy could contribute to the choice of a personalized therapeutic strategy.
In the observational study NCT06702891, the researchers are looking for specific biomarkers and potential therapeutic targets that can be used to develop new diagnostic tools and treatment strategies for GcC. GcC is a type of stomach cancer that begins in the mucus-producing cells in the lining of the gastric cardia (the part of the stomach closest to the esophagus). In this study, clinical information is collected from multiple biological samples, such as serum and tissue, which will be analyzed using an integrative analysis consisting of exosome-mediated single-cell transcriptomics and proteomics.
Additional clinical trials are currently running, for which only a very limited number of data is available. We recall here the following. NCT06224166 is a multicenter study in HNC patients, where miRNA detection from blood and saliva samples is being used as a non-invasive strategy to detect HNC recurrence. NCT06001099 is a prospective study to validate miRNAs, extracted from blood samples, for the early diagnosis of gynecological tumors. The aim of the NCT05901376 trial is to verify whether miR-20a, miR-21, miR-106b, miR-199a, and miR-22, extracted from blood samples, are upregulated in GC patients compared to healthy volunteers, to be effectively used as diagnostic biomarkers. The NCT06240195 trial is a prospective, multicenter study aimed at identifying predictive biomarkers of the efficacy/tolerability of Sacituzumab–Govitecan in the treatment of patients with metastatic TNBC. Sacituzumab–Govitecan is an antibody–drug conjugate composed of an antibody targeting human trophoblast cell surface antigen 2 (Trop-2), expressed in most breast tumors, coupled to SN-38 (a topoisomerase I inhibitor) through a hydrolysable linker [241].
The current recruitment status of the 2023 trial NCT05697224 is “not yet recruiting”. NCT05697224 aims to evaluate a Schistosoma haematobium (Sch)-specific miRNA, Sha-miR-71a, as a potential marker for the early diagnosis and prognosis of bilharzial BlC. An analysis of urine from patients with bilharzial BlC shows higher levels of Sha-miR-71a compared to both those observed in BlC not associated with bilharziasis (schistosomiasis) and in benign bladder cystitis associated with schistosomiasis [242]. This observation suggests that Sha-miR-71a can be used in the identification of BlC associated with infection. Schistosoma haematobium is a trematode worm that causes parasitic disease. In particular, the Sch eggs trapped in tissues release antigens that induce an immune response called schistosomiasis, or bilharzias/bilharziasis, in honor of the German surgeon Theodore Bilharz, who first identified the etiological agent Sch in 1851. The persistent immune response leads to the formation of granulomas, a compact assembly of inflammatory and resident cells, e.g., T cells, macrophages, and eosinophils, which form a well-defined structure surrounding the parasite eggs. A granulomatous reaction can result in organ damage [243]. In addition, urinary bladder infection due to Sch is correlated with the induction of BlC; in fact, the International Agency for Research on Cancer considers Sch a biologically carcinogenic agent [244].
The CT NCT05746858 aims to identify biomarkers that will predict the outcomes of standard and targeted therapies in patients with relapsed/refractory DLBCL.
Finally, the last CT, started in 2023, is NCT06320184. The goal of this CT is to refine LC risk assessment using blood biomarkers, including circulating miRNAs, in combination with artificial intelligence (AI)-integrated low-dose computed tomography to further implement LDCT screening strategies.
5. siRNAs
5.1. Biogenesis of siRNAs
siRNAs’ biogenesis can start from either exogenous or endogenous sources. Exogenous sources are predominantly foreign nucleic acids such as cytoplasmic dsRNAs derived from viral genome replication upon infection or from RNA secondary structures within viral genomes. Endogenous sources are specific genomic regions, such as repetitive DNA or TE, whose transcription gives rise to siRNA precursors. Mature siRNAs that originate from an endogenous source are called endogenous siRNAs or endo-siRNAs. Both endogenous and exogenous sources initially generate a long dsRNA precursor that will be processed to produce the mature siRNA.
Although the genomic origin of endo-siRNAs has been described in many model organisms, including Caenorhabditis elegans [245,246], Drosophila melanogaster [247,248,249,250], and Mus musculus [251,252,253,254,255,256], in Homo sapiens, the genomic origin of siRNAs remains elusive. However, the results presented in the work of Chen and collaborators suggest that the expression of the LINE-1 TE is regulated by endo-siRNAs. Indeed, in human BrC cells, both the significant depletion of endo-siRNAs and the increased activity of LINE-1 are observed compared to normal breast cells. The overexpression of endo-siRNAs in BrC is correlated with the silencing of endogenous LINE-1 expression [257]. Additionally, Jing and coworkers, building a small RNA deep sequencing data set, identified endo-siRNAs derived from tandem Alu SINE TE within the intron of the gon-4-like (GON4L) gene [258].
The long dsRNA precursor of siRNA is processed similarly to miR (Figure 2); in the cytoplasm, Dicer cleaves the precursor into a smaller dsRNA molecule known as a siRNA. siRNAs are approximately 21–23 nt long, with two-nucleotide overhangs at the 3′ end. The siRNA is then loaded into the RISC, where the endonuclease AGO2 cleaves the passenger strand of the siRNA, while the guide strand remains associated with the RISC. The siRNA guide strand directs the active RISC to its target mRNA for cleavage by AGO2 [259].
5.2. Functional Role of siRNAs
The functional mechanism of siRNAs shares some elements with the miRNA pathway, such as the involvement of the AGO2 protein and the formation of the siRNA–RISC complex, which induce the consequent endonucleolytic cleavage of siRNA targets (Figure 2). The sharing of some elements between the siRNA and miRNA pathways implies that the use of siRNAs as drugs requires the optimization of their concentrations to avoid the saturation of the RISC and the alteration of the endogenous mechanisms controlled by miRNAs [260,261,262]. The main functional difference between miRNAs and siRNAs is that the latter exploit fully complementary base pairing with their targets, i.e., mRNAs, lncRNAs, and circRNAs [263,264]. This feature allows siRNAs to recognize only a specific target.
In addition to the mentioned post-transcriptional silencing occurring in the cytoplasm, siRNAs and some components of their pathway, such as AGO2 and Dicer, have also been found in the nucleus. Nuclear siRNAs bind to promoter regions and mediate chromatin remodeling and histone modifications, resulting in transcriptional silencing [265]. Synthetic siRNAs are commercially available and have been widely adopted in RNAi technology. The use of RNAi with siRNAs allows the silencing of specific targets, thus representing another strategy to modulate overexpressed RNA molecules in cancer [266].
5.3. Therapeutic Applications of siRNAs
In the context of clinical applications, siRNAs and miRNAs share not only the same limitations, such as poor stability in vivo, delivery challenges, immune responses, and off-target effects, but also strengths, as their nature allows the development of similar strategies aimed at enhancing their efficacy and specificity, thus reducing off-target effects. siRNAs are generally introduced into cells via transfection, but their effects are transient because they are rapidly degraded. However, it has been observed that some chemical modifications, such as the substitution of the ribose 2′-OH group with other chemical groups (including 2′-O-methyl (2′-O-Me), 2′-fluoro (2′-F)) or the use of siRNAs composed of LNAs, increase their stability [207,267,268]. Furthermore, siRNAs can cause immune responses in both sequence-independent and -dependent manners. In the first case, through the activation of protein kinase R, many genes belonging to the interferon pathway are stimulated, which is part of the defense mechanism against viral infection, resulting in non-specific mRNA degradation and apoptosis. In the second case, specific immunostimulatory sequences induce the activation of the immune response by activating the transmembrane receptors TLR 7 and TLR 8 present in the endosomes of immune cells. Possible solutions to these problems include, for example, the use of delivery agents that exclude the endosomal release of siRNAs (electroporation), the replacement of immunostimulatory sequences with other sequences that do not induce such a response, and the use of chemically modified immunostimulatory sequences that do not allow their recognition by TLR receptors [269]. However, the modifications made to siRNAs can have toxic effects or render the molecule less efficient; therefore, the use of these substances in the clinical setting must be carefully evaluated to avoid adverse effects in patients [270].
A strategy to improve siRNAs’ efficiency is the use of short hairpin RNAs (shRNAs), which are expressed in the nucleus through delivery by viral vectors (Figure 4).
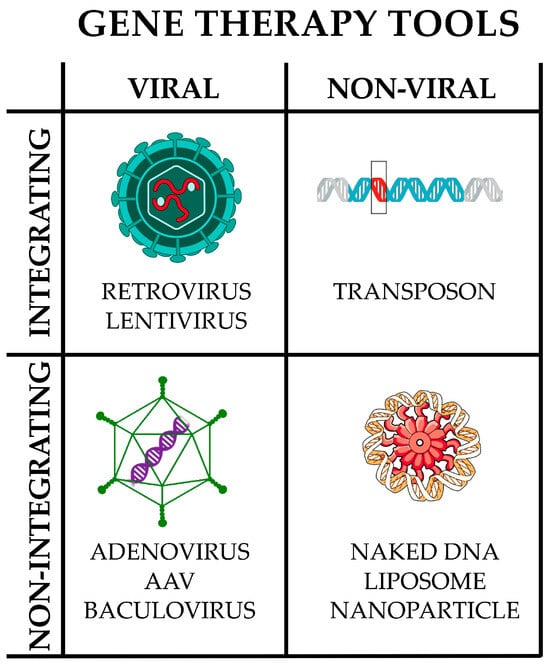
Figure 4.
Main gene therapy tools. They can be broadly divided into viral and non-viral; in turn, each may or may not integrate into the host genome. Only representative examples are reported; the list is not intended to be comprehensive. Image partially built using freely available resources at NIH BioArt (https://bioart.niaid.nih.gov/).
Within the nucleus, shRNAs are transcribed by RNA polymerase III, bypass Drosha processing, and are exported to the cytoplasm via XPO5 as pre-miRNA-like molecules. In the cytoplasm, they enter the physiological siRNA processing pathway, which culminates with loading into the RISC [271]. The viral presentation method, which includes the use of lentiviruses, adenoviruses, and adeno-associated viruses [272,273,274], guarantees not only very high efficiency in transferring the shRNA vectors into the nucleus but also high shRNA expression. The use of lentiviruses, which can integrate into the host genome, allows for shRNA-based therapies that are more persistent than siRNA-mediated ones, which require repeated administration because they are dependent on the rate of cell division. However, integration into the host genome increases the risk of insertional mutagenesis [275,276]. Furthermore, the use of viruses, particularly adenoviruses, is associated with high immunogenicity [277]. Recently, Alsing and collaborators developed a novel system, used for retinal gene therapy, that uses lentiviral vectors to present an expression cassette transcribed by polymerase III consisting of an RNAi construct (VEGFA-RNAi) in an Ago2-dependent shRNA (agshRNA) vector. The agshRNA vector is designed to be processed by Ago2 and produces a single guide strand, while classical shRNAs are processed by Dicer into a guide strand and a passenger strand. Since agshRNAs produce no passenger strand activity, a decrease in undesirable cellular responses is observed. Furthermore, agshRNAs show increased specificity and safety compared to shRNAs [278,279].
5.4. siRNA-Based Clinical Studies
The first CT involving the use of siRNAs dates back to 2004. In that trial, siRNA-027 was used for the treatment of age-related macular degeneration [280]. Meanwhile, the first clinical study using siRNAs for the treatment of cancer began in 2008 (NCT00689065) [281,282,283], with a phase 1 CT for the treatment of several solid tumors. The trial evaluated not only the safety, toxicity, and MTD but also the tumor response to the drug CALAA-01, a nanocomplex consisting of four main components: a siRNA duplex, a polymer, a stabilizing agent, and a targeting agent. The siRNA duplex used was not chemically modified and was designed to reduce the expression of the Ribonucleotide Reductase M2 subunit (RRM2), which participates in nucleotide metabolism and catalyzes the conversion of nucleotides to deoxynucleotides, maintaining dNTP pools for DNA biosynthesis, repair, and replication [284,285]. The other three components (the polymer, the stabilizing agent, and the targeting agent) form a nanoparticle of approximately 100 nanometers in diameter, inside which the siRNA is transported, thus ensuring its protection against nuclease degradation. The polymer is the basic constituent of the particle and it is a cyclodextrin-based polymer. The stabilizing agent is a hydrophilic polymer, PEG, used to promote nanoparticle stability in biological fluids. Finally, the targeting agent is constituted by human transferrin protein (Tf), which is exposed on the surface of the nanoparticle. Tf is recognized by Tf receptors (TfR) on the surfaces of cancer cells that overexpress the receptor. When CALAA-01 reaches the target cell, Tf binds to the TfRs on the cell surface, inducing drug endocytosis. Inside the cell, the nanoparticle releases a siRNA, which can exert interfering effects on RRM2. Data obtained from the trial revealed slight liver toxicity due to the chemical nature of the nanoparticle constituents but not to the siRNA used. Toxicity was alleviated by using a predosing hydration protocol (500 mL of 5% (wt/vol) dextrose in water before CALAA-01 infusions) and by advising patients to drink 2–3 L/day of fluids during treatment. However, this study was terminated early because 7 of 24 patients enrolled in the trial (29%) experienced disease progression characterized by an increase in tumor size [281].
Nowadays, there are many clinical studies using siRNAs as a therapeutic tool in oncology, but, in this case, similarly to what was described for miRNAs, the preferred targets are the overexpressed mRNAs of cancer-associated proteins [219,221].
Meanwhile, the use of siRNAs directed against specific ncRNAs is still in the preclinical stage. In particular, siRNAs are used to reduce the expression of overexpressed ncRNAs in tumors and evaluate the effects of silencing in relation to both the response to pharmacological treatment and improvements in tumor hallmarks (proliferation, migration, invasion, apoptosis, epithelial–mesenchymal transition).
However, there are some particularly interesting preclinical studies, such as the works of Liu [286] and Connerty [287], which used siRNAs to silence lncRNAs or circRNAs; these may soon be evaluated in CTs. Liu et al. tested a trivalent N-acetylgalactosamine (GalNAc)-conjugated siRNA construct, named GalNAc-silncRNA16 or Nano-silncRNA16, to perform the silencing of lncRNA16. An analysis of the serum lncRNA16 levels in NSCLC patients suggested that patients with elevated lncRNA16 values showed a poor response to chemotherapy, implying that lncRNA16 is a possible therapeutic target. Preclinical studies in mouse models of NSCLC suggest that silencing lncRNA16 with GalNAc-silncRNA16 restores chemosensitivity and results in tumor growth inhibition. Furthermore, GalNAc-silncRNA16 is specific and without detectable toxicity [286]. In the study conducted by Connerty et al., an LNP formulation, D-Lin-MC3-DMA, was used to deliver a siRNA for the treatment of t(8;21) pediatric ALL. This construct silences LINC01257, which is an oncogenic lncRNA that is overexpressed in AML cells, resulting in rigorous tumor growth and differentiation. The silencing of LINC01257 reduced tumor growth and had limited cytotoxicity [287].
Recently, Miao et al. used an LNP-encapsulated siRNA (LNP-siRNA) to silence Hsa_circ_0136666 in GC. Hsa_circ_0136666 competitively regulates PRKDC (a DNA-PK catalytic subunit) expression by sponging miR-375-3p. This results in the phosphorylation of PD-L1 (an immune checkpoint protein), which prevents its degradation. The phosphorylation of PD-L1 suppresses its immune function, thereby impairing the immune response to cancer. The use of the LNP-siRNA improved the efficacy of the anti-PDL1 drug and inhibited immune escape [288].
Epigallocatechin-3-gallate-lysozyme (EGCG-LYS) fibrils represent a novel siRNA delivery system for circMAP2K2 silencing, described by Dong et al. circMAP2K2 is abundantly expressed in GC, where it mediates the activation of the AKT/GSK3β/EMT signaling pathway. Furthermore, it enhances the proliferation and metastatic capacity of GC cells. The authors suggest that this novel delivery method has good circulatory stability, excellent biosafety, and in vivo anti-tumor capacity [289].
You et al. synthesized a novel siRNA delivery system, PEG-PCL (polycaprolactone)–PEI C14 (polyethyleneimine derivative)–SPION (PPPCS), based on superparamagnetic iron oxide nanoparticles (SPIONs). The intravenous injection of the PPPCS/siRNA complex silenced circ_0058051, resulting in the inhibition of tumor growth. Furthermore, the nanocomposite was nontoxic to the organs of nude mice [290].
6. piRNAs
6.1. Biogenesis of piRNAs
piRNA biogenesis is a complicated process, and many description models are based on Drosophila and mice, the best-characterized systems to date. Although some variation in piRNA biogenesis has emerged among the studied species, the process is generally conserved in its core components [291,292].
piRNAs are sncRNAs, approximately 24/25 to 31/32 nt in length, expressed mainly in the germ cells of animals. The majority of piRNA genes are organized into clusters at specific genomic loci. In these clusters, piRNAs align end to end or slightly overlap [293].
According to their genomic origin, piRNAs are divided into the following subclasses: transposon-derived [26], mRNA-derived [294], lncRNA-derived [294,295], snoRNA-derived [296,297], and processed transfer RNA (tRNA)-derived [32]. The piRNAs produced by TEs and repetitive sequences can be bidirectionally transcribed, generating sense and antisense piRNAs, with the latter being complementary to the DNA template. mRNA-derived piRNAs can be processed from full-length mRNAs [298] or from introns, exons, or the 3′ UTR of the pre-mRNA and recognize the same mRNA from which they originated [299,300,301].
piRNA biogenesis occurs in two phases, called the “primary” and “secondary” amplification cycles (also described as the “ping-pong cycle”) [302,303], which occur in somatic and germ cells or only in germ cells, respectively [304,305].
With primary amplification, the transcription of a piRNA cluster occurs by RNA polymerase II. The transcript formed is the piRNA precursor, which is a long ssRNA with a 5′ cap and a 3′ polyadenylated tail, which is then exported into the cytoplasm. There, secondary structures are resolved and the piRNA precursor is cleaved by MitoPLD and its co-factors into individual pre-piRNAs. The 5′ pre-piRNA is recognized and then loaded onto PIWI proteins, which are members of the highly conserved Argonaute protein family. After loading, further 3′ cleavage and concomitant methylation at the 3′ ends are required for the maturation of the piRNA molecule. In humans, mature piRNAs are expressed in germline and somatic tissues and generally have a uracil at the 5′ end, position +1, and an adenine at the +10 position, and they are 2′-O methylated at the 3′ end [299,306].
Once piRNA processing is completed, the mature piRNA–PIWI complex is formed and enters the nucleus, where it promotes TGS, or remains in the cytoplasm, where it can promote both PTGS and multiprotein interactions [307,308,309].
The secondary pathway is a mechanism that takes place in the cytoplasm and allows the rapid amplification of piRNAs. The antisense strands of piRNAs produced by primary amplification are bound by Aubergine (Aub) proteins; these are RNA-binding proteins that, in Drosophila, belong to the Piwi clade, formed by the Piwi, Aubergine, and Argonaute 3 (Ago3) proteins. The Piwi clade is part of the Argonaute protein family. In humans, the ortholog of Drosophila Aub is PIWIL1 [310].
The Aub–antisense-strand piRNA complex recognizes and cleaves the sense strand of a piRNA precursor. From this cleavage, a sense strand is generated, which is bound by Ago3. In contrast, Ago3 binds to sense-strand piRNAs and cleaves antisense piRNA precursors. In this case, an antisense piRNA is produced, which is loaded onto Aub proteins. However, Ago3 can only be loaded with secondary (Aub-generated) piRNAs, and the ping-pong cycle is initiated only by the piRNA–Aub complex. These piRNAs are then bound by PIWI proteins, forming the mature piRNA–PIWI complex [311,312,313].
6.2. Functional Role of piRNAs
piRNAs were first discovered in the testis of Drosophila melanogaster in 2001 [314] and were initially considered novel long siRNAs. Subsequently, piRNAs have been identified in about 44 species [315], including humans [316], for which there are more than 30,000 known piRNAs listed in the available piRNA-based databases. piRNAs show a tissue-specific expression profile, suggesting that they play important functional roles [317], but, for the most part, they are still unknown [318].
Functionally, piRNAs, like miRNAs, act through imperfect base pairing but, unlike the latter, piRNAs have considerable interspecific diversity and hence limited sequence conservation [303,319]. piRNAs are mainly involved in TGS and PTGS. Concerning TGS, the first reported function of piRNAs was the silencing of TE mobilization in fly germline cells [320], which is essential to maintain genome integrity. The inhibitory activity on TEs was later verified in other organisms, including humans [321]. Furthermore, it has been observed that piRNAs and Piwi proteins directly modify the chromatin structure and histone proteins in the nucleus by repressing the transcription of both TEs [322,323] and target genes [324].
In particular, piRNAs and Piwi proteins can repress transcription either by guiding DNMTs and promoting the methylation of CpG islands in promoter regions [324,325] or by interacting with the histone methylation machinery by regulating histone H3 lysine 9 (H3K9) [326] and lysine 4 (H3K4) [327,328] modification.
At the post-transcriptional level, piRNAs and Piwi proteins function similarly to miRNAs. The interaction with target molecules occurs by base pairing at the 5′ end of piRNAs and involves only a part of the piRNA sequence: 2–11 nt for strict base pairing and 12–21 nt for less strict base pairing [329]. piRNAs can bind to different RNA molecules, including transcribed pseudogenes [330], lncRNAs [331], and mRNAs [332]. The piRNA–mRNA interaction can occur by pairing in the 3′ UTR of mRNAs to promote deadenylation, with subsequent degradation via the mRNA decay machinery [291,332,333].
piRNAs are involved in post-transcriptional regulation also through the modulation of epigenetic m6A reversible modifications of RNAs [334]. Participating in this mechanism are “writer” methyltransferases, such as methyltransferase-like 3 (METTL3) and Wilms tumor 1-associated protein (WTAP), and “eraser” demethylases, such as AlkB homolog H5 (ALKBH5) and fat mass and obesity (FTO). The m6A modification of a mRNA affects the stability of the mRNA and regulates both the initiation and the elongation of its translation, but the precise fate of the mRNA depends on the functions of the different “readers” that bind the RNA molecule [335]. In general, the m6A modification can cause the destabilization of transcripts, which accelerates their degradation [336]. However, it has been described that the piRNA CHAPIR blocks the METTL3-mediated m6A methylation of Parp10 mRNA transcripts, which leads to an increase in the stability of the Parp10 mRNA, thus increasing in its expression [337]. Additionally, piRNA-30473 induces the upregulation of WTAP, which in turn increases the hexokinase 2 (HK2) m6A level, resulting in increased protein expression. Increased HK2 expression correlates with tumorigenesis in patients with diffuse large B-cell lymphoma (DLBCL) [338].
Finally, there are studies suggesting that piRNAs are also involved in the regulation of post-translational modifications such as phosphorylation [339,340].
A large body of literature suggests that piRNAs, as well as the associated Piwi pathway, may contribute to oncogenesis and tumor progression in various ways: (I) the anomalous expression of piRNAs is related to the development of different tumor hallmarks and to chemotherapy resistance [341,342,343,344,345,346,347,348]; (II) the deregulation of the piRNA–PIWI pathway can influence epigenetic mechanisms and lead to the altered regulation of gene expression with the consequent development of tumors [349,350,351,352]; and (III) impaired TE silencing may contribute to genomic instability, which is one of the hallmarks of cancer [353,354].
6.3. Therapeutic Applications of piRNAs
Numerous observations show that piRNAs’ abnormal expression is frequent in different types of cancer, but the exact mechanism behind their deregulation is still under investigation. To date, there are many preclinical studies on different tumors, including HCC [355,356,357], GC [358,359,360,361], CRC [362,363,364,365], osteosarcoma [366], LC [367,368,369,370], BrC [347,371,372,373], PrC [374,375], RCC [344,348,376], HNC [377], OC and CC [378,379], PaC [380], ALL [381], testicular cancer [382], ThC [383], and tongue squamous cell carcinoma [384].
These studies have explored piRNAs for their possible use as potential markers and provide new opportunities for cancer diagnosis, prognosis, or therapeutic approaches. Compared with other types of ncRNAs, piRNAs are a relatively new type of sncRNA, and aspects such as the complicated mechanisms through which they are generated, the difficulty in their identification, and the current lack of knowledge of their regulatory mechanisms represent limitations to their use in clinical practice. Furthermore, the role of piRNAs in the immune response and drug resistance in tumors is still largely unexplored. However, despite the many challenges in their clinical use, the interest in piRNAs and the availability of multiomics and sequencing technologies are leading to an improved understanding of their biological roles and their use as cancer therapy drugs.
6.4. piRNA-Based CTs
Research in this area is in its infancy; unfortunately, to date, there are no interventional CTs using piRNAs as therapeutic targets. However, several papers have suggested that piRNAs may be used as diagnostic or prognostic biomarkers in several types of cancer.
A recent study by Saha and collaborators suggests that human piR-23246, piR-32858, and piR-9137 may be used as biomarkers to diagnose PaC [385]. Xue et al. performed a meta-analysis on 27 studies to identify molecules in EV that could be used as non-invasive biomarkers for early GC diagnosis. From this study, several RNA molecules with diagnostic value emerge, including three piRNAs: piR-018569, piR-004918, piR-019308 [386]. Rui and coworkers identified five significantly upregulated exosome-derived piRNAs, piR-1029, piR-15254, piR-35395, piR-32132, and piR-43597, which could be used as biomarkers for HCC diagnosis [387]. Li et al. showed that serum exosomal piR-26925 and piR-5444 could be potential biomarkers for the diagnosis of LAC [388]. Nayak profiled miRNAs, piRNAs, and genes in GB U-87 MG cells and identified the targets related to progression and survival in GB patients. Among the identified targets, there are also some piRNAs that could be used as biomarkers in GB [389]. Peng found that the expression of piR-349843, piR-382289, piR-158533, and piR-002468 in urinary EV was significantly increased in PrC patients compared with a healthy control group, suggesting their use as diagnostic biomarkers [390]. Chang et al. observed that piR-13643 and piR-21238 were significantly upregulated in human PTC and suggested that they are promising novel biomarkers for the accurate detection of PTC [391]. Finally, Wang analyzed the serum of CRC patients and found that piRNAs piR-020619 and piR-020450 were upregulated compared to the controls. The authors indicate that the serum levels of the analyzed piRNAs show potential as specific early detection biomarkers for CRC [392].
In this context, there are only two observational CTs based on piRNA molecules: NCT06320418 and NCT04835454 (Table 3). NCT06320418 is a trial that started in 2022, and the current recruitment status is “active, not recruiting”. This trial aimed to investigate the regulatory role played by piR-823 in OC. Compared to non-tumor control tissues, piR-823 was deregulated in several cancers. Specifically, it was upregulated in HCC, CRC, and BC and downregulated in GC, showing that the mechanisms in which it is involved are tumor-specific. In fact, in HCC, it is involved in the pathophysiology of the tumor through the upregulation of the protein transforming growth factor b1 (TGF-b1) [355]. The TGF-b1 gene is frequently upregulated in tumor cells, and the protein regulates cell proliferation, differentiation, and growth [393]. In CRC, the upregulation of piR-823 promotes proliferation and inhibits apoptosis. Moreover, piR-823 upregulates heat shock transcription factor 1 (HSF1) expression by enhancing HSF1’s transcriptional activity through post-translational modification [340]. In BC, piR-823 promotes malignant cell proliferation, and its increased expression during cancer development may be associated with hormone levels [394]. In GC, piR-823’s downregulation is correlated with tumor growth inhibition [395]. However, the functional mechanisms of pir-823 in OC have not been investigated, so the above trial seeks to evaluate the possible use of pir-823 as a prognostic agent or as a potential target for drug development.
NCT04835454 is a trial that started in 2021, and the current recruitment status is not reported. The aim of the trial was to identify new biomarkers in PrC diagnosis. The potential biomarkers analyzed in CTs include different biological compounds, such as piRNAs, amino acids, and small nuclear RNAs. Currently, the early detection of PrC is based on two methodologies: digital rectal examination and the determination of the PSA levels in the blood. PSA testing seems not to be sufficiently specific for PrC as it often gives false positives [396], suggesting the need to identify new biomarkers.
7. circRNAs
7.1. Biogenesis of circRNAs
circRNAs are a type of ncRNA consisting of a ssRNA that forms a covalently closed circular structure between the 3′ and 5′ ends of the strand. Depending on the genomic origin, circRNAs can consist of a single exon or multiple contiguous or non-contiguous exons [397], from truncated forms of exons [398], from introns only [399], or from a combination of exons and introns [400,401] (Figure 5). Some circRNAs can originate from mitochondrial RNAs or from intron self-splicing that occurs during the maturation process of some constitutive ncRNAs, such as snRNAs, rRNAs, and tRNAs [27,28,29,30,31].
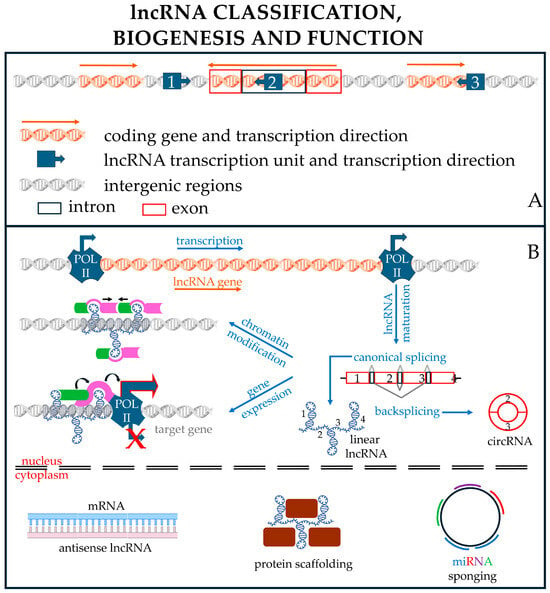
Figure 5.
lncRNA biology. (A) Classification based on lncRNA gene position in the genome, which can be outside a coding region (intergenic lncRNA, sometimes named lincRNA) (1) or inside the intron of a coding gene (intragenic lncRNA) (2). Sometimes, the lncRNA is encoded on the complementary strand of a coding gene, resulting in an antisense lncRNA (3) with translation regulation functions. (B) Transcription and function of lncRNAs. Genes encoding lncRNAs are usually transcribed by Pol II and, in many cases, undergo maturation (i.e., capping, polyadenylation, and splicing) as their mRNA counterparts. Some transcripts can undergo a particular splicing mechanism called backsplicing, which generates circular RNAs (circRNA). lncRNAs may have roles either inside the nucleus or in the cytoplasm. In the figure, some examples of these roles are reported. Inside the nucleus, the lncRNA can modify the chromatin structure (e.g., modifying nucleosome positioning to achieve more compact chromatin, black arrows), or it can recruit proteins (green and pink elements), which can alter the gene expression profile (e.g., transcription factors or methylases), possibly causing chromatin modification and either enhancing (Pol II, top) or repressing (Pol II bottom, with the red X indicating the inhibition of transcription) target gene expression. Curved arrows indicate that the green protein interacting with the lncRNA can recruit the pink protein, which, in turn, interacts with Pol II to modify target gene expression. In the cytoplasm, lncRNAs (either linear or circular) may interact with target mRNAs (antisense) or with proteins (scaffolds) or may sponge microRNAs (either different or multiple copies of the same miRNA). Image partially built using freely available resources at NIH BioArt (https://bioart.niaid.nih.gov/).
The biogenesis of a circRNA starts in the cell nucleus from a precursor (pre-mRNA) of protein-coding genes synthesized by RNA polymerase II (RNA Pol II), which are spliced using non-canonical mechanisms such as backsplicing and its variants, including lariat-driven circularization, intron-pairing-driven circularization, and RBP-driven circularization [402].
The backsplicing mechanism is regulated by both cis-regulatory elements (e.g., splice sites, enhancers, silencers, inverted Alu repeats) [403] and trans-acting factors (e.g., spliceosome factors, RNA helicases, and RNA-binding proteins) [404]. In this type of splicing, the 3′ splice site of the exon of a pre-mRNA is joined to the 5′ splice site of an upstream exon of the same mRNA molecule. Backsplicing allows the head-to-tail closure of a molecule because a downstream splice donor (5′ splice site) joins backwards to an upstream splice acceptor (3′ splice site).
The canonical splicing of pre-mRNAs usually removes introns between adjacent exons. However, sometimes, as in exon skipping, introns are removed from the pre-mRNA along with the exons, forming a structure called a lariat. The lariat can be further spliced to form a circRNA from either the exon alone or the intron alone [405,406]. In the first case, the circRNA is formed through a phosphodiester bond between the 3′ hydroxyl of the 3′ exon and the 5′ phosphate of the 5′ exon [407], while, in the second case, the circRNA is formed through a phosphodiester bond between the 2′ hydroxyl of the 5′ intron and the 5′ phosphate of the 3′-intron [399].
Intron pairing is a mechanism that occurs through the complementary pairing of flanking introns on both sides of the exons. The pairing of introns allows the formation of a ds stem. At the end of the stem, on one side, there is a downstream splice donor site, and, on the other, there is the upstream splice acceptor site of the pre-circRNA. The proximity of these sites favors reverse splicing events that remove the intron stem, allowing the joining of the sites (donors and acceptor) and the circularization of the RNA. The intron-pairing mechanism is strictly dependent on the pairing ability of complementary intron sequences (CIS), which are often derived from TEs that are inverted repeats, such as Alu elements [408,409]. The deletion of CIS from endogenous gene loci alters the formation of circRNAs [410,411], while some chromosomal translocations present in cancer cells create new CIS, promoting the generation of new circRNAs [412,413].
RBPs represent a category of proteins whose elements act as trans-acting factors in the regulation of circRNAs’ biogenesis [414]. Using high-throughput screening, approximately 1500 RBPs have been identified in the human genome, of which approximately 9% are likely involved in carcinogenesis [415,416,417]. There are two main types of RBPs, dsRBPs and ssRBPs. The former have dsRNA-binding domains and can regulate circRNA formation by affecting the RNA-pairing stability. ssRBPs do not have dsRNA-binding domains, recognize specific motifs present in the intronic sequence, and can dimerize with each other. RBPs promote contact between donor and acceptor splice sites. However, there are also RBPs that inhibit the formation of circRNAs, such as adenosine deaminase RNA-specific 1 (ADAR1). ADAR1 binds dsRNA to mediate adenosine-to-inosine (A-to-I) RNA editing, which is a post-transcriptional modification of the RNA sequence. ADAR1-guided editing modifies the circRNA precursor sequence, thereby altering the base pairing between complementary sequences in flanking introns, resulting in the negative regulation of circRNA biogenesis. Conversely, it has been reported that ADAR1 depletion can upregulate the formation of circRNAs [418,419]. In general, circRNA biogenesis is a very complex process, and one aspect that further complicates the understanding of this mechanism is that individual gene loci can generate multiple circRNAs (alternative circularization) [400]. This is possible by using different splice sites to form the backsplicing junction (a process called an alternative backsplicing event) and by using various types of alternative splicing [397,420].
The nucleotide sequences of many circRNAs can be post-transcriptionally modified, influencing different biological aspects of the molecule, such as its half-life, translation, nucleocytoplasmic export, and localization. The most commonly observed modifications are A-to-I editing and the m6A modification [421,422]. The m6A modification is particularly frequent, and its effect depends on “reader” proteins that recognize the modification sites [422].
Mature circRNAs can have different fates: they can carry out their activity in the nucleus, they can localize in the cytoplasm or in organelles such as mitochondria [30,423] or ribosomes [424], or they can be loaded into exosomes—thus acting outside the cells from which they originate. The nucleus contains mainly exon–intron or intron-only circRNAs. The mechanisms by which circRNAs are retained in the nucleus have not been fully elucidated. The cytoplasmic transport mechanism may depend on several adaptor proteins and some nuclear exportins. Adaptor proteins bind the circRNA molecule in relation to the length of the molecule. Huang and collaborators demonstrated that, in human cells, circRNAs of different sizes are transported into the cytoplasm by binding to different proteins, such as UAP56 and URH49, which regulate the nucleo-cytoplasmic transport of long (>1298 nt) and short (<356 nt) circRNAs, respectively [425]. However, other factors may be related to cytoplasmic transport, such as m6A modifications [426] or RNA duplex structures within circRNAs [427]. In both cases, however, the involved molecular mechanisms are not yet characterized. The circRNA-bound adaptor proteins are then exported into the cytoplasm through the involvement of nuclear exportins such XPO4 [428] or XPO2 [429]. Exosome RNA sorting involves specialized RNA sequences and/or secondary structures associated with RBPs. The precise mechanism by which circRNAs are sorted into exosomes remains undetermined [430,431].
Studies demonstrating that some circRNAs contain open reading frames that could be translated into small peptides are continually increasing. However, since circRNAs possess neither a 5′-cap nor a 3′-poly(A) tail, they adopt strategies that are different from canonical cap-dependent translation, such as the use of IRES plus additional regulatory sequence elements [432], mechanisms associated with A-to-I RNA editing [433] or m6A modification [434,435,436,437,438], the use of an exon junction complex [439,440,441], and rolling circle translation [442,443]. Many of the peptides produced by these mechanisms are implicated in carcinogenesis, suggesting their potential use as therapeutic targets.
7.2. Functional Role of circRNAs
The history of circRNA discovery began in 1976 with the description of a pathogenic viroid containing a covalently closed ssRNA [444]. Then, at the beginning of the 1990s, the pioneering works of Nigro et al. [445], Cocquerelle et al. [446], and Capel et al. [447] were published, which demonstrated the presence of circRNAs also in eukaryotes, including humans. However, an increase in circRNA research occurred in only around 2010, thanks to improvements in RNA-seq technologies, together with the development of specialized computational pipelines. Nowadays, it is estimated that human circRNAs amount to over 100,000. This is an impressive estimation considering that the circular nature of the molecule and the low abundance of some circRNAs often make their identification challenging. However, over the years, in addition to methods such as Northern blotting or RT-qPCR, high-sensitivity and -throughput strategies have been developed, such as rolling circle amplification [448,449], RNA sequencing (RNA-seq) coupled with NanoString technologies [450], microarrays [451], fluorescence in situ hybridization [452], and RNA-seq [453].
The analysis of circRNAs has also allowed us to improve our knowledge of the structural elements related to the functions that characterize the molecules. In general, a circRNA sequence may possess one or more of the following structural elements: short, inverted repeats and hairpin structures, MREs, or IRES.
The presence of inverted repeats and hairpin structures confers protein-binding properties to circRNAs. Protein interaction has several biological implications, including protein stabilization [454], degradation [455], localization [456,457], translation regulation [458,459], or scaffold functions [460,461].
Some circRNAs possess MRE sites that allow competitive binding to miRNAs. This binding inhibits the repressive function of miRNAs and consequently allows the expression of miRNA targets. This mechanism is called RNA sponging. A typical miRNA sponge consists of multiple miRNA-binding sites, each containing mismatches at intermediate positions to prevent the activation of the endoribonuclease function of Ago2 [462]. circRNAs can contain either multiple binding sites for the same miRNA or multiple miRNA-binding sites for different miRNAs. For example, circZNF91 contains 24 miR-23b-3p target sites [463], while circCCDC66, whose elevated expression in CRC is associated with poor prognosis, possesses multiple miRNA-binding sites, including those for miR-33b, miR-93, and miR-185 [464].
There are numerous examples in the literature implicating the deregulation of the miRNA sponge function of circRNAs in the development of numerous pathologies, including cancer. In this context, circRNAs can either repress or promote oncogenesis, and, in general, the alteration of the circRNA–miRNA–mRNA axis (composed of ceRNAs) influences every aspect of carcinogenesis, including the response to treatment [465,466,467,468,469,470].
CircRNAs, in addition to their function as miRNA sponges to regulate the activity of miRNAs, also have additional roles, including miRNA storage (e.g., CDR1-AS) [471] and transport (e.g., circDlc1(2)) [472]. The cerebellar degeneration-related 1 (CDR1) gene expresses an antisense circular transcript, known as CDR1-AS, which interacts with several miRNAs, including miRNA-671 and miRNA-7. CDR1-AS binds miRNA-7, for which it has 70 MREs, and transports it to a specific site. Subsequently, binding to miRNA-671 induces the degradation of the circRNA and the release of miRNA-7. The regulation of this mechanism is not fully understood but it has been hypothesized that it could depend on spatial–temporal signals [473]. circDlc1(2) binds some mRNAs associated with glutamate receptor signaling (gluRNAs) and allows the correct localization of miR-130b-5p at synaptic regions, where a gluRNA is localized. An interesting aspect of this circRNA is that the binding to the miRNA does not induce its inhibition. In fact, the control of the subcellular localization of the miRNA promotes its activity, suggesting a new mechanism compared to the canonical one of miRNA inhibition [474].
Nuclear circRNAs may be involved in the regulation of gene expression by modulating the activity of RNA polymerase II, epigenetic mechanisms, mRNA splicing, and rRNA processing. Some circRNAs, such as circEIF3J and circPAIP2, have been described to bind U1 spliceosomal RNA through an RNA–RNA interaction. U1 spliceosomal RNA is the snRNA component of U1 snRNP, a complex involved in the assembly of the spliceosome. The circRNA–U1 snRNP interaction could regulate the Pol II transcription complex at the promoters (cis via) of parental genes to enhance gene expression [418]. Furthermore, some circRNAs, such as circFECR1, can activate the transcription of their parental genes through the modulation of epigenetic mechanisms. circFECR1 is an FLI1 exonic circRNA, and, by inducing DNA hypomethylation in the CpG islands of the FLI1 promoter, it enhances its transcription. In addition, in BrC, FLI1 regulates the formation of metastases through the epigenetic regulation promoted by its exonic circRNA [475]. Other nuclear circRNAs, such as circ-MBL [476] and circSMARCA5 [477], regulate mRNA splicing. circ-MBL works through competitive binding with the splicing machinery, while circSMARCA5 has binding motifs for several RBPs, including the splicing factors SRSF1, SRSF3, and PTB2. circANRIL, through interaction with nucleolar protein 14 (NOP14), blocks pre-rRNA processing and ribosome biosynthesis, inducing cell apoptosis [478].
Finally, some circRNAs, through binding to specific proteins, such as transcription factors, allow their nuclear translocation. For example, the interaction of circ-Amotl1 with c-myc not only allows the protection of c-myc from degradation but also the translocation of the protein into the nucleus, where it can perform its function [479].
Currently, numerous manuscripts have highlighted that the alteration of circRNA pathways is responsible for the development of many pathologies, including cancer. Unfortunately, the majority of the works on circRNAs focus on the consequences of the alteration of the expression levels of a specific circRNA and not on the causes. However, despite the few available studies, it is possible to describe some of the factors that lead to the deregulation of circRNAs.
Some alterations could depend on genomic mutations that directly affect intronic flanking regions, leading to the generation of new oncogenic circRNAs. An example is the SLC34A2–ROS1 (solute carrier family 34 member 2 and ROS proto-oncogene 1) gene fusion that results from a chromosomal translocation, creating a new flanking complementary sequence with canonical splicing sites. This involves the biogenesis of two new circRNAs: F-circSR1 and F-circSR2. Both promote cell migration in NSCLC and are important for cancer progression [412]. circMLL is also a byproduct of a chromosomal translocation and promotes transcriptional pausing, proteasome inhibition, chromatin reorganization, and DNA breakage in the early stages of the development of acute leukemias [480].
Qiu and collaborators, through the screening of circRNA expression in relation to CNV in LAC, identified circPRKCI. CNV represents one of the main causes of structural variation in the genome, involving both duplications and deletions of chromosome fragments, and is involved in tumorigenesis. circPRKCI is a proto-oncogenic circRNA derived from the LAC 3q26.2 amplicon. circPRKCI is overexpressed in tumors and promotes cell growth and migration by functioning as a sponge for both miR-545 and miR-589 [481].
Altered epigenetic mechanisms can influence the biogenesis of circRNAs. For example, some circRNAs are transcriptionally silenced through hypermethylation or altered histone modifications at their host gene promoters, leading to a subsequent reduction in circRNA expression [482,483,484].
Alterations in splicing factors or the production of abnormally alternative splicing isoforms is a condition often detected in tumors [485,486]. Kong et al. report that, in PDAC, the upregulation of circARFGEF2 is dependent on the KRASG12D mutation. This mutation is responsible for the overexpression of the RNA splicing factor Quaking (QKI)-5, which, in turn, facilitates cir-cARFGEF2 biogenesis by binding the QKI-binding motifs and reverse complement sequence in introns 3 and 6 of the ARFGEF2 pre-mRNA. Thus, the overexpression of circ-ARFGEF2 induces lymph node metastasis in PDAC [487,488]. The biogenesis of a circRNA depends on the correct execution of the backsplicing mechanisms and its variants, which is guaranteed by the action of cis-acting elements and trans-acting splice factors. Thus, alterations at the level of cis- and trans-elements can influence circRNA biogenesis [489]. Fernandez and collaborators performed circRNA-seq on lymphoid and myeloid cell lines expressing the most common splicing factor (SF) mutations, showing the general upregulation of circRNAs. In addition, each mutant SF is characterized by its own set of upregulated circRNAs [490]. The RNA-binding protein QKI is involved in alternative splicing and binds QKI-binding motifs present on some introns, contributing to the biogenesis of circRNAs. Conn and collaborators demonstrated that, during EMT in immortalized human mammary epithelial (HMLE) cells, hundreds of circRNAs are upregulated in a QKI-dependent manner [491]. Additionally, the depletion or inhibition of core spliceosome components, including the U1 and U2 snRNPs, results in circRNA upregulation [404,492].
The stabilization of the molecule by some post-transcriptional modifications, such as m6A, could also contribute to the accumulation of the circRNA, as observed for circRPS6KC1 in PrC [493].
Finally, some circRNAs can be loaded into exosomes. The incorrect loading of circRNAs could have a dual effect: on the one hand, the cell will experience a decrease in its circRNA load; on the other hand, exosomes lead to the enrichment of incorrect circRNAs in inappropriate locations.
Mechanistically, the altered expression level of circRNAs, both endogenous and exosomal, observed in cancer cells has been associated with the deregulation of key cellular signaling pathways, such as PI3K/AKT/mTOR, Wnt, notch and hippo, p53/Bcl-2, and TGF-β/Smad, which in turn, in the context of carcinogenesis, influences cell proliferation, EMT, invasion, metastasis, apoptosis, angiogenesis, and the pharmacological response [494,495].
In recent years, the study of circRNAs has produced a large amount of data, which has made it necessary to organize them into databases. There are different types of databases that facilitate study and consultation for those working in the field. For example, MiOncoCirc provides information about the associations between circRNAs and cancer [496], and Lnc2Cancer 3.0 contains not only circRNA–cancer associations but also information on the regulatory mechanisms, biological functions, and clinical applications of circRNAs in cancer [497]. CircFunBase [498], deepBase [499], and circBank [500] are three additional examples of databases that collect data on the interactions of circRNAs with RNAs and proteins. Finally, the circVAR database collects SNPs and small insertions and deletions (INDELs) in putative circRNA regions. The use of circRNA variants in GWAS allows the identification of many cancer-based somatic variants, suggesting novel mechanisms for cancer development [501].
7.3. Therapeutic Applications of circRNAs
Therapeutic applications are mainly limited by the poor characterization of the biological functions performed by circRNAs. However, the understanding of RNAs’ structure and function and advances in nucleic acid synthesis/modification technology have led to the development of synthetic circRNAs that could be used as therapeutic agents [502,503]. This approach enables the synthesis of circRNAs and protein expression at multiple levels, from the specific cellular compartment or within cells and tissues, with the aim of pursuing targeted therapy. For the therapeutic use of synthetic circRNAs, some technical aspects must be optimized. In fact, the synthesis of this molecule can lead to the incorporation of exogenous noncoding sequences or to a final product that is not sufficiently purified and can activate a patient’s immune response after administration. In this context, the choice of the vector to be used must also be made with particular care to avoid adverse responses. Currently, the delivery vectors can be broadly categorized into viral (adenoviral, retroviral, and AAV vectors) and nonviral vectors (physical methods, chemical methods, and biologically derived vectors), with the latter being preferred [502] (Figure 3).
Regarding the expression of circRNAs in disease, upregulation is more common than downregulation. This aspect has led to the use of different oligonucleotide-based strategies that are designed to achieve circRNA knockdown by targeting the backsplicing junction site, allowing one to restore circRNA expression to a healthy level. These strategies include the use of siRNA [504], shRNA [505], ASO [506], or CRISPR/Cas systems [506], which represent a powerful gene editing tool that specifically knocks down a circRNA without interfering with its homologous mRNA [424].
7.4. CircRNA-Based CTs
The use of circRNAs in clinical practice is currently limited to their use as biological markers for early diagnosis. For example, PrC diagnosis was significantly improved in clinical studies using the combination of circRNA detection and PSA testing [507]. The use of circRNAs as biological markers depends on some particularly advantageous characteristics that they possess. The closed structure of a circRNA ensures greater stability than linear RNAs. This feature depends on the ability to resist exonuclease-based degradation and allows circRNAs to have a longer half-life (ranging from 19 h to 24 h) than linear transcripts with identical nucleotide sequences (ranging from 4 h to 7 h) [508].
circRNAs present in exosomes are also protected from degradation, thus allowing them to exert their action far from the cell of origin. Furthermore, exosomes are not only easily detectable but also present in many human biofluids, such as urine, saliva, gastric juice, cerebrospinal fluid, ascites, and blood, and could also be used for liquid biopsies [495], allowing for fast and minimally invasive detection procedures.
circRNA-based therapies are currently limited to preclinical studies only. At present, a search on the ClinicalTrials.gov website produces only 10 CTs based on circRNAs: nine observational trials evaluating the use of circRNAs as markers (Table 4) and one interventional trial (phase 1) evaluating the use of circFAM53B-219aa DC in cancer vaccine monotherapy.

Table 4.
Summary of oncology CTs evaluating lncRNAs as biomarkers.
The trial NCT06649253 started in 2025, and the current status is “not yet recruiting”. The aim of this CT was to analyze the network of lncRNA/circRNA/miRNA/mRNAs involved in the regulation of CD9 in B-ALL. B-ALL is the most common cancer in children and is characterized by a rather high percentage of patients with relapse. CD9 is a transmembrane protein associated with B-ALL relapse [509,510], and understanding the mechanisms that regulate its expression could be useful to identify relapse markers.
NCT06617585 started in 2024, and the current status is “not yet recruiting”. The aim of the trial was to evaluate the possible diagnostic role of circDENND4C [511] in OC.
The trial NCT06042842 started in 2023, and the current status is “not yet recruiting”. This trial is evaluating hsa_circ_0004001 [512] as a diagnostic biomarker for HCC.
The trial NCT05934045 started in 2023, and the current status is “active, not recruiting”. This trial aims to evaluate the possibility of using circRNAs as circulating prognostic and/or predictive biomarkers in ALCL, an aggressive T-cell pediatric lymphoma.
The trial NCT05771337 started in 2023, and the current status is “not yet recruiting”. The aim of this trial is to evaluate hsa_circ_0001785 (Circ-ELP3) and hsa_circ_100219 (Circ-FAF1) [513] in serum samples from BrC patients as possible diagnostic and prognostic biomarkers.
The trial NCT05377736 started in 2022, and the current status is “enrolling by invitation”. This pilot study aims to describe the distribution of molecular alterations in benign and malignant thyroid nodules at the DNA and RNA levels, including circRNAs, in the Danish population.
The trial NCT04464122 started in 2020, and the current status is “recruiting”. The goal of this study is to identify novel biomarkers, including circRNAs, from tumor-educated platelets (TEPs) in the diagnosis and assessment of the treatment response in pulmonary and gastro-entero-pancreatic NENs. Platelets are involved in the processes of tumorigenesis; during their life cycle, in the bloodstream, they absorb and enrich substances produced by the tumor—hence, they are named TEPs [514]. NENs are a group of heterogeneous tumors with neuroendocrine differentiation that can arise from cells distributed within the neuroendocrine system [515].
The trial NCT04584996 started in 2020, and the current status is “unknown”, meaning that the trial has passed its completion date and the status has not been verified in the last 2 years. Studies with an unknown status are considered closed studies. In this trial, the aim was to define the circRNA expression profile in PDAC and to find, among the dysregulated ones, a candidate for a new, clinically relevant diagnostic or prognostic biomarker.
The trial NCT03334708 started in 2017, and the current status is “recruiting”. In this trial, blood samples are being analyzed for molecules, such as specific circRNAs, that could be used as biomarkers to diagnose PaC in the early stages of the disease and monitor the response to treatment.
The trial NCT06530082 started in 2024, and the current status is “not yet recruiting”. This trial is phase 1 and is aimed at evaluating the safety, efficacy, and tolerability of circFAM53B-219aa DC [516] vaccine monotherapy and its combination with camrelizumab in the treatment of HER2-negative advanced BrC.
8. lncRNAs
8.1. Biogenesis of lncRNAs
lncRNAs are a heterogeneous class of RNA molecules longer than 200 nucleotides that do not encode proteins. In the last few decades, they have emerged as key regulators of gene expression and play significant roles in various biological processes and diseases, including cancer. Their biosynthesis involves transcription by RNA polymerase II, similar to what happens to mRNAs. Likewise, their maturation follows comparable steps, including capping at the 5′ end, splicing, and polyadenylation at the 3′ end. However, unlike mRNAs, lncRNAs often exhibit lower expression levels and higher tissue specificity [65,517]. The transcription of lncRNAs is tightly regulated by various transcription factors and epigenetic modifications, which ensure their precise expression in different cellular environments (Figure 5).
8.2. Functional Role of lncRNAs
lncRNAs regulate gene expression at multiple levels, including chromatin remodeling, transcriptional control, and post-transcriptional processing [518]. They can interact with DNA, RNA, and proteins to modulate various cellular processes, such as cell cycle progression, apoptosis, and differentiation [519]. Some lncRNAs act as molecular scaffolds, bringing together different proteins to form functional complexes [520], while others serve as decoys to sequester regulatory molecules [517]. Furthermore, lncRNAs can influence mRNA stability and translation by binding to complementary sequences or interacting with RNA-binding proteins [521]. They also play roles in the formation of nuclear bodies and the maintenance of the nuclear architecture [522]. lncRNAs have gained attention for their potential as biomarkers in cancer diagnosis and prognosis due to their specific expression patterns in different cancer types. For instance, the lncRNA PCA3 is used as a biomarker for PrC [471]. lncRNAs can be detected also in body fluids, making them suitable for non-invasive diagnostic tests [472,523,524,525]. Additionally, they are involved in cancer progression by regulating oncogenes and tumor suppressor genes, thus representing potential therapeutic targets [526].
8.3. Therapeutic Applications of lncRNAs
Therapeutic strategies targeting lncRNAs include antisense oligonucleotides, siRNAs, and small molecules designed to modulate their function. Recent studies have shown that lncRNAs can also mediate the resistance to chemotherapy and radiotherapy, highlighting their importance in personalized cancer treatment.
Pseudogenes are a particular class of lncRNAs that resemble functional genes. They arise through gene duplication or retrotransposition but accumulate mutations that prevent them from encoding functional proteins. Despite being considered “junk DNA”, some pseudogenes are transcribed into RNAs and can regulate gene expression by acting as decoys for miRNAs or by generating regulatory RNAs. Pseudogenes can also contribute to genomic instability and cancer development by serving as sources of genetic variation [527,528].
Thus, lncRNAs and pseudogenes represent significant components of the noncoding genome with crucial roles in gene regulation and disease. Their unique properties and functions offer promising avenues for the development of novel diagnostic and therapeutic strategies in cancer.
8.4. lncRNA-Based CTs
There is excellent potential for the development of cancer therapies targeting lncRNAs, but, currently, these are only in the early stages of development. Instead, CTs evaluating their use as diagnostic, prognostic, or predictive markers (detection of relapse, response to treatment, monitoring of therapeutic efficacy) are more numerous (Table 4).
Unfortunately, the results of the completed CTs have not yet been published. However, from the CTs presented in Table 4, two interesting observations emerge. The first is that approximately 46% of the trials (12 out of 26 CTs) have started in the last three years, suggesting a greater interest in experimentation; the second is that the most frequently evaluated lncRNAs in CTs are lnc-GC1 (NCT05397548, NCT05647941, NCT05334849), H19 (NCT05943093, NCT04767750), and HOTTIP (NCT06544005, NCT04729855), which, taken together, constitute about 27% (7 out of 26 CTs) of the listed trials. These lncRNAs are studied in GC (lnc-GC1), in HCC (H19, HOTTIP), in ALL (H19), and in CRC (HOTTIP), and, according to data communicated for the year 2022 by the GCO (a platform curated by the WHO and the IARC), they are among the tumors with the highest incidence and mortality in the world.
Sun et al. demonstrated that GC-associated long noncoding RNA1 (lncRNA-GC1) is upregulated in GC, where it influences several aspects of carcinogenesis, such as the tumor size, metastasis, and prognosis. The authors suggest that, mechanistically, lncRNA-GC1 acts as a scaffold for two protein factors involved in histone modifications: WDR5 (a histone methyltransferase) and KAT2A (a histone acetyltransferase). The action of lncRNA-GC1 allows the correct localization of the two proteins, thus ensuring their normal function on the target genes. The abnormal expression of lncRNA-GC1 affects the functions of WDR5 and KAT2A, inducing altered gene expression that leads to the formation of GC [529]. Subsequent studies have identified lncRNA-GC1 as a highly expressed GC-specific lncRNA in both GC cells and exosomes. In particular, the work of Guo et al. demonstrated a correlation between lncRNA-GC1 levels and the GC stage [530]. This implies that, in GC patients, the detection of circulating exosomal lncRNA-GC1 provides clinically important diagnostic and prognostic information [531].
H19 is a lncRNA involved in many regulatory cellular functions [532], and its altered expression has been observed in many tumors [533]. Depending on the type analyzed, H19 can behave as an oncogene or a tumor suppressor [534]. For example, in ALL (both B-ALL and T-ALL) [535] and HCC [536], H19 behaves as an oncogene. Specifically, in ALL, the observed expression level of H19 was significantly higher than in controls [535], and it was correlated with an unfavorable prognosis [537]. Zhao et al. suggest a mechanism in which increased H19 could competitively bind miR-19a and miR-19b, resulting in the upregulation of inhibitor of DNA binding 2 (ID2), which is an important regulator in cell proliferation and differentiation [538,539]. In HCC, H19 is involved in carcinogenesis and the recurrence, metastasis, and chemoresistance of tumors. In addition, it was observed that H19 expression increased in the liver cancer stem cells, tissue, and plasma of patients with HCC, while it decreased after a partial/complete therapeutic response [536]. Although the biological function of H19 in HCC remains to be elucidated, Nokkeaw et al. suggest that H19 may act as a molecular sponge for miR-107 to promote cyclin-dependent kinase 6 (CDK6) expression and cell cycle progression; this axis may explain the correlation between H19 overexpression and increased cell proliferation in HCC [540].
The lncRNA HOTTIP (homeobox A (HOXA) transcribed at the distal tip) is encoded by a genomic region at the 5′ tip of the HOXA locus, which plays a key role in embryologic development. HOTTIP binds the histone methyltransferase complex, allowing its correct localization to specific genomic loci, including the HOXA gene cluster. HOTTIP is an example of the cis regulation of gene expression by a lncRNA, since its action allows the trimethylation of histone H3 at lysine 4 (H3K4me3) and the activation of HOXA genes [541]. HOTTIP expression levels are altered in many tumors [542,543,544], including HCC [545] and CRC [546]. In HCC patients, HOTTIP upregulation is correlated with advanced tumor stages and a poorer prognosis. The mechanisms by which HOTTIP is involved in HCC carcinogenesis are multiple. For example, Wang and coworkers suggest that HOTTIP promotes the activation of HOXA genes, which, in turn, is correlated with cell proliferation, migration, and invasion [541,545]. Wei et al. indicate that HOTTIP promotes HCC by regulating glutamine metabolism [547]. The regulation of glutamine metabolism is a type of metabolic reprogramming that is considered a hallmark of cancer cells [548]. In the literature, there are several works that exploit the properties of HOTTIP, such as its stability in serum and the high expression levels in HCC patients compared to healthy controls, to evaluate a possible role as a tumor marker. In this context, Bao et al. and Kim et al. indicate HOTTIP as a potential marker for the early diagnosis of HCC [549,550]; in addition, Bao and coauthors correlate high serum HOTTIP expression levels with increased metastasis formation in HCC patients [550]. HOTTIP is also upregulated in human primary CRC tissues, where it promotes cell proliferation, migration, and invasion [546]. Several preclinical studies have shown that HOTTIP can be used as a possible marker for the early diagnosis of CRC [551], as a drug response prediction marker [552], as a predictive marker of risk, and as a prognostic marker [553].
9. The Role of Mitochondrial ncRNAs in Cancer
Despite its small genome size and limited protein production capacity, the mitochondrion has a very complex ncRNA profile. Indeed, like its nuclear counterpart, mtDNA encodes several ncRNA species (mt-ncRNAs), including piRNAs, miRNAs, and sncRNAs derived from lncRNAs or from the processing of tRNA and rRNA genes. Increasing evidence suggests that mt-ncRNAs play important roles in cellular homeostasis, and their alteration induces mitochondrial dysfunction associated with the development of several diseases, including cancer. Although the discovery of mt-ncRNAs is quite recent and their biological functions are poorly understood [554], the importance of some of them is emerging since they modulate processes such as cell division, apoptosis, cell proliferation, and metastasis formation [555].
9.1. SncmtRNA, ASncmtRNA-1, and ASncmtRNA-2
Three lncRNAs originate from the mitochondrial rRNA 16S (mt-RNR2) gene: a mt-ncRNA (SncmtRNA) and two antisense transcripts, ASncmtRNA-1 and ASncmtRNA-2. SncmtRNA is expressed in proliferating and normal tumor cells but not in non-proliferating control cells, suggesting a role in cell division. ASncmtRNA-1 and ASncmtRNA-2 are expressed in normal proliferating human cells, are downregulated in tumor tissue, and are not expressed in non-dividing cells [556,557]. The expression pattern of ASncmtRNAs suggests two important considerations: (I) they may act as tumor suppressors and be involved in malignant cell transformation processes; (II) the different expression profile observed allows us to specifically distinguish between normal and tumoral cells, and this aspect can be used in a therapeutic strategy.
9.2. LIPCAR
Long intergenic noncoding RNA predicting cardiac remodeling (LIPCAR) is a chimeric mitochondrial transcript in which the 5′ half maps to the antisense strand of the mitochondrial gene lncCyt b and the 3′ half maps to the antisense strand of the mitochondrial gene COX2. Although the literature on LIPCAR is mainly focused on its role in cardiac diseases, its involvement in cell proliferation and carcinogenesis has been recently explored in the process of phenotypic switching. This process allows cells to change their phenotype to a “highly synthetic phenotype”, which in turn permits cells to acquire various characteristics, such as proliferative and migratory abilities, increased protein synthesis, and the secretion of collagen, elastin, and matrix metalloproteinases [558]. Alterations in the switching process have been observed in cancer and influence malignant transformation by promoting invasion, metastasis, and tumor growth [559,560].
LIPCAR is upregulated in both HCC patients and the HCC HepG2 cell line. LIPCAR’s overexpression in HepG2 induces the inhibition of apoptosis and increases cell proliferation, migration, and invasiveness through increased levels of N-cadherin, Vimentin, and Claudin and decreased levels of E-cadherin. Furthermore, BALB/c nude mice injected with LIPCAR-overexpressing HepG2 cells formed HCC tumors and showed greater metastasis. In this system, LIPCAR overexpression promotes tumor growth and metastasis via the activation of the EMT process [561].
Finally, a positive correlation between LIPCAR and the TGF-β/Smad pathway in atrial muscle tissues was demonstrated [562]. This pathway is involved in cell growth, differentiation, motility, and apoptosis, and its alteration has been associated with both the development and progression of many tumors, as well as with drug resistance. Thus, it is plausible to speculate that the mechanisms of action that correlate LIPCAR with the TGF-β/Smad pathway may also function in carcinogenesis; however, further studies are needed to support this hypothesis.
9.3. lncCytB and mcPGK1
The mitochondrial antisense strand of the CytB gene transcribes two lncRNAs called lncCytB and mitochondrial circRNA for translocating phosphoglycerate kinase 1 (mcPGK1). Studies comparing the localization of lncCytB in HL7702 (normal hepatic cells) and in HepG2 (hepatoma cells) show that, in HL7702, lncCytB is primarily localized in the mitochondria, while, in HepG2, it is still localized in the mitochondrion but mainly in the nucleus. Although the nuclear function has not been fully clarified, the authors suggest the aberrant shuttling of lncCytB in the HepG2 nucleus, where it may function in the epigenetic regulation of genes involved in carcinogenesis [563]. Zhang et al., using both mouse models and human cell lines, such as AC16 (human cardiomyocytes), HL-1 (mouse cardiac muscle cells), and neonatal mouse cardiomyocytes (NMCM), suggest that lncCytB could function as a ceRNA in the lncCytB/miR-103-3p/PTEN/AKT axis. This mechanism is particularly interesting for several reasons: (I) the role of miR-103-3p in carcinogenesis has been described in various tumors [564,565]; (II) in NSCLC, the miR-103-3p/PTEN interaction regulates AKT-inducing cell proliferation and invasion [566]; (III) miR-103-3p sponging by lncRNAs is involved in the regulation of different hallmarks of cancer [567,568]. Although yet to be demonstrated, these considerations suggest that the lncCytB/miR-103-3p/PTEN/AKT axis may be involved in carcinogenesis.
In another report, Chen et al. separated liver tumor-initiating cells (TICs) from non-TICs from primary liver cancer and analyzed the circRNAs differentially expressed between the two cell types. Circular mcPGK1 was more enriched in TICs than in non-TICs, and subsequent investigations suggested its involvement in the molecular circuit, which inhibits OXPHOS and promotes the Warburg effect [569].
9.4. MDL1 and MDL1AS
The mitochondrial D-loop encodes two lncRNAs called mitochondrial D-loop 1 (MDL1), which originates from the sense strand, and mitochondrial D-loop 1 antisense (MDL1AS), which originates from the antisense strand.
An analysis of A549 LC cells reveals that MDL1 participates in retrograde signaling to mediate the regulatory control of nuclear genes involved in cell cycle regulation. In particular, MDL1 mediates the formation of a ternary complex, MDL1/p53/Tid1, that inhibits p53 nuclear translocation [570]. Protein p53 is a transcription factor with hundreds of targets; therefore, the inhibition of p53 nuclear translocation has a broad regulatory effect on the expression of nuclear genes and influences the expression of numerous genes involved in apoptosis, cell cycle arrest, autophagy, metabolism, DNA repair, and feedback mechanisms [571].
Garrido et al. analyzed the expression levels of MDL1 and MDL1AS in different tumor tissues (colon, rectum, breast, and larynx cancer) and compared them to those in normal surrounding tissues. The authors found no differences in the MDL1 levels between tumor and control samples, while the MDL1AS levels were variable and tumor-type-dependent. In fact, the MDL1AS levels were lower in CRC and higher in BrC and LarC. The authors explored the role of MDL1AS using ds interfering RNA (DsiRNA) to downregulate MDL1AS in colon (HCT-116) and breast (MCF7 and MDA-MB-231) cancer cell lines. The results showed that, in HCT-116 cells, the downregulation of MDL1AS reduced both cell growth and migration, while the opposite behavior was observed in MDA-MB-231 cells. A molecular analysis was subsequently performed using both markers of apoptosis (BAD, BAX, BCL2) and the cell cycle (CDK4, CDKN1A, CCNA1). The results showed that BCL2, CDK4, CDKN1A, and CCNA1 were downregulated in both cell lines upon MDL1AS downregulation, compared to control cells not treated with DsiRNA. In contrast, BAD and BAX showed downregulation only in HTC-116 and MDA-MB-231, respectively, when compared to control cells not treated with DsiRNA [572].
9.5. circ-COX2
circ-COX2 is a new circRNA generated by backsplicing from the transcription of the mitochondrial COX2 gene. An analysis of exosomes isolated from the plasma of patients with CLL showed the enrichment of circ-COX2 compared to normal controls, suggesting a role of circ-COX2 in CLL. A functional analysis performed on different CLL cell lines showed that the downregulation of circ-COX2 affects mitochondrial function and induces both the suppression of cell proliferation and increased apoptosis, while the upregulation of circ-COX2 is correlated with disease progression and reduced survival [573].
9.6. circ-ND5 and ND6
Liu and coworkers identified two circRNAs, circ-ND1 and circ-ND5, that were upregulated in HCC compared to controls, i.e., normal adjacent cancer tissues. circ-ND1 and circ-ND5 are transcribed from the mitochondrial ND1 and ND5 genes, respectively, and appear to be involved in facilitating mitochondrial protein import. However, their potential role in HCC pathogenesis needs further exploration [574].
9.7. Mitochondrial ncRNA-Based Therapies in CTs
Due to their central role in many cellular processes, mitochondria are suitable therapeutic targets for the treatment of various diseases, including cancer. Molecules designed to inhibit proteins/enzymes required for their function are often tested [575,576], as well as those that target mtDNA replication, which has a critical role in the formation of new mitochondria [577]. The molecules evaluated in most CTs target protein/enzyme components of mitochondria; however, to date, there are at least two CTs that have investigated mt-ncRNAs, namely ASncmtRNA-1 and ASncmtRNA-2, as therapeutic targets.
ASncmtRNAs were chosen in several preclinical studies that have highlighted two key factors for this purpose. First, the analysis of the expression pattern of ASncmtRNAs allows us to specifically distinguish between normal and tumoral cells. This implies that a therapeutic agent targeting ASncmtRNAs could preferentially reach abnormal cells, thus increasing the specificity of the treatment and decreasing its side effects. Second, several studies have used an ASO, namely Andes-1537, complementary to the loop region of both ASncmtRNAs. The use of this construct has contributed to understanding the biological roles of both ASncmtRNAs and, consequently, their potential as oncological therapeutic targets. Using cells derived from primary and metastatic ccRCC and an orthotopic xenograft model of ccRCC, both treated with Andes-1537, an increase in apoptosis and a decrease in proliferative capacity and metastasis formation were generally observed. At the molecular level, ASncmtRNAs knockdown induces a reduction in cellular proliferation through the reduction of cyclins B1 and D1 and the inhibition of metastasis formation through the reduction of N-cadherin [578]. The works of Fitzpatrick [579] and Bendek [580] show that the knockdown of ASncmtRNAs influences the function of some key factors required for cell cycle progression and ensures genomic integrity. In particular, they showed the downregulation of CDK1, CDK4, survivin, aurora kinase A (AURKA), and topoisomerase IIα (TOPO2A). In addition, they also demonstrated that Andes-1537 induced the upregulation of both mitochondrial encoded hsa-miR-4485-3p and hsa-miR-1973, together with nuclear hsa-miR-5096 and hsa-miR-3609. The miRNA hsa-miR-4485-3p induces the downregulation of cyclins B1 and D1, while hsa-miR-1973 seems to have no effect on the regulation of cyclins. Instead, an analysis of nuclear hsa-miR-5096 and hsa-miR-3609 using the TargetScanHuman prediction tool identified the canonical binding sites on the CDK1 mRNA [579].
Preclinical studies led to the design of a phase I CT (NCT02508441). This study was completed in 2018, with the aim to determine the safety, tolerability, and MTD of Andes-1537. Twenty-two patients with advanced unresectable solid tumors participated in this study. The results were encouraging because the authors not only demonstrated the safety of the drug and established the MTD (600 mg injected subcutaneously every two weeks), but also showed that the treatment with Andes-1537 stabilized the disease progression beyond 6 months for one patient with PaC and one patient with ChC [227]. Andes-1537 was also tested in an additional CT (NCT03985072), which started in 2019 (Table 2). In this CT, 67 patients were enrolled, with different advanced solid tumors, such as GBTC, CC, GC, PaC, and CRC. Although completed in 2022, the results of this trial have not yet been published.
10. Conclusions
In recent years, the interest in ncRNAs has grown significantly, and this has contributed to a better understanding of the complex cellular mechanisms in which they are involved and their importance in ensuring cellular homeostasis. In fact, in every analyzed tumor, it is possible to find an altered ncRNA expression pattern. The alteration of this pattern depends mainly on ncRNAs of nuclear origin, but, recently, a role has been highlighted also for ncRNAs of mitochondrial origin. In fact, despite the small size of the mitochondrial genome compared to its nuclear counterpart, this organelle can transcribe an intricate and varied network of ncRNAs. Although several aspects of mitochondrial function in cancer are still far from being elucidated, the study of mtDNA-encoded ncRNAs could further expand the list of possible anti-cancer targets. In this context, it will be interesting to continue the evaluation in CTs of Andes-1537, which is the only ASO directed against a mt-ncRNA.
The role that nuclear and mitochondrial ncRNAs play in oncogenesis and in the various aspects that lead to tumor progression and development suggests huge potential for the development of cancer therapies targeting ncRNAs, but these are currently only in the early development phase. There are still many challenges to overcome for their use in clinical practice. One of these concerns their functional characterization, which is very fragmented. To date, there is an evident imbalance between the number of various classes of ncRNAs annotated in their specific databases, all of which are constantly updated, and the number of ncRNAs analyzed at the bench. In addition, ncRNAs often have multiple cellular targets that include RNAs, DNA, and proteins, forming intricate interaction networks that are not known in detail. Even the genetic/molecular mechanisms that regulate ncRNAs’ biogenesis and cellular functions are often poorly understood. With the current knowledge, the use of ncRNAs as therapeutic targets could contribute to several side effects, such as off-target deregulation, the alteration of healthy cellular molecular pathways, toxicity, and adverse immune responses. In addition to the issues related to the functional knowledge of ncRNAs, there are further challenges to overcome, such as efficient and targeted delivery systems that, in addition to ensuring the stability, bioavailability, and correct dosage at the cellular level of the ncRNA, allow the delivery of the molecules specifically to tumor cells. Finally, the half-lives of these drugs and their vectors also need to be strongly regulated to avoid their depletion in target cells and ensure their correct excretion after exerting the desired effect. Although the therapeutic use of ncRNAs is at the early stages, these molecules are also being investigated as exciting biomarkers in different CTs for their diagnostic, prognostic, and predictive potential, as well as their therapeutic efficacy or for the monitoring of the pathology over time. These new biomarkers could improve the current oncological clinical protocols, with a significant effect in terms of improving patients’ prognosis, thanks, for example, to early diagnosis, as well as contributing to the choice of the best therapy to be performed. As such, in the near future, they will likely be included in panels for the molecular characterization of patients and their diseases. Overall, despite the challenges, ncRNAs are increasingly emerging as promising tools for a new era of precision and personalized medicine in cancer therapy.
Author Contributions
Conceptualization, R.P. and S.S.; writing—original draft preparation, R.P. and S.S.; writing—review and editing, R.P. and S.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer Statistics for the Year 2020: An Overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef]
- What Is Cancer. Available online: https://www.cancer.gov/about-cancer/understanding/what-is-cancer (accessed on 22 December 2024).
- Hatton, I.A.; Galbraith, E.D.; Merleau, N.S.C.; Miettinen, T.P.; Smith, B.M.; Shander, J.A. The Human Cell Count and Size Distribution. Proc. Natl. Acad. Sci. USA 2023, 120, e2303077120. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Lin, W.; Zhu, L. Targeted Drug Delivery for the Treatment of Blood Cancers. Molecules 2022, 27, 1310. [Google Scholar] [CrossRef] [PubMed]
- Sharma, I.; Son, M.J.; Motamedi, S.; Hoeft, A.; Teller, C.; Hamby, T.; Ray, A. Utilization of Genomic Tumor Profiling in Pediatric Liquid Tumors: A Clinical Series. Hematol. Rep. 2023, 15, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Siddika, A.; Chowdhury, S.; Hasan, M.R.; Moniruzzaman, M.; Been Sayeed, S.K.J.; Tabassum, T.; Chowduary, M.; Tabassum, T.; Islam, A.; Rahman, M.M. Clinicopathological Patterns of Malignant Solid Tumors in Adult Patients: A Hospital-Based Study From Bangladesh. Cureus 2023, 15, e34925. [Google Scholar] [CrossRef]
- Cancers by Body Location/System. Available online: http://www.cancer.gov/types/by-body-location (accessed on 22 December 2024).
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer Statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef]
- Ostroverkhova, D.; Przytycka, T.M.; Panchenko, A.R. Cancer Driver Mutations: Predictions and Reality. Trends Mol. Med. 2023, 29, 554–566. [Google Scholar] [CrossRef]
- Doane, A.S.; Elemento, O. Alterations in Transcriptional Networks in Cancer: The Role of Noncoding Somatic Driver Mutations. Curr. Opin. Genet. Dev. 2022, 75, 101919. [Google Scholar] [CrossRef]
- Rheinbay, E.; Nielsen, M.M.; Abascal, F.; Wala, J.A.; Shapira, O.; Tiao, G.; Hornshøj, H.; Hess, J.M.; Juul, R.I.; Lin, Z.; et al. Analyses of Non-Coding Somatic Drivers in 2,658 Cancer Whole Genomes. Nature 2020, 578, 102–111. [Google Scholar] [CrossRef]
- Juul, M.; Bertl, J.; Guo, Q.; Nielsen, M.M.; Świtnicki, M.; Hornshøj, H.; Madsen, T.; Hobolth, A.; Pedersen, J.S. Non-Coding Cancer Driver Candidates Identified with a Sample- and Position-Specific Model of the Somatic Mutation Rate. eLife 2017, 6, e21778. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Li, S.; Xu, B.; Luo, H. Cancer Evolution: A Means by Which Tumors Evade Treatment. Biomed. Pharmacother. 2021, 133, 111016. [Google Scholar] [CrossRef]
- Alison, M.R. The Cellular Origins of Cancer with Particular Reference to the Gastrointestinal Tract. Int. J. Exp. Pathol. 2020, 101, 132–151. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.S.; Amend, S.R.; Austin, R.H.; Gatenby, R.A.; Hammarlund, E.U.; Pienta, K.J. Updating the Definition of Cancer. Mol. Cancer Res. 2023, 21, 1142–1147. [Google Scholar] [CrossRef]
- Ciriello, G.; Magnani, L.; Aitken, S.J.; Akkari, L.; Behjati, S.; Hanahan, D.; Landau, D.A.; Lopez-Bigas, N.; Lupiáñez, D.G.; Marine, J.-C.; et al. Cancer Evolution: A Multifaceted Affair. Cancer Discov. 2024, 14, 36–48. [Google Scholar] [CrossRef] [PubMed]
- Stepankiw, N.; Yang, A.W.H.; Hughes, T.R. The Human Genome Contains over a Million Autonomous Exons. Genome Res. 2023, 33, 1865–1878. [Google Scholar] [CrossRef]
- Cannon, M.E.; Mohlke, K.L. Deciphering the Emerging Complexities of Molecular Mechanisms at GWAS Loci. Am. J. Hum. Genet. 2018, 103, 637–653. [Google Scholar] [CrossRef]
- Edwards, S.L.; Beesley, J.; French, J.D.; Dunning, A.M. Beyond GWASs: Illuminating the Dark Road from Association to Function. Am. J. Hum. Genet. 2013, 93, 779–797. [Google Scholar] [CrossRef]
- Tan, K.-T.; Slevin, M.K.; Leibowitz, M.L.; Garrity-Janger, M.; Shan, J.; Li, H.; Meyerson, M. Neotelomeres and Telomere-Spanning Chromosomal Arm Fusions in Cancer Genomes Revealed by Long-Read Sequencing. Cell Genom. 2024, 4, 100588. [Google Scholar] [CrossRef]
- Liehr, T. Repetitive Elements in Humans. Int. J. Mol. Sci. 2021, 22, 2072. [Google Scholar] [CrossRef]
- Cosby, R.L.; Judd, J.; Zhang, R.; Zhong, A.; Garry, N.; Pritham, E.J.; Feschotte, C. Recurrent Evolution of Vertebrate Transcription Factors by Transposase Capture. Science 2021, 371, eabc6405. [Google Scholar] [CrossRef]
- Liao, X.; Zhu, W.; Zhou, J.; Li, H.; Xu, X.; Zhang, B.; Gao, X. Repetitive DNA Sequence Detection and Its Role in the Human Genome. Commun. Biol. 2023, 6, 954. [Google Scholar] [CrossRef] [PubMed]
- Petri, R.; Brattås, P.L.; Sharma, Y.; Jönsson, M.E.; Pircs, K.; Bengzon, J.; Jakobsson, J. LINE-2 Transposable Elements Are a Source of Functional Human MicroRNAs and Target Sites. PLoS Genet. 2019, 15, e1008036. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Ben, S.; Xin, J.; Li, S.; Zheng, R.; Wang, H.; Fan, L.; Du, M.; Zhang, Z.; Wang, M. The Biogenesis and Biological Function of PIWI-Interacting RNA in Cancer. J. Hematol. Oncol. 2021, 14, 93. [Google Scholar] [CrossRef]
- Clark, C.G.; Cross, G.A.M. Circular Ribosomal RNA Genes Are a General Feature of Schizopyrenid Amoebae. J. Protozool. 1988, 35, 326–329. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.A.; Giusto, J.D.; Bao, A.; Hopper, A.K.; Matera, A.G. Molecular Determinants of Metazoan TricRNA Biogenesis. Nucleic Acids Res. 2019, 47, 6452–6465. [Google Scholar] [CrossRef]
- Tang, X.; Ren, H.; Guo, M.; Qian, J.; Yang, Y.; Gu, C. Review on Circular RNAs and New Insights into Their Roles in Cancer. Comput. Struct. Biotechnol. J. 2021, 19, 910–928. [Google Scholar] [CrossRef]
- Zhao, Q.; Liu, J.; Deng, H.; Ma, R.; Liao, J.-Y.; Liang, H.; Hu, J.; Li, J.; Guo, Z.; Cai, J.; et al. Targeting Mitochondria-Located CircRNA SCAR Alleviates NASH via Reducing MROS Output. Cell 2020, 183, 76–93.e22. [Google Scholar] [CrossRef]
- Yang, L.; Wilusz, J.E.; Chen, L.-L. Biogenesis and Regulatory Roles of Circular RNAs. Annu. Rev. Cell Dev. Biol. 2022, 38, 263–289. [Google Scholar] [CrossRef]
- Karpagavalli, M.; Sivagurunathan, S.; Panda, T.S.; Srikakulam, N.; Arora, R.; Dohadwala, L.; Tiwary, B.K.; Sadras, S.R.; Arunachalam, J.P.; Pandi, G.; et al. PiRNAs in the Human Retina and Retinal Pigment Epithelium Reveal a Potential Role in Intracellular Trafficking and Oxidative Stress. Mol. Omics 2024, 20, 248–264. [Google Scholar] [CrossRef]
- Malebary, S.J.; Khan, Y.D. Evaluating Machine Learning Methodologies for Identification of Cancer Driver Genes. Sci. Rep. 2021, 11, 12281. [Google Scholar] [CrossRef] [PubMed]
- Nourbakhsh, M.; Saksager, A.; Tom, N.; Chen, X.S.; Colaprico, A.; Olsen, C.; Tiberti, M.; Papaleo, E. A Workflow to Study Mechanistic Indicators for Driver Gene Prediction with Moonlight. Brief. Bioinform. 2023, 24, bbad274. [Google Scholar] [CrossRef] [PubMed]
- Amaral, P.; Carbonell-Sala, S.; De La Vega, F.M.; Faial, T.; Frankish, A.; Gingeras, T.; Guigo, R.; Harrow, J.L.; Hatzigeorgiou, A.G.; Johnson, R.; et al. The Status of the Human Gene Catalogue. Nature 2023, 622, 41–47. [Google Scholar] [CrossRef]
- Nurk, S.; Koren, S.; Rhie, A.; Rautiainen, M.; Bzikadze, A.V.; Mikheenko, A.; Vollger, M.R.; Altemose, N.; Uralsky, L.; Gershman, A.; et al. The Complete Sequence of a Human Genome. Science 2022, 376, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Shen, X. Noncoding RNA-Chromatin Association: Functions and Mechanisms. Fundam. Res. 2023, 3, 665–675. [Google Scholar] [CrossRef]
- Yuan, W.; Zhang, R.; Lyu, H.; Xiao, S.; Guo, D.; Zhang, Q.; Ali, D.W.; Michalak, M.; Chen, X.-Z.; Zhou, C.; et al. Dysregulation of TRNA Methylation in Cancer: Mechanisms and Targeting Therapeutic Strategies. Cell Death Discov. 2024, 10, 327. [Google Scholar] [CrossRef]
- Babaian, A.; Rothe, K.; Girodat, D.; Minia, I.; Djondovic, S.; Milek, M.; Spencer Miko, S.E.; Wieden, H.-J.; Landthaler, M.; Morin, G.B.; et al. Loss of M1acp3Ψ Ribosomal RNA Modification Is a Major Feature of Cancer. Cell Rep. 2020, 31, 107611. [Google Scholar] [CrossRef]
- Kang, J.; Brajanovski, N.; Chan, K.T.; Xuan, J.; Pearson, R.B.; Sanij, E. Ribosomal Proteins and Human Diseases: Molecular Mechanisms and Targeted Therapy. Signal Transduct. Target. Ther. 2021, 6, 323. [Google Scholar] [CrossRef]
- Tao, Y.; Zhang, Q.; Wang, H.; Yang, X.; Mu, H. Alternative Splicing and Related RNA Binding Proteins in Human Health and Disease. Signal Transduct. Target. Ther. 2024, 9, 26. [Google Scholar] [CrossRef]
- Huang, Z.-H.; Du, Y.-P.; Wen, J.-T.; Lu, B.-F.; Zhao, Y. SnoRNAs: Functions and Mechanisms in Biological Processes, and Roles in Tumor Pathophysiology. Cell Death Discov. 2022, 8, 259. [Google Scholar] [CrossRef]
- Chauhan, W.; Sudharshan, S.J.; Kafle, S.; Zennadi, R. SnoRNAs: Exploring Their Implication in Human Diseases. Int. J. Mol. Sci. 2024, 25, 7202. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Sun, Y.-M.; Pan, Q.; Fang, K.; Chen, X.-T.; Zeng, Z.-C.; Chen, T.-Q.; Zhu, S.-X.; Huang, L.-B.; Luo, X.-Q.; et al. The SnoRNA-like LncRNA LNC-SNO49AB Drives Leukemia by Activating the RNA-Editing Enzyme ADAR1. Cell Discov. 2022, 8, 117. [Google Scholar] [CrossRef] [PubMed]
- Ender, C.; Krek, A.; Friedländer, M.R.; Beitzinger, M.; Weinmann, L.; Chen, W.; Pfeffer, S.; Rajewsky, N.; Meister, G. A Human SnoRNA with MicroRNA-Like Functions. Mol. Cell 2008, 32, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Ono, M.; Yamada, K.; Avolio, F.; Scott, M.S.; van Koningsbruggen, S.; Barton, G.J.; Lamond, A.I. Analysis of Human Small Nucleolar RNAs (SnoRNA) and the Development of SnoRNA Modulator of Gene Expression Vectors. Mol. Biol. Cell 2010, 21, 1569–1584. [Google Scholar] [CrossRef]
- Fan, Z.; Chen, Y.; Yan, D.; Li, Q. Effects of Differentially Methylated CpG Sites in Enhancer and Promoter Regions on the Chromatin Structures of Target LncRNAs in Breast Cancer. Int. J. Mol. Sci. 2024, 25, 11048. [Google Scholar] [CrossRef]
- Glaich, O.; Parikh, S.; Bell, R.E.; Mekahel, K.; Donyo, M.; Leader, Y.; Shayevitch, R.; Sheinboim, D.; Yannai, S.; Hollander, D.; et al. DNA Methylation Directs MicroRNA Biogenesis in Mammalian Cells. Nat. Commun. 2019, 10, 5657. [Google Scholar] [CrossRef]
- Saviana, M.; Le, P.; Micalo, L.; Del Valle-Morales, D.; Romano, G.; Acunzo, M.; Li, H.; Nana-Sinkam, P. Crosstalk between MiRNAs and DNA Methylation in Cancer. Genes 2023, 14, 1075. [Google Scholar] [CrossRef]
- Tuna, M.; Machado, A.S.; Calin, G.A. Genetic and Epigenetic Alterations of Micro RNAs and Implications for Human Cancers and Other Diseases. Genes Chromosomes Cancer 2016, 55, 193–214. [Google Scholar] [CrossRef]
- Saito, Y.; Liang, G.; Egger, G.; Friedman, J.M.; Chuang, J.C.; Coetzee, G.A.; Jones, P.A. Specific Activation of MicroRNA-127 with Downregulation of the Proto-Oncogene BCL6 by Chromatin-Modifying Drugs in Human Cancer Cells. Cancer Cell 2006, 9, 435–443. [Google Scholar] [CrossRef]
- Vijay, A.; Jha, P.K.; Garg, I.; Sharma, M.; Ashraf, M.Z.; Kumar, B. Micro-RNAs Dependent Regulation of DNMT and HIF1α Gene Expression in Thrombotic Disorders. Sci. Rep. 2019, 9, 4815. [Google Scholar] [CrossRef]
- Wu, H.; Zhang, W.; Wu, Z.; Liu, Y.; Shi, Y.; Gong, J.; Shen, W.; Liu, C. MiR-29c-3p Regulates DNMT3B and LATS1 Methylation to Inhibit Tumor Progression in Hepatocellular Carcinoma. Cell Death Dis. 2019, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Xu, X.; Wang, Z.; Wang, F.; Wu, W.; Geng, J.; Liu, X. Methyl-CpG Binding Domain Protein 2 Inhibits the Malignant Characteristic of Lung Adenocarcinoma through the Epigenetic Modulation of 10 to 11 Translocation 1 and MiR-200s. Am. J. Pathol. 2019, 189, 1065–1076. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Wu, J.; Zhang, Y.; Song, J.; Shi, Z.; Chang, H. LINC00518 Promotes Cell Proliferation by Regulating the Cell Cycle of Lung Adenocarcinoma Through MiR-185-3p Targeting MECP2. Front. Oncol. 2021, 11, 646559. [Google Scholar] [CrossRef]
- Chaput, G.; Sumar, N. Endocrine Therapies for Breast and Prostate Cancers. Can. Fam. Physician 2022, 68, 271–276. [Google Scholar] [CrossRef]
- Debela, D.T.; Muzazu, S.G.; Heraro, K.D.; Ndalama, M.T.; Mesele, B.W.; Haile, D.C.; Kitui, S.K.; Manyazewal, T. New Approaches and Procedures for Cancer Treatment: Current Perspectives. SAGE Open Med. 2021, 9, 20503121211034366. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; MacRae, I.J. Regulation of MicroRNA Function in Animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef]
- Wen, K.; Chen, X.; Gu, J.; Chen, Z.; Wang, Z. Beyond Traditional Translation: NcRNA Derived Peptides as Modulators of Tumor Behaviors. J. Biomed. Sci. 2024, 31, 63. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, J.; Zhao, Y.; Wang, H.; Liu, T.; Li, Y.; Cui, T.; Li, W.; Feng, Y.; Luo, J.; et al. CncRNAdb: A Manually Curated Resource of Experimentally Supported RNAs with Both Protein-Coding and Noncoding Function. Nucleic Acids Res. 2021, 49, D65–D70. [Google Scholar] [CrossRef]
- Postic, G.; Tav, C.; Platon, L.; Zehraoui, F.; Tahi, F. IRSOM2: A Web Server for Predicting Bifunctional RNAs. Nucleic Acids Res. 2023, 51, W281–W288. [Google Scholar] [CrossRef]
- Cao, Y.; Liu, H.; Lu, S.S.; Jones, K.A.; Govind, A.P.; Jeyifous, O.; Simmons, C.Q.; Tabatabaei, N.; Green, W.N.; Holder, J.L.; et al. RNA-Based Translation Activators for Targeted Gene Upregulation. Nat. Commun. 2023, 14, 6827. [Google Scholar] [CrossRef]
- Dragomir, M.P.; Manyam, G.C.; Ott, L.F.; Berland, L.; Knutsen, E.; Ivan, C.; Lipovich, L.; Broom, B.M.; Calin, G.A. FuncPEP: A Database of Functional Peptides Encoded by Non-Coding RNAs. Noncoding RNA 2020, 6, 41. [Google Scholar] [CrossRef] [PubMed]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long Non-Coding RNAs: Definitions, Functions, Challenges and Recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Bhat, A.A.; Younes, S.N.; Raza, S.S.; Zarif, L.; Nisar, S.; Ahmed, I.; Mir, R.; Kumar, S.; Sharawat, S.K.; Hashem, S.; et al. Role of Non-Coding RNA Networks in Leukemia Progression, Metastasis and Drug Resistance. Mol. Cancer 2020, 19, 57. [Google Scholar] [CrossRef]
- Srijyothi, L.; Ponne, S.; Prathama, T.; Ashok, C.; Baluchamy, S. Roles of Non-Coding RNAs in Transcriptional Regulation. In Transcriptional and Post-Transcriptional Regulation; InTech: Rijeka, Croatia, 2018. [Google Scholar]
- Bure, I.V.; Nemtsova, M. V Mutual Regulation of NcRNAs and Chromatin Remodeling Complexes in Normal and Pathological Conditions. Int. J. Mol. Sci. 2023, 24, 7848. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The Noncoding RNA Revolution—Trashing Old Rules to Forge New Ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Kaneda, H.; Hayashi, J.; Takahama, S.; Taya, C.; Lindahl, K.F.; Yonekawa, H. Elimination of Paternal Mitochondrial DNA in Intraspecific Crosses during Early Mouse Embryogenesis. Proc. Natl. Acad. Sci. USA 1995, 92, 4542–4546. [Google Scholar] [CrossRef]
- Sutovsky, P.; Moreno, R.D.; Ramalho-Santos, J.; Dominko, T.; Simerly, C.; Schatten, G. Ubiquitin Tag for Sperm Mitochondria. Nature 1999, 402, 371–372. [Google Scholar] [CrossRef]
- Zhang, Y.; Qu, Y.; Gao, K.; Yang, Q.; Shi, B.; Hou, P.; Ji, M. High Copy Number of Mitochondrial DNA (MtDNA) Predicts Good Prognosis in Glioma Patients. Am. J. Cancer Res. 2015, 5, 1207–1216. [Google Scholar]
- Mortimer, D. The Functional Anatomy of the Human Spermatozoon: Relating Ultrastructure and Function. MHR Basic Sci. Reprod. Med. 2018, 24, 567–592. [Google Scholar] [CrossRef]
- Ankel-Simons, F.; Cummins, J.M. Misconceptions about Mitochondria and Mammalian Fertilization: Implications for Theories on Human Evolution. Proc. Natl. Acad. Sci. USA 1996, 93, 13859–13863. [Google Scholar] [CrossRef]
- Cummins, J.M. Epigenetic and Experimental Modifications in Early Mammalian Development: Part I: Mitochondria: Potential Roles in Embryogenesis and Nucleocytoplasmic Transfer. Hum. Reprod. Update 2001, 7, 217–228. [Google Scholar] [CrossRef][Green Version]
- Degli Esposti, D.; Hamelin, J.; Bosselut, N.; Saffroy, R.; Sebagh, M.; Pommier, A.; Martel, C.; Lemoine, A. Mitochondrial Roles and Cytoprotection in Chronic Liver Injury. Biochem. Res. Int. 2012, 2012, 1–16. [Google Scholar] [CrossRef]
- D’Erchia, A.M.; Atlante, A.; Gadaleta, G.; Pavesi, G.; Chiara, M.; De Virgilio, C.; Manzari, C.; Mastropasqua, F.; Prazzoli, G.M.; Picardi, E.; et al. Tissue-Specific MtDNA Abundance from Exome Data and Its Correlation with Mitochondrial Transcription, Mass and Respiratory Activity. Mitochondrion 2015, 20, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Moran, J.C.; Brivanlou, A.; Brischigliaro, M.; Fontanesi, F.; Rouskin, S.; Barrientos, A. The Human Mitochondrial MRNA Structurome Reveals Mechanisms of Gene Expression. Science 2024, 385, eadm9238. [Google Scholar] [CrossRef] [PubMed]
- Vučković, A.; Freyer, C.; Wredenberg, A.; Hillen, H.S. The Molecular Machinery for Maturation of Primary MtDNA Transcripts. Hum. Mol. Genet. 2024, 33, R19–R25. [Google Scholar] [CrossRef]
- Guo, B.; Zhai, D.; Cabezas, E.; Welsh, K.; Nouraini, S.; Satterthwait, A.C.; Reed, J.C. Humanin Peptide Suppresses Apoptosis by Interfering with Bax Activation. Nature 2003, 423, 456–461. [Google Scholar] [CrossRef]
- Ha, C.P.; Hua, T.N.M.; Vo, V.T.A.; Om, J.; Han, S.; Cha, S.-K.; Park, K.-S.; Jeong, Y. Humanin Activates Integrin AV–TGFβ Axis and Leads to Glioblastoma Progression. Cell Death Dis. 2024, 15, 464. [Google Scholar] [CrossRef]
- Peña Agudelo, J.A.; Pidre, M.L.; Garcia Fallit, M.; Pérez Küper, M.; Zuccato, C.; Nicola Candia, A.J.; Marchesini, A.; Vera, M.B.; De Simone, E.; Giampaoli, C.; et al. Mitochondrial Peptide Humanin Facilitates Chemoresistance in Glioblastoma Cells. Cancers 2023, 15, 4061. [Google Scholar] [CrossRef]
- Cheng, J.; Li, M.; Motta, E.; Barci, D.; Song, W.; Zhou, D.; Li, G.; Zhu, S.; Yang, A.; Vaillant, B.D.; et al. Myeloid Cells Coordinately Induce Glioma Cell-Intrinsic and Cell-Extrinsic Pathways for Chemoresistance via GP130 Signaling. Cell Rep. Med. 2024, 5, 101658. [Google Scholar] [CrossRef]
- Yuan, Y.; Ju, Y.S.; Kim, Y.; Li, J.; Wang, Y.; Yoon, C.J.; Yang, Y.; Martincorena, I.; Creighton, C.J.; Weinstein, J.N.; et al. Comprehensive Molecular Characterization of Mitochondrial Genomes in Human Cancers. Nat. Genet. 2020, 52, 342. [Google Scholar] [CrossRef]
- Kim, M.; Mahmood, M.; Reznik, E.; Gammage, P.A. Mitochondrial DNA Is a Major Source of Driver Mutations in Cancer. Trends Cancer 2022, 8, 1046–1059. [Google Scholar] [CrossRef]
- Gorelick, A.N.; Kim, M.; Chatila, W.K.; La, K.; Hakimi, A.A.; Berger, M.F.; Taylor, B.S.; Gammage, P.A.; Reznik, E. Respiratory Complex and Tissue Lineage Drive Recurrent Mutations in Tumour MtDNA. Nat. Metab. 2021, 3, 558–570. [Google Scholar] [CrossRef] [PubMed]
- Triska, P.; Kaneva, K.; Merkurjev, D.; Sohail, N.; Falk, M.J.; Triche, T.J.; Biegel, J.A.; Gai, X. Landscape of Germline and Somatic Mitochondrial DNA Mutations in Pediatric Malignancies. Cancer Res. 2019, 79, 1318–1330. [Google Scholar] [CrossRef]
- Wei, W.; Tuna, S.; Keogh, M.J.; Smith, K.R.; Aitman, T.J.; Beales, P.L.; Bennett, D.L.; Gale, D.P.; Bitner-Glindzicz, M.A.K.; Black, G.C.; et al. Germline Selection Shapes Human Mitochondrial DNA Diversity. Science 2019, 364, eaau6520. [Google Scholar] [CrossRef] [PubMed]
- Jónsson, H.; Sulem, P.; Kehr, B.; Kristmundsdottir, S.; Zink, F.; Hjartarson, E.; Hardarson, M.T.; Hjorleifsson, K.E.; Eggertsson, H.P.; Gudjonsson, S.A.; et al. Parental Influence on Human Germline de Novo Mutations in 1,548 Trios from Iceland. Nature 2017, 549, 519–522. [Google Scholar] [CrossRef] [PubMed]
- Whitehall, J.C.; Smith, A.L.M.; Greaves, L.C. Mitochondrial DNA Mutations and Ageing. In Biochemistry and Cell Biology of Ageing: Part III Biomedical Science; Springer International Publishing: Cham, Switzerland, 2023; pp. 77–98. [Google Scholar]
- Sprason, C.; Tucker, T.; Clancy, D. MtDNA Deletions and Aging. Front. Aging 2024, 5. [Google Scholar] [CrossRef]
- Krasich, R.; Copeland, W.C. DNA Polymerases in the Mitochondria: A Critical Review of the Evidence. Front. Biosci. (Landmark Ed.) 2017, 22, 692–709. [Google Scholar] [CrossRef]
- Saneto, R.P.; Naviaux, R.K. Polymerase Gamma Disease through the Ages. Dev. Disabil. Res. Rev. 2010, 16, 163–174. [Google Scholar] [CrossRef]
- Wisnovsky, S.; Jean, S.R.; Liyanage, S.; Schimmer, A.; Kelley, S.O. Mitochondrial DNA Repair and Replication Proteins Revealed by Targeted Chemical Probes. Nat. Chem. Biol. 2016, 12, 567–573. [Google Scholar] [CrossRef]
- Arana, M.E.; Seki, M.; Wood, R.D.; Rogozin, I.B.; Kunkel, T.A. Low-Fidelity DNA Synthesis by Human DNA Polymerase Theta. Nucleic Acids Res. 2008, 36, 3847–3856. [Google Scholar] [CrossRef] [PubMed]
- Rong, Z.; Tu, P.; Xu, P.; Sun, Y.; Yu, F.; Tu, N.; Guo, L.; Yang, Y. The Mitochondrial Response to DNA Damage. Front. Cell Dev. Biol. 2021, 9, 669379. [Google Scholar] [CrossRef]
- Yin, P.-H.; Wu, C.-C.; Lin, J.-C.; Chi, C.-W.; Wei, Y.-H.; Lee, H.-C. Somatic Mutations of Mitochondrial Genome in Hepatocellular Carcinoma. Mitochondrion 2010, 10, 174–182. [Google Scholar] [CrossRef]
- Pinheiro, M.; Veiga, I.; Pinto, C.; Afonso, L.; Sousa, O.; Fragoso, M.; Santos, L.; Lopes, P.; Pais, I.; Lopes, C.; et al. Mitochondrial Genome Alterations in Rectal and Sigmoid Carcinomas. Cancer Lett. 2009, 280, 38–43. [Google Scholar] [CrossRef]
- Weerts, M.J.A.; Timmermans, E.C.; van de Stolpe, A.; Vossen, R.H.A.M.; Anvar, S.Y.; Foekens, J.A.; Sleijfer, S.; Martens, J.W.M. Tumor-Specific Mitochondrial DNA Variants Are Rarely Detected in Cell-Free DNA. Neoplasia 2018, 20, 687–696. [Google Scholar] [CrossRef]
- Koshikawa, N.; Akimoto, M.; Hayashi, J.-I.; Nagase, H.; Takenaga, K. Association of Predicted Pathogenic Mutations in Mitochondrial ND Genes with Distant Metastasis in NSCLC and Colon Cancer. Sci. Rep. 2017, 7, 15535. [Google Scholar] [CrossRef]
- Yusnita, Y.; Norsiah, M.D.; Rahman, A.J. Mutations in Mitochondrial NADH Dehydrogenase Subunit 1 (MtND1) Gene in Colorectal Carcinoma. Malays. J. Pathol. 2010, 32, 103–110. [Google Scholar]
- Kassem, A.M.; El-Guendy, N.; Tantawy, M.; Abdelhady, H.; El-Ghor, A.; Abdel Wahab, A.H. Mutational Hotspots in the Mitochondrial D-Loop Region of Cancerous and Precancerous Colorectal Lesions in Egyptian Patients. DNA Cell Biol. 2011, 30, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Akouchekian, M.; Houshmand, M.; Akbari, M.H.H.; Kamalidehghan, B.; Dehghan, M. Analysis of Mitochondrial ND1 Gene in Human Colorectal Cancer. J. Res. Med. Sci. 2011, 16, 50–55. [Google Scholar]
- Sun, W.; Zhou, S.; Chang, S.S.; McFate, T.; Verma, A.; Califano, J.A. Mitochondrial Mutations Contribute to HIF1α Accumulation via Increased Reactive Oxygen Species and Up-Regulated Pyruvate Dehydrogenease Kinase 2 in Head and Neck Squamous Cell Carcinoma. Clin. Cancer Res. 2009, 15, 476–484. [Google Scholar] [CrossRef]
- Yuan, Y.; Wang, W.; Li, H.; Yu, Y.; Tao, J.; Huang, S.; Zeng, Z. Nonsense and Missense Mutation of Mitochondrial ND6 Gene Promotes Cell Migration and Invasion in Human Lung Adenocarcinoma. BMC Cancer 2015, 15, 346. [Google Scholar] [CrossRef]
- Beadnell, T.C.; Scheid, A.D.; Vivian, C.J.; Welch, D.R. Roles of the Mitochondrial Genetics in Cancer Metastasis: Not to Be Ignored Any Longer. Cancer Metastasis Rev. 2018, 37, 615–632. [Google Scholar] [CrossRef]
- Guo, Z.; Jin, C.; Yao, Z.; Wang, Y.; Xu, B. Analysis of the Mitochondrial 4977 Bp Deletion in Patients with Hepatocellular Carcinoma. Balk. J. Med. Genet. 2017, 20, 81–86. [Google Scholar] [CrossRef]
- Shao, J.-Y. Quantitative Detection of Common Deletion of Mitochondrial DNA in Hepatocellular Carcinoma and Hepatocellular Nodular Hyperplasia. World J. Gastroenterol. 2004, 10, 1560. [Google Scholar] [CrossRef] [PubMed]
- Dimberg, J.; Hong, T.T.; Skarstedt, M.; Löfgren, S.; Zar, N.; Matussek, A. Novel and Differential Accumulation of Mitochondrial DNA Deletions in Swedish and Vietnamese Patients with Colorectal Cancer. Anticancer Res. 2014, 34, 147–152. [Google Scholar]
- Shen, L.; Fang, H.; Chen, T.; He, J.; Zhang, M.; Wei, X.; Xin, Y.; Jiang, Y.; Ding, Z.; Ji, J.; et al. Evaluating Mitochondrial DNA in Cancer Occurrence and Development. Ann. N. Y. Acad. Sci. 2010, 1201, 26–33. [Google Scholar] [CrossRef]
- Mohamed Yusoff, A.A.; Mohd Khair, S.Z.N.; Abd Radzak, S.M.; Idris, Z.; Lee, H.-C. Prevalence of Mitochondrial DNA Common Deletion in Patients with Gliomas and Meningiomas: A First Report from a Malaysian Study Group. J. Chin. Med. Assoc. 2020, 83, 838–844. [Google Scholar] [CrossRef]
- Mennuni, M.; Wilkie, S.E.; Michon, P.; Alsina, D.; Filograna, R.; Lindberg, M.; Sanin, D.E.; Rosenberger, F.; Schaaf, A.; Larsson, E.; et al. High Mitochondrial DNA Levels Accelerate Lung Adenocarcinoma Progression. Sci. Adv. 2024, 10, adp3481. [Google Scholar] [CrossRef]
- Al-awadhi, R.; Alroomy, M.; Al-Waheeb, S.; Alwehaidah, M.S. Altered Mitochondrial DNA Copy Number in Cervical Exfoliated Cells among High-risk HPV-positive and HPV-negative Women. Exp. Ther. Med. 2023, 26, 521. [Google Scholar] [CrossRef]
- Alwehaidah, M.S.; Al-Awadhi, R.; Roomy, M.A.; Baqer, T. Al Mitochondrial DNA Copy Number and Risk of Papillary Thyroid Carcinoma. BMC Endocr. Disord. 2024, 24, 138. [Google Scholar] [CrossRef]
- Zhuang, F.; Huang, S.; Liu, L. PYCR3 Modulates MtDNA Copy Number to Drive Proliferation and Doxorubicin Resistance in Triple-Negative Breast Cancer. Int. J. Biochem. Cell Biol. 2024, 171, 106581. [Google Scholar] [CrossRef] [PubMed]
- Weerts, M.J.A.; Sieuwerts, A.M.; Smid, M.; Look, M.P.; Foekens, J.A.; Sleijfer, S.; Martens, J.W.M. Mitochondrial DNA Content in Breast Cancer: Impact on in Vitro and in Vivo Phenotype and Patient Prognosis. Oncotarget 2016, 7, 29166–29176. [Google Scholar] [CrossRef]
- Yu, M.; Zhou, Y.; Shi, Y.; Ning, L.; Yang, Y.; Wei, X.; Zhang, N.; Hao, X.; Niu, R. Reduced Mitochondrial DNA Copy Number Is Correlated with Tumor Progression and Prognosis in Chinese Breast Cancer Patients. IUBMB Life 2007, 59, 450–457. [Google Scholar] [CrossRef]
- Yu, M.; Wan, Y.; Zou, Q. Decreased Copy Number of Mitochondrial DNA in Ewing’s Sarcoma. Clin. Chim. Acta 2010, 411, 679–683. [Google Scholar] [CrossRef]
- Reznik, E.; Miller, M.L.; Şenbabaoğlu, Y.; Riaz, N.; Sarungbam, J.; Tickoo, S.K.; Al-Ahmadie, H.A.; Lee, W.; Seshan, V.E.; Hakimi, A.A.; et al. Mitochondrial DNA Copy Number Variation across Human Cancers. eLife 2016, 5, e10769. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Nomoto, S.; Fujii, T.; Kaneko, T.; Takeda, S.; Inoue, S.; Kanazumi, N.; Nakao, A. Correlation between Copy Number of Mitochondrial DNA and Clinico-pathologic Parameters of Hepatocellular Carcinoma. Eur. J. Surg. Oncol. (EJSO) 2006, 32, 303–307. [Google Scholar] [CrossRef]
- Cui, H.; Huang, P.; Wang, Z.; Zhang, Y.; Zhang, Z.; Xu, W.; Wang, X.; Han, Y.; Guo, X. Association of Decreased Mitochondrial DNA Content with the Progression of Colorectal Cancer. BMC Cancer 2013, 13, 110. [Google Scholar] [CrossRef]
- Wang, Y.; He, S.; Zhu, X.; Qiao, W.; Zhang, J. High Copy Number of Mitochondrial DNA Predicts Poor Prognosis in Patients with Advanced Stage Colon Cancer. Int. J. Biol. Markers 2016, 31, 382–388. [Google Scholar] [CrossRef]
- Harutyunyan, T. The Known Unknowns of Mitochondrial Carcinogenesis: De Novo NUMTs and Intercellular Mitochondrial Transfer. Mutagenesis 2024, 39, 1–12. [Google Scholar] [CrossRef]
- Singh, K.K.; Choudhury, A.R.; Tiwari, H.K. Numtogenesis as a Mechanism for Development of Cancer. Semin. Cancer Biol. 2017, 47, 101–109. [Google Scholar] [CrossRef]
- Farberov, L.; Weissglas-Volkov, D.; Shapira, G.; Zoabi, Y.; Schiff, C.; Kloeckener-Gruissem, B.; Neidhardt, J.; Shomron, N. MRNA Splicing Is Modulated by Intronic MicroRNAs. iScience 2023, 26, 107723. [Google Scholar] [CrossRef]
- Melamed, Z.; Levy, A.; Ashwal-Fluss, R.; Lev-Maor, G.; Mekahel, K.; Atias, N.; Gilad, S.; Sharan, R.; Levy, C.; Kadener, S.; et al. Alternative Splicing Regulates Biogenesis of MiRNAs Located across Exon-Intron Junctions. Mol. Cell 2013, 50, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Pianigiani, G.; Licastro, D.; Fortugno, P.; Castiglia, D.; Petrovic, I.; Pagani, F. Microprocessor-Dependent Processing of Splice Site Overlapping MicroRNA Exons Does Not Result in Changes in Alternative Splicing. RNA 2018, 24, 1158–1171. [Google Scholar] [CrossRef] [PubMed]
- Mercuri, R.L.V.; Conceição, H.B.; Guardia, G.D.A.; Goldstein, G.; Vibranovski, M.D.; Hinske, L.C.; Galante, P.A.F. Retro-MiRs: Novel and Functional MiRNAs Originating from MRNA Retrotransposition. Mob. DNA 2023, 14, 12. [Google Scholar] [CrossRef]
- Downie Ruiz Velasco, A.; Parsons, A.L.; Heatley, M.C.; Martin, A.R.G.; Smart, A.D.; Shah, N.; Jopling, C.L. MicroRNA Biogenesis Is Broadly Disrupted by Inhibition of the Splicing Factor SF3B1. Nucleic Acids Res. 2024, 52, 9210–9229. [Google Scholar] [CrossRef]
- Sundaram, G.M.; Common, J.E.A.; Gopal, F.E.; Srikanta, S.; Lakshman, K.; Lunny, D.P.; Lim, T.C.; Tanavde, V.; Lane, E.B.; Sampath, P. ‘See-Saw’ Expression of MicroRNA-198 and FSTL1 from a Single Transcript in Wound Healing. Nature 2013, 495, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-M.; Jin, S.W.; Jang, B.; Ko, Y.K.; Gim, J.-A. Transposable Elements-Derived MicroRNA Expression Patterns in TCGA Dataset for 10 Species. Evol. Bioinform. 2023, 19, 11769343231194020. [Google Scholar] [CrossRef]
- Mustafin, R.N.; Khusnutdinova, E. Perspective for Studying the Relationship of MiRNAs with Transposable Elements. Curr. Issues Mol. Biol. 2023, 45, 3122–3145. [Google Scholar] [CrossRef]
- Szelągowski, A.; Kozakiewicz, M. A Glance at Biogenesis and Functionality of MicroRNAs and Their Role in the Neuropathogenesis of Parkinson’s Disease. Oxid. Med. Cell. Longev. 2023, 2023, 7759053. [Google Scholar] [CrossRef]
- Partin, A.C.; Zhang, K.; Jeong, B.-C.; Herrell, E.; Li, S.; Chiu, W.; Nam, Y. Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA. Mol. Cell 2020, 78, 411–422.e4. [Google Scholar] [CrossRef]
- Nguyen, T.A.; Jo, M.H.; Choi, Y.-G.; Park, J.; Kwon, S.C.; Hohng, S.; Kim, V.N.; Woo, J.-S. Functional Anatomy of the Human Microprocessor. Cell 2015, 161, 1374–1387. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.C.; Nguyen, T.A.; Choi, Y.-G.; Jo, M.H.; Hohng, S.; Kim, V.N.; Woo, J.-S. Structure of Human DROSHA. Cell 2016, 164, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.I.; Katsura, A.; Yasuda, T.; Ueno, T.; Mano, H.; Sugimoto, K.; Miyazono, K. Small-RNA Asymmetry Is Directly Driven by Mammalian Argonautes. Nat. Struct. Mol. Biol. 2015, 22, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-W.; Noland, C.; Siridechadilok, B.; Taylor, D.W.; Ma, E.; Felderer, K.; Doudna, J.A.; Nogales, E. Structural Insights into RNA Processing by the Human RISC-Loading Complex. Nat. Struct. Mol. Biol. 2009, 16, 1148–1153. [Google Scholar] [CrossRef]
- Frank, F.; Sonenberg, N.; Nagar, B. Structural Basis for 5′-Nucleotide Base-Specific Recognition of Guide RNA by Human AGO2. Nature 2010, 465, 818–822. [Google Scholar] [CrossRef]
- Lee, S.; Jee, D.; Srivastava, S.; Yang, A.; Ramidi, A.; Shang, R.; Bortolamiol-Becet, D.; Pfeffer, S.; Gu, S.; Wen, J.; et al. Promiscuous Splicing-Derived Hairpins Are Dominant Substrates of Tailing-Mediated Defense of MiRNA Biogenesis in Mammals. Cell Rep. 2023, 42, 112111. [Google Scholar] [CrossRef]
- Kakumani, P.K.; Ko, Y.; Ramakrishna, S.; Christopher, G.; Dodgson, M.; Shrinet, J.; Harvey, L.-M.; Shin, C.; Simard, M.J. CSDE1 Promotes MiR-451 Biogenesis. Nucleic Acids Res. 2023, 51, 9385–9396. [Google Scholar] [CrossRef]
- Yoda, M.; Cifuentes, D.; Izumi, N.; Sakaguchi, Y.; Suzuki, T.; Giraldez, A.J.; Tomari, Y. Poly(A)-Specific Ribonuclease Mediates 3′-End Trimming of Argonaute2-Cleaved Precursor MicroRNAs. Cell Rep. 2013, 5, 715–726. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From MicroRNA Sequences to Function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Fromm, B.; Høye, E.; Domanska, D.; Zhong, X.; Aparicio-Puerta, E.; Ovchinnikov, V.; Umu, S.U.; Chabot, P.J.; Kang, W.; Aslanzadeh, M.; et al. MirGeneDB 2.1: Toward a Complete Sampling of All Major Animal Phyla. Nucleic Acids Res. 2022, 50, D204–D210. [Google Scholar] [CrossRef]
- Wang, D.; Sun, X.; Wei, Y.; Liang, H.; Yuan, M.; Jin, F.; Chen, X.; Liu, Y.; Zhang, C.-Y.; Li, L.; et al. Nuclear MiR-122 Directly Regulates the Biogenesis of Cell Survival OncomiR MiR-21 at the Posttranscriptional Level. Nucleic Acids Res. 2018, 46, 2012–2029. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Zhang, X.; Nie, X.; Li, H.; Yuan, S.; Dai, B.; Zhan, J.; Wen, Z.; Jiang, J.; Chen, C.; et al. Nuclear MiR-665 Aggravates Heart Failure via Suppressing Phosphatase and Tensin Homolog Transcription. Sci. China Life Sci. 2020, 63, 724–736. [Google Scholar] [CrossRef]
- Liang, Y.; Zou, Q.; Yu, W. Steering Against Wind: A New Network of NamiRNAs and Enhancers. Genom. Proteom. Bioinform. 2017, 15, 331–337. [Google Scholar] [CrossRef]
- Liang, Y.; Xu, P.; Zou, Q.; Luo, H.; Yu, W. An Epigenetic Perspective on Tumorigenesis: Loss of Cell Identity, Enhancer Switching, and NamiRNA Network. Semin. Cancer Biol. 2019, 57, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Zhang, Y.; Huang, Q.-Q.; Qian, M.-M.; Li, Z.-X.; Li, Y.-J.; Li, B.-P.; Qiu, Z.-L.; Yue, J.-J.; Guo, Z.-Y. Genome-Wide Identification and Analysis of Enhancer-Regulated MicroRNAs Across 31 Human Cancers. Front. Genet. 2020, 11, 644. [Google Scholar] [CrossRef]
- Odame, E.; Chen, Y.; Zheng, S.; Dai, D.; Kyei, B.; Zhan, S.; Cao, J.; Guo, J.; Zhong, T.; Wang, L.; et al. Enhancer RNAs: Transcriptional Regulators and Workmates of NamiRNAs in Myogenesis. Cell. Mol. Biol. Lett. 2021, 26, 4. [Google Scholar] [CrossRef]
- Naeli, P.; Winter, T.; Hackett, A.P.; Alboushi, L.; Jafarnejad, S.M. The Intricate Balance between MicroRNA-Induced MRNA Decay and Translational Repression. FEBS J. 2023, 290, 2508–2524. [Google Scholar] [CrossRef] [PubMed]
- Kirstein, N.; Dokaneheifard, S.; Cingaram, P.R.; Valencia, M.G.; Beckedorff, F.; Gomes Dos Santos, H.; Blumenthal, E.; Tayari, M.M.; Gaidosh, G.S.; Shiekhattar, R. The Integrator Complex Regulates MicroRNA Abundance through RISC Loading. Sci. Adv. 2023, 9, eadf0597. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of MicroRNA Biogenesis and Its Crosstalk with Other Cellular Pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA Circles Function as Efficient MicroRNA Sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Karagkouni, D.; Karavangeli, A.; Paraskevopoulou, M.D.; Hatzigeorgiou, A.G. Characterizing MiRNA–LncRNA Interplay. In Long Non-Coding RNAs: Methods and Protocols; Humana: New York, NY, USA, 2021; pp. 243–262. [Google Scholar]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The Multilayered Complexity of CeRNA Crosstalk and Competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Moreno-García, L.; López-Royo, T.; Calvo, A.C.; Toivonen, J.M.; de la Torre, M.; Moreno-Martínez, L.; Molina, N.; Aparicio, P.; Zaragoza, P.; Manzano, R.; et al. Competing Endogenous Rna Networks as Biomarkers in Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 9582. [Google Scholar] [CrossRef]
- Chen, X.; Wan, L.; Wang, W.; Xi, W.-J.; Yang, A.-G.; Wang, T. Re-Recognition of Pseudogenes: From Molecular to Clinical Applications. Theranostics 2020, 10, 1479–1499. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhou, J.; Zhang, Z.; Li, J.; Wang, F.; Ma, L.; Tian, X.; Mao, Z.; Yang, Y. Exosomal MiR-485-3p Derived from Pancreatic Ductal Epithelial Cells Inhibits Pancreatic Cancer Metastasis through Targeting PAK1. Chin. Med. J. 2022, 135, 2326–2337. [Google Scholar] [CrossRef]
- Mustafa, A.; Shabbir, M.; Badshah, Y.; Khan, K.; Abid, F.; Trembley, J.H.; Afsar, T.; Almajwal, A.; Razak, S. Genetic Polymorphism in Untranslated Regions of PRKCZ Influences MRNA Structure, Stability and Binding Sites. BMC Cancer 2024, 24, 1147. [Google Scholar] [CrossRef]
- Toledo-Stuardo, K.; Ribeiro, C.H.; Campos, I.; Tello, S.; Latorre, Y.; Altamirano, C.; Dubois-Camacho, K.; Molina, M.C. Impact of MICA 3′UTR Allelic Variability on MiRNA Binding Prediction, a Bioinformatic Approach. Front. Genet. 2023, 14, 1273296. [Google Scholar] [CrossRef]
- Tian, Y.-F.; Huang, C.-J.; Liu, C.-Y.; Yang, S.-H.; Hung, C.-S.; Lin, K.-Y.; Lai, C.-L.; Chang, C.-C. MicroRNA-24 Alleviates Colorectal Cancer Progression via a Rs28382740 Single Nucleotide Polymorphism in the Long Noncoding Region of X-linked Inhibitor of Apoptosis Protein. Oncol. Lett. 2024, 28, 591. [Google Scholar] [CrossRef]
- Homberg, N.; Galvão Ferrarini, M.; Gaspin, C.; Sagot, M.-F. MicroRNA Target Identification: Revisiting Accessibility and Seed Anchoring. Genes 2023, 14, 664. [Google Scholar] [CrossRef]
- Broughton, J.P.; Lovci, M.T.; Huang, J.L.; Yeo, G.W.; Pasquinelli, A.E. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol. Cell 2016, 64, 320–333. [Google Scholar] [CrossRef]
- Kim, D.; Sung, Y.M.; Park, J.; Kim, S.; Kim, J.; Park, J.; Ha, H.; Bae, J.Y.; Kim, S.; Baek, D. General Rules for Functional MicroRNA Targeting. Nat. Genet. 2016, 48, 1517–1526. [Google Scholar] [CrossRef]
- McGeary, S.E.; Bisaria, N.; Pham, T.M.; Wang, P.Y.; Bartel, D.P. MicroRNA 3′-Compensatory Pairing Occurs through Two Binding Modes, with Affinity Shaped by Nucleotide Identity and Position. eLife 2022, 11, e69803. [Google Scholar] [CrossRef] [PubMed]
- Kosek, D.M.; Banijamali, E.; Becker, W.; Petzold, K.; Andersson, E.R. Efficient 3′-Pairing Renders MicroRNA Targeting Less Sensitive to MRNA Seed Accessibility. Nucleic Acids Res. 2023, 51, 11162–11177. [Google Scholar] [CrossRef] [PubMed]
- Shin, C.; Nam, J.-W.; Farh, K.K.-H.; Chiang, H.R.; Shkumatava, A.; Bartel, D.P. Expanding the MicroRNA Targeting Code: Functional Sites with Centered Pairing. Mol. Cell 2010, 38, 789–802. [Google Scholar] [CrossRef]
- Shang, R.; Lee, S.; Senavirathne, G.; Lai, E.C. MicroRNAs in Action: Biogenesis, Function and Regulation. Nat. Rev. Genet. 2023, 24, 816–833. [Google Scholar] [CrossRef]
- Ma, F.; Liu, X.; Li, D.; Wang, P.; Li, N.; Lu, L.; Cao, X. MicroRNA-466l Upregulates IL-10 Expression in TLR-Triggered Macrophages by Antagonizing RNA-Binding Protein Tristetraprolin-Mediated IL-10 MRNA Degradation. J. Immunol. 2010, 184, 6053–6059. [Google Scholar] [CrossRef] [PubMed]
- Long, J.M.; Maloney, B.; Rogers, J.T.; Lahiri, D.K. Novel Upregulation of Amyloid-β Precursor Protein (APP) by MicroRNA-346 via Targeting of APP MRNA 5′-Untranslated Region: Implications in Alzheimer’s Disease. Mol. Psychiatry 2019, 24, 345–363. [Google Scholar] [CrossRef]
- Vasudevan, S. Posttranscriptional Upregulation by MicroRNAs. WIREs RNA 2012, 3, 311–330. [Google Scholar] [CrossRef]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from Repression to Activation: MicroRNAs Can Up-Regulate Translation. Science 2007, 318, 1931–1934. [Google Scholar] [CrossRef]
- Vasudevan, S.; Steitz, J.A. AU-Rich-Element-Mediated Upregulation of Translation by FXR1 and Argonaute 2. Cell 2007, 128, 1105–1118. [Google Scholar] [CrossRef]
- Bukhari, S.I.A.; Truesdell, S.S.; Lee, S.; Kollu, S.; Classon, A.; Boukhali, M.; Jain, E.; Mortensen, R.D.; Yanagiya, A.; Sadreyev, R.I.; et al. A Specialized Mechanism of Translation Mediated by FXR1a-Associated MicroRNP in Cellular Quiescence. Mol. Cell 2016, 61, 760–773. [Google Scholar] [CrossRef]
- Truesdell, S.S.; Mortensen, R.D.; Seo, M.; Schroeder, J.C.; Lee, J.H.; LeTonqueze, O.; Vasudevan, S. MicroRNA-Mediated MRNA Translation Activation in Quiescent Cells and Oocytes Involves Recruitment of a Nuclear MicroRNP. Sci. Rep. 2012, 2, 842. [Google Scholar] [CrossRef]
- Ørom, U.A.; Nielsen, F.C.; Lund, A.H. MicroRNA-10a Binds the 5′UTR of Ribosomal Protein MRNAs and Enhances Their Translation. Mol. Cell 2008, 30, 460–471. [Google Scholar] [CrossRef]
- Li, J.; Chen, J.; Wang, S.; Li, P.; Zheng, C.; Zhou, X.; Tao, Y.; Chen, X.; Sun, L.; Wang, A.; et al. Blockage of Transferred Exosome-shuttled MiR-494 Inhibits Melanoma Growth and Metastasis. J. Cell. Physiol. 2019, 234, 15763–15774. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Ji, X.; Liu, K.; Shi, Y.; Wang, C.; Li, Y.; Zhang, T.; He, Y.; Xiang, M.; Zhao, R. Exosomal MiR-200c-3p Negatively Regulates the Migration and Invasion of Lipopolysaccharide (LPS)-Stimulated Colorectal Cancer (CRC). BMC Mol. Cell Biol. 2020, 21, 48. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Yang, X.; Yang, H.; Lv, M.; Sun, X.; Zhou, B. Exosomal MiR-338-3p Suppresses Non-Small-Cell Lung Cancer Cells Metastasis by Inhibiting CHL1 through the MAPK Signaling Pathway. Cell Death Dis. 2021, 12, 1030. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Qu, L.; Yang, J.; Xu, J.; Sun, L.; Wei, X.; Qu, X.; Bai, T.; Guo, Z.; Zhu, Y. Exosome–Transmitted MicroRNA-133b Inhibited Bladder Cancer Proliferation by Upregulating Dual-specificity Protein Phosphatase 1. Cancer Med. 2020, 9, 6009–6019. [Google Scholar] [CrossRef]
- Hunter, S.; Nault, B.; Ugwuagbo, K.; Maiti, S.; Majumder, M. Mir526b and Mir655 Promote Tumour Associated Angiogenesis and Lymphangiogenesis in Breast Cancer. Cancers 2019, 11, 938. [Google Scholar] [CrossRef]
- Shojaei, S.; Moradi-Chaleshtori, M.; Paryan, M.; Koochaki, A.; Sharifi, K.; Mohammadi-Yeganeh, S. Mesenchymal Stem Cell-Derived Exosomes Enriched with MiR-218 Reduce the Epithelial–Mesenchymal Transition and Angiogenesis in Triple-Negative Breast Cancer Cells. Eur. J. Med. Res. 2023, 28, 516. [Google Scholar] [CrossRef]
- Tang, Y.-T.; Huang, Y.-Y.; Li, J.-H.; Qin, S.-H.; Xu, Y.; An, T.-X.; Liu, C.-C.; Wang, Q.; Zheng, L. Alterations in Exosomal MiRNA Profile upon Epithelial-Mesenchymal Transition in Human Lung Cancer Cell Lines. BMC Genom. 2018, 19, 802. [Google Scholar] [CrossRef]
- Tai, Y.; Chen, K.; Hsieh, J.; Shen, T. Exosomes in Cancer Development and Clinical Applications. Cancer Sci. 2018, 109, 2364–2374. [Google Scholar] [CrossRef]
- Zhao, Y.; Jin, L.-J.; Zhang, X.-Y. Exosomal MiRNA-205 Promotes Breast Cancer Chemoresistance and Tumorigenesis through E2F1. Aging 2021, 13, 18498–18514. [Google Scholar] [CrossRef]
- Qin, X.; Yu, S.; Zhou, L.; Shi, M.; Hu, Y.; Xu, X.; Shen, B.; Liu, S.; Yan, D.; Feng, J. Cisplatin-Resistant Lung Cancer Cell–Derived Exosomes Increase Cisplatin Resistance of Recipient Cells in Exosomal MiR-100–5p-Dependent Manner. Int. J. Nanomed. 2017, 12, 3721–3733. [Google Scholar] [CrossRef]
- Liu, S.; Wang, W.; Ning, Y.; Zheng, H.; Zhan, Y.; Wang, H.; Yang, Y.; Luo, J.; Wen, Q.; Zang, H.; et al. Exosome-Mediated MiR-7-5p Delivery Enhances the Anticancer Effect of Everolimus via Blocking MNK/EIF4E Axis in Non-Small Cell Lung Cancer. Cell Death Dis. 2022, 13, 129. [Google Scholar] [CrossRef]
- Díez-Sainz, E.; Lorente-Cebrián, S.; Aranaz, P.; Riezu-Boj, J.I.; Martínez, J.A.; Milagro, F.I. Potential Mechanisms Linking Food-Derived MicroRNAs, Gut Microbiota and Intestinal Barrier Functions in the Context of Nutrition and Human Health. Front. Nutr. 2021, 8, 586564. [Google Scholar] [CrossRef]
- Deveci, G.; Capasso, R.; Ağagündüz, D. Xeno-MiRs and Circulating MiRNAs as Novel Biomarkers in Certain Diseases. Biologics 2022, 3, 1. [Google Scholar] [CrossRef]
- Norouzi, M.; Bakhtiarizadeh, M.R.; Salehi, A. Investigation of the Transability of Dietary Small Non-Coding RNAs to Animals. Front. Genet. 2022, 13, 933709. [Google Scholar] [CrossRef]
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in Health and Diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent Deletions and Down-Regulation of Micro- RNA Genes MiR15 and MiR16 at 13q14 in Chronic Lymphocytic Leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Liu, C.; Tang, D.G. MicroRNA Regulation of Cancer Stem Cells. Cancer Res. 2011, 71, 5950–5954. [Google Scholar] [CrossRef]
- Lou, Y.; Yang, X.; Wang, F.; Cui, Z.; Huang, Y. MicroRNA-21 Promotes the Cell Proliferation, Invasion and Migration Abilities in Ovarian Epithelial Carcinomas through Inhibiting the Expression of PTEN Protein. Int. J. Mol. Med. 2010, 26, 819–827. [Google Scholar] [CrossRef]
- Bader, A.G.; Brown, D.; Stoudemire, J.; Lammers, P. Developing Therapeutic MicroRNAs for Cancer. Gene Ther. 2011, 18, 1121–1126. [Google Scholar] [CrossRef]
- van Rooij, E.; Marshall, W.S.; Olson, E.N. Toward MicroRNA–Based Therapeutics for Heart Disease. Circ. Res. 2008, 103, 919–928. [Google Scholar] [CrossRef]
- Quemener, A.M.; Bachelot, L.; Forestier, A.; Donnou-Fournet, E.; Gilot, D.; Galibert, M.D. The powerful world of antisense oligonucleotides: From bench to bedside. Wiley Interdiscip. Rev. RNA 2020, 11, e1594. [Google Scholar] [CrossRef] [PubMed]
- Koziolkiewicz, M.; Gendaszewska, E.; Maszewska, M.; Stein, C.A.; Stec, W.J. The Mononucleotide-Dependent, Nonantisense Mechanism of Action of Phosphodiester and Phosphorothioate Oligonucleotides Depends upon the Activity of an Ecto-5′-Nucleotidase. Blood 2001, 98, 995–1002. [Google Scholar] [CrossRef] [PubMed]
- Nicolussi, A.; D’Inzeo, S.; Capalbo, C.; Giannini, G.; Coppa, A. The Role of Peroxiredoxins in Cancer. Mol. Clin. Oncol. 2017, 6, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Monia, B.P.; Lesnik, E.A.; Gonzalez, C.; Lima, W.F.; McGee, D.; Guinosso, C.J.; Kawasaki, A.M.; Cook, P.D.; Freier, S.M. Evaluation of 2′-Modified Oligonucleotides Containing 2′-Deoxy Gaps as Antisense Inhibitors of Gene Expression. J. Biol. Chem. 1993, 268, 14514–14522. [Google Scholar]
- Hagedorn, P.H.; Persson, R.; Funder, E.D.; Albæk, N.; Diemer, S.L.; Hansen, D.J.; Møller, M.R.; Papargyri, N.; Christiansen, H.; Hansen, B.R.; et al. Locked Nucleic Acid: Modality, Diversity, and Drug Discovery. Drug Discov. Today 2018, 23, 101–114. [Google Scholar] [CrossRef]
- Elmén, J.; Lindow, M.; Silahtaroglu, A.; Bak, M.; Christensen, M.; Lind-Thomsen, A.; Hedtjärn, M.; Hansen, J.B.; Hansen, H.F.; Straarup, E.M.; et al. Antagonism of MicroRNA-122 in Mice by Systemically Administered LNA-AntimiR Leads to up-Regulation of a Large Set of Predicted Target MRNAs in the Liver. Nucleic Acids Res. 2008, 36, 1153–1162. [Google Scholar] [CrossRef]
- Grünweller, A.; Hartmann, R.K. Locked Nucleic Acid Oligonucleotides. BioDrugs 2007, 21, 235–243. [Google Scholar] [CrossRef]
- Braasch, D.A.; Liu, Y.; Corey, D.R. Antisense Inhibition of Gene Expression in Cells by Oligonucleotides Incorporating Locked Nucleic Acids: Effect of MRNA Target Sequence and Chimera Design. Nucleic Acids Res. 2002, 30, 5160–5167. [Google Scholar] [CrossRef]
- Frieden, M.; Orum, H. Locked Nucleic Acid Holds Promise in the Treatment of Cancer. Curr. Pharm. Des. 2008, 14, 1138–1142. [Google Scholar] [CrossRef]
- Elmén, J.; Thonberg, H.; Ljungberg, K.; Frieden, M.; Westergaard, M.; Xu, Y.; Wahren, B.; Liang, Z.; Ørum, H.; Koch, T.; et al. Locked Nucleic Acid (LNA) Mediated Improvements in SiRNA Stability and Functionality. Nucleic Acids Res. 2005, 33, 439–447. [Google Scholar] [CrossRef]
- Roberts, J.; Palma, E.; Sazani, P.; Ørum, H.; Cho, M.; Kole, R. Efficient and Persistent Splice Switching by Systemically Delivered LNA Oligonucleotides in Mice. Mol. Ther. 2006, 14, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Fackenthal, J.D. Alternative MRNA Splicing and Promising Therapies in Cancer. Biomolecules 2023, 13, 561. [Google Scholar] [CrossRef] [PubMed]
- Obad, S.; dos Santos, C.O.; Petri, A.; Heidenblad, M.; Broom, O.; Ruse, C.; Fu, C.; Lindow, M.; Stenvang, J.; Straarup, E.M.; et al. Silencing of MicroRNA Families by Seed-Targeting Tiny LNAs. Nat. Genet. 2011, 43, 371–378. [Google Scholar] [CrossRef]
- Krützfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of MicroRNAs in Vivo with ‘Antagomirs’. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, Y.; Zhang, Y.; Li, X.; Fang, M.; Qian, D. Targeting Exosomes Enveloped EBV-miR-BART1-5p-antagomiRs for NPC Therapy through Both Anti-vasculogenic Mimicry and Anti-angiogenesis. Cancer Med. 2023, 12, 12608–12621. [Google Scholar] [CrossRef]
- Bader, A.G.; Brown, D.; Winkler, M. The Promise of MicroRNA Replacement Therapy. Cancer Res. 2010, 70, 7027–7030. [Google Scholar] [CrossRef]
- Fu, Y.; Chen, J.; Huang, Z. Recent Progress in MicroRNA-Based Delivery Systems for the Treatment of Human Disease. ExRNA 2019, 1, 24. [Google Scholar] [CrossRef]
- Garreau, M.; Weidner, J.; Hamilton, R.; Kolosionek, E.; Toki, N.; Stavenhagen, K.; Paris, C.; Bonetti, A.; Czechtizky, W.; Gnerlich, F.; et al. Chemical Modification Patterns for MicroRNA Therapeutic Mimics: A Structure-Activity Relationship (SAR) Case-Study on MiR-200c. Nucleic Acids Res. 2024, 52, 2792–2807. [Google Scholar] [CrossRef]
- Petrek, H.; Batra, N.; Ho, P.Y.; Tu, M.-J.; Yu, A.-M. Bioengineering of a Single Long Noncoding RNA Molecule That Carries Multiple Small RNAs. Appl. Microbiol. Biotechnol. 2019, 103, 6107–6117. [Google Scholar] [CrossRef] [PubMed]
- van Rooij, E.; Kauppinen, S. Development of Micro RNA Therapeutics Is Coming of Age. EMBO Mol. Med. 2014, 6, 851–864. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Gu, S.; Chen, B.-F.; Shen, W.-L.; Yin, Z.; Xu, G.-W.; Hu, J.-J.; Zhu, T.; Li, G.; Wan, C.; et al. Nanoparticle Delivery of Stable MiR-199a-5p Agomir Improves the Osteogenesis of Human Mesenchymal Stem Cells via the HIF1a Pathway. Biomaterials 2015, 53, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Grillone, K.; Caridà, G.; Luciano, F.; Cordua, A.; Di Martino, M.T.; Tagliaferri, P.; Tassone, P. A Systematic Review of Non-Coding RNA Therapeutics in Early Clinical Trials: A New Perspective against Cancer. J. Transl. Med. 2024, 22, 731. [Google Scholar] [CrossRef]
- Çakan, E.; Lara, O.D.; Szymanowska, A.; Bayraktar, E.; Chavez-Reyes, A.; Lopez-Berestein, G.; Amero, P.; Rodriguez-Aguayo, C. Therapeutic Antisense Oligonucleotides in Oncology: From Bench to Bedside. Cancers 2024, 16, 2940. [Google Scholar] [CrossRef]
- Bartolucci, D.; Pession, A.; Hrelia, P.; Tonelli, R. Precision Anti-Cancer Medicines by Oligonucleotide Therapeutics in Clinical Research Targeting Undruggable Proteins and Non-Coding RNAs. Pharmaceutics 2022, 14, 1453. [Google Scholar] [CrossRef]
- Tassone, P.; Di Martino, M.T.; Arbitrio, M.; Fiorillo, L.; Staropoli, N.; Ciliberto, D.; Cordua, A.; Scionti, F.; Bertucci, B.; Salvino, A.; et al. Safety and Activity of the First-in-Class Locked Nucleic Acid (LNA) MiR-221 Selective Inhibitor in Refractory Advanced Cancer Patients: A First-in-Human, Phase 1, Open-Label, Dose-Escalation Study. J. Hematol. Oncol. 2023, 16, 68. [Google Scholar] [CrossRef]
- Querfeld, C.; Foss, F.M.; Kim, Y.H.; Pinter-Brown, L.; William, B.M.; Porcu, P.; Pacheco, T.; Haverkos, B.M.; DeSimone, J.; Guitart, J.; et al. Phase 1 Trial of Cobomarsen, an Inhibitor of Mir-155, in Cutaneous T Cell Lymphoma. Blood 2018, 132, 2903. [Google Scholar] [CrossRef]
- Hong, D.S.; Kang, Y.-K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.-L.; Kim, T.-Y.; et al. Phase 1 Study of MRX34, a Liposomal MiR-34a Mimic, in Patients with Advanced Solid Tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef]
- Reid, G.; Kao, S.C.; Pavlakis, N.; Brahmbhatt, H.; MacDiarmid, J.; Clarke, S.; Boyer, M.; van Zandwijk, N. Clinical Development of TargomiRs, a MiRNA Mimic-Based Treatment for Patients with Recurrent Thoracic Cancer. Epigenomics 2016, 8, 1079–1085. [Google Scholar] [CrossRef]
- van den Bosch, M.T.J.; Yahyanejad, S.; Alemdehy, M.F.; Telford, B.J.; de Gunst, T.; den Boer, H.C.; Vos, R.M.; Stegink, M.; van Pinxteren, L.A.H.; Schaapveld, R.Q.J.; et al. Transcriptome-Wide Analysis Reveals Insight into Tumor Suppressor Functions of 1B3, a Novel Synthetic MiR-193a-3p Mimic. Mol. Ther. Nucleic Acids 2021, 23, 1161–1171. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, M.S.; Aggarwal, R.R.; Boyd, E.; Comerford, K.; Zhang, J.; Méndez, B.; Valenzuela, P.; Grabowsky, J.; Thomas, S.; Munster, P.N. Phase 1 Study of ANDES-1537: A Novel Antisense Oligonucleotide against Non-Coding Mitochondrial DNA in Advanced Solid Tumors. J. Clin. Oncol. 2018, 36, 2557-2557. [Google Scholar] [CrossRef]
- Seto, A.G.; Beatty, X.; Lynch, J.M.; Hermreck, M.; Tetzlaff, M.; Duvic, M.; Jackson, A.L. Cobomarsen, an Oligonucleotide Inhibitor of MiR-155, Co-ordinately Regulates Multiple Survival Pathways to Reduce Cellular Proliferation and Survival in Cutaneous T-cell Lymphoma. Br. J. Haematol. 2018, 183, 428–444. [Google Scholar] [CrossRef]
- miRagen Therapeutics, Inc. PRISM: Efficacy and Safety of Cobomarsen (MRG-106) in Subjects with Mycosis Fungoides Who Have Completed the SOLAR Study (PRISM). Available online: https://clinicaltrials.gov/study/NCT03837457 (accessed on 16 January 2025).
- James, A.M.; Ruckman, J.; Pestano, L.A.; Hopkins, R.D.; Rodgers, R.C.; Marshall, W.S.; Rubin, P.; Escolar, D. SOLAR: A phase 2, global, randomized, active comparator study to investigate the efficacy and safety of cobomarsen in subjects with mycosis fungoides (MF). Hematol. Oncol. 2019, 37, 562–563. [Google Scholar] [CrossRef]
- Peltier, H.J.; Kelnar, K.; Bader, A.G. Effects of MRX34, a Liposomal MiR-34 Mimic, on Target Gene Expression in Human White Blood Cells (HWBCs): QRT-PCR Results from a First-in-Human Trial of MicroRNA Cancer Therapy. J. Clin. Oncol. 2016, 34, e14090. [Google Scholar] [CrossRef]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.-K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I Study of MRX34, a Liposomal MiR-34a Mimic, Administered Twice Weekly in Patients with Advanced Solid Tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef]
- van Zandwijk, N.; Pavlakis, N.; Kao, S.C.; Linton, A.; Boyer, M.J.; Clarke, S.; Huynh, Y.; Chrzanowska, A.; Fulham, M.J.; Bailey, D.L.; et al. Safety and Activity of MicroRNA-Loaded Minicells in Patients with Recurrent Malignant Pleural Mesothelioma: A First-in-Man, Phase 1, Open-Label, Dose-Escalation Study. Lancet Oncol. 2017, 18, 1386–1396. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Khoshbakht, T.; Hussen, B.M.; Abdullah, S.T.; Taheri, M.; Samadian, M. A Review on the Role of Mir-16-5p in the Carcinogenesis. Cancer Cell Int. 2022, 22, 342. [Google Scholar] [CrossRef]
- Kook, E.; Lee, J.; Kim, D.-H. YES1 as a Potential Target to Overcome Drug Resistance in EGFR-Deregulated Non-Small Cell Lung Cancer. Arch. Toxicol. 2024, 98, 1437–1455. [Google Scholar] [CrossRef]
- Chia, P.L.; Parakh, S.; Russell, P.; Gan, H.K.; Asadi, K.; Gebski, V.; Murone, C.; Walkiewicz, M.; Liu, Z.; Thapa, B.; et al. Expression of EGFR and Conformational Forms of EGFR in Malignant Pleural Mesothelioma and Its Impact on Survival. Lung Cancer 2021, 153, 35–41. [Google Scholar] [CrossRef]
- Perrino, M.; De Vincenzo, F.; Cordua, N.; Borea, F.; Aliprandi, M.; Santoro, A.; Zucali, P.A. Immunotherapy with Immune Checkpoint Inhibitors and Predictive Biomarkers in Malignant Mesothelioma: Work Still in Progress. Front. Immunol. 2023, 14, 1121557. [Google Scholar] [CrossRef]
- Telford, B.J.; Yahyanejad, S.; de Gunst, T.; den Boer, H.C.; Vos, R.M.; Stegink, M.; van den Bosch, M.T.; Alemdehy, M.F.; van Pinxteren, L.A.; Schaapveld, R.Q.; et al. Multi-Modal Effects of 1B3, a Novel Synthetic MiR-193a-3p Mimic, Support Strong Potential for Therapeutic Intervention in Oncology. Oncotarget 2021, 12, 422–439. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Croce, C.M. MicroRNA: Trends in Clinical Trials of Cancer Diagnosis and Therapy Strategies. Exp. Mol. Med. 2023, 55, 1314–1321. [Google Scholar] [CrossRef]
- Sur, D.; Advani, S.; Braithwaite, D. MicroRNA Panels as Diagnostic Biomarkers for Colorectal Cancer: A Systematic Review and Meta-Analysis. Front. Med. 2022, 9, 915226. [Google Scholar] [CrossRef]
- Caputo, R.; Buono, G.; Piezzo, M.; Martinelli, C.; Cianniello, D.; Rizzo, A.; Pantano, F.; Staropoli, N.; Cangiano, R.; Turano, S.; et al. Sacituzumab Govitecan for the Treatment of Advanced Triple Negative Breast Cancer Patients: A Multi-Center Real-World Analysis. Front. Oncol. 2024, 14, 1362641. [Google Scholar] [CrossRef]
- Gaber, D.A.; Wassef, R.M.; El-Ayat, W.M.; El-Moazen, M.I.; Montasser, K.A.; Swar, S.A.; Amin, H.A.A. Role of a Schistosoma Haematobium Specific MicroRNA as a Predictive and Prognostic Tool for Bilharzial Bladder Cancer in Egypt. Sci. Rep. 2020, 10, 18844. [Google Scholar] [CrossRef]
- Malta, K.K.; Palazzi, C.; Neves, V.H.; Aguiar, Y.; Silva, T.P.; Melo, R.C.N. Schistosomiasis Mansoni-Recruited Eosinophils: An Overview in the Granuloma Context. Microorganisms 2022, 10, 2022. [Google Scholar] [CrossRef]
- Zaghloul, M.S.; Zaghloul, T.M.; Bishr, M.K.; Baumann, B.C. Urinary Schistosomiasis and the Associated Bladder Cancer: Update. J. Egypt. Natl. Cancer Inst. 2020, 32, 44. [Google Scholar] [CrossRef]
- Gajic, Z.; Kaur, D.; Ni, J.; Zhu, Z.; Zhebrun, A.; Gajic, M.; Kim, M.; Hong, J.; Priyadarshini, M.; Frøkjær-Jensen, C.; et al. Target-Dependent Suppression of SiRNA Production Modulates the Levels of Endogenous SiRNAs in the Caenorhabditis elegans Germline. Development 2022, 149, dev200692. [Google Scholar] [CrossRef]
- Ruby, J.G.; Jan, C.; Player, C.; Axtell, M.J.; Lee, W.; Nusbaum, C.; Ge, H.; Bartel, D.P. Large-Scale Sequencing Reveals 21U-RNAs and Additional MicroRNAs and Endogenous SiRNAs in C. elegans. Cell 2006, 127, 1193–1207. [Google Scholar] [CrossRef]
- Okamura, K.; Chung, W.-J.; Ruby, J.G.; Guo, H.; Bartel, D.P.; Lai, E.C. The Drosophila Hairpin RNA Pathway Generates Endogenous Short Interfering RNAs. Nature 2008, 453, 803–806. [Google Scholar] [CrossRef]
- Okamura, K.; Balla, S.; Martin, R.; Liu, N.; Lai, E.C. Two Distinct Mechanisms Generate Endogenous SiRNAs from Bidirectional Transcription in Drosophila Melanogaster. Nat. Struct. Mol. Biol. 2008, 15, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Ghildiyal, M.; Seitz, H.; Horwich, M.D.; Li, C.; Du, T.; Lee, S.; Xu, J.; Kittler, E.L.W.; Zapp, M.L.; Weng, Z.; et al. Endogenous SiRNAs Derived from Transposons and MRNAs in Drosophila Somatic Cells. Science 2008, 320, 1077–1081. [Google Scholar] [CrossRef]
- Czech, B.; Malone, C.D.; Zhou, R.; Stark, A.; Schlingeheyde, C.; Dus, M.; Perrimon, N.; Kellis, M.; Wohlschlegel, J.A.; Sachidanandam, R.; et al. An Endogenous Small Interfering RNA Pathway in Drosophila. Nature 2008, 453, 798–802. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Pasolli, H.A.; Landthaler, M.; Hafner, M.; Ojo, T.; Sheridan, R.; Sander, C.; O’Carroll, D.; Stoffel, M.; Tuschl, T.; et al. DGCR8-Dependent MicroRNA Biogenesis Is Essential for Skin Development. Proc. Natl. Acad. Sci. USA 2009, 106, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, C.; Romero, Y.; Warnefors, M.; Bilican, A.; Borel, C.; Smith, L.B.; Kotaja, N.; Kaessmann, H.; Nef, S. Germ Cell-Specific Targeting of DICER or DGCR8 Reveals a Novel Role for Endo-SiRNAs in the Progression of Mammalian Spermatogenesis and Male Fertility. PLoS ONE 2014, 9, e107023. [Google Scholar] [CrossRef]
- Song, R.; Hennig, G.W.; Wu, Q.; Jose, C.; Zheng, H.; Yan, W. Male Germ Cells Express Abundant Endogenous SiRNAs. Proc. Natl. Acad. Sci. USA 2011, 108, 13159–13164. [Google Scholar] [CrossRef]
- Watanabe, T.; Totoki, Y.; Toyoda, A.; Kaneda, M.; Kuramochi-Miyagawa, S.; Obata, Y.; Chiba, H.; Kohara, Y.; Kono, T.; Nakano, T.; et al. Endogenous SiRNAs from Naturally Formed DsRNAs Regulate Transcripts in Mouse Oocytes. Nature 2008, 453, 539–543. [Google Scholar] [CrossRef]
- Tam, O.H.; Aravin, A.A.; Stein, P.; Girard, A.; Murchison, E.P.; Cheloufi, S.; Hodges, E.; Anger, M.; Sachidanandam, R.; Schultz, R.M.; et al. Pseudogene-Derived Small Interfering RNAs Regulate Gene Expression in Mouse Oocytes. Nature 2008, 453, 534–538. [Google Scholar] [CrossRef]
- Babiarz, J.E.; Ruby, J.G.; Wang, Y.; Bartel, D.P.; Blelloch, R. Mouse ES Cells Express Endogenous ShRNAs, SiRNAs, and Other Microprocessor-Independent, Dicer-Dependent Small RNAs. Genes Dev. 2008, 22, 2773–2785. [Google Scholar] [CrossRef]
- Chen, L.; Dahlstrom, J.E.; Lee, S.-H.; Rangasamy, D. Naturally Occurring Endo-SiRNA Silences LINE-1 Retrotransposons in Human Cells through DNA Methylation. Epigenetics 2012, 7, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Joyce, C.E.; Bowcock, A.M.; Zhang, W. Noncanonical MicroRNAs and Endogenous SiRNAs in Normal and Psoriatic Human Skin. Hum. Mol. Genet. 2013, 22, 737–748. [Google Scholar] [CrossRef]
- Liu, J.; Carmell, M.A.; Rivas, F.V.; Marsden, C.G.; Thomson, J.M.; Song, J.-J.; Hammond, S.M.; Joshua-Tor, L.; Hannon, G.J. Argonaute2 Is the Catalytic Engine of Mammalian RNAi. Science 2004, 305, 1437–1441. [Google Scholar] [CrossRef]
- Grimm, D.; Streetz, K.L.; Jopling, C.L.; Storm, T.A.; Pandey, K.; Davis, C.R.; Marion, P.; Salazar, F.; Kay, M.A. Fatality in Mice Due to Oversaturation of Cellular MicroRNA/Short Hairpin RNA Pathways. Nature 2006, 441, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Grimm, D. The Dose Can Make the Poison: Lessons Learned from Adverse in Vivo Toxicities Caused by RNAi Overexpression. Silence 2011, 2, 8. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.L.; Linsley, P.S. Recognizing and Avoiding SiRNA Off-Target Effects for Target Identification and Therapeutic Application. Nat. Rev. Drug Discov. 2010, 9, 57–67. [Google Scholar] [CrossRef]
- Cao, S.; Chen, G.; Yan, L.; Li, L.; Huang, X. Contribution of Dysregulated CircRNA_100876 to Proliferation and Metastasis of Esophageal Squamous Cell Carcinoma. OncoTargets Ther. 2018, 11, 7385–7394. [Google Scholar] [CrossRef]
- Lee, K.; Jang, B.; Lee, Y.; Suh, E.; Yoo, J.; Lee, M.; Lee, J.; Lee, H. The Cutting-Edge Technologies of SiRNA Delivery and Their Application in Clinical Trials. Arch. Pharm. Res. 2018, 41, 867–874. [Google Scholar] [CrossRef]
- Ouvrard, J.; Muniz, L.; Nicolas, E.; Trouche, D. Small Interfering RNAs Targeting a Chromatin-Associated RNA Induce Its Transcriptional Silencing in Human Cells. Mol. Cell Biol. 2022, 42, e00271-22. [Google Scholar] [CrossRef]
- Mahmoodi Chalbatani, G.; Dana, H.; Gharagouzloo, E.; Grijalvo, S.; Eritja, R.; Logsdon, C.D.; Memari, F.; Miri, S.R.; Rezvani Rad, M.; Marmari, V. Small Interfering RNAs (SiRNAs) in Cancer Therapy: A Nano-Based Approach. Int. J. Nanomed. 2019, 14, 3111–3128. [Google Scholar] [CrossRef]
- Vinnikov, I.A.; Hajdukiewicz, K.; Reymann, J.; Beneke, J.; Czajkowski, R.; Roth, L.C.; Novak, M.; Roller, A.; Dorner, N.; Starkuviene, V.; et al. Hypothalamic MiR-103 Protects from Hyperphagic Obesity in Mice. J. Neurosci. 2014, 34, 10659–10674. [Google Scholar] [CrossRef] [PubMed]
- Mook, O.R.; Baas, F.; de Wissel, M.B.; Fluiter, K. Evaluation of Locked Nucleic Acid–Modified Small Interfering RNA in Vitro and in Vivo. Mol. Cancer Ther. 2007, 6, 833–843. [Google Scholar] [CrossRef] [PubMed]
- Bramsen, J.B.; Kjems, J. Engineering Small Interfering RNAs by Strategic Chemical Modification. In siRNA Design: Methods and Protocols; Humana Press: Totowa, NJ, USA, 2013; pp. 87–109. [Google Scholar]
- Jackson, A.L.; Burchard, J.; Schelter, J.; Chau, B.N.; Cleary, M.; Lim, L.; Linsley, P.S. Widespread SiRNA “off-Target” Transcript Silencing Mediated by Seed Region Sequence Complementarity. RNA 2006, 12, 1179–1187. [Google Scholar] [CrossRef]
- Paddison, P.J.; Caudy, A.A.; Bernstein, E.; Hannon, G.J.; Conklin, D.S. Short Hairpin RNAs (ShRNAs) Induce Sequence-Specific Silencing in Mammalian Cells. Genes Dev. 2002, 16, 948–958. [Google Scholar] [CrossRef] [PubMed]
- Borel, F.; Kay, M.A.; Mueller, C. Recombinant AAV as a Platform for Translating the Therapeutic Potential of RNA Interference. Mol. Ther. 2014, 22, 692–701. [Google Scholar] [CrossRef]
- Brandt, M.R.G.; Kirste, A.G.; Pozzuto, T.; Schubert, S.; Kandolf, R.; Fechner, H.; Bock, C.-T.; Kurreck, J. Adenovirus Vector-Mediated RNA Interference for the Inhibition of Human Parvovirus B19 Replication. Virus Res. 2013, 176, 155–160. [Google Scholar] [CrossRef]
- Kasar, S.; Salerno, E.; Yuan, Y.; Underbayev, C.; Vollenweider, D.; Laurindo, M.F.; Fernandes, H.; Bonci, D.; Addario, A.; Mazzella, F.; et al. Systemic in Vivo Lentiviral Delivery of MiR-15a/16 Reduces Malignancy in the NZB de Novo Mouse Model of Chronic Lymphocytic Leukemia. Genes Immun. 2012, 13, 109–119. [Google Scholar] [CrossRef]
- Baum, C.; Kustikova, O.; Modlich, U.; Li, Z.; Fehse, B. Mutagenesis and Oncogenesis by Chromosomal Insertion of Gene Transfer Vectors. Hum. Gene Ther. 2006, 17, 253–263. [Google Scholar] [CrossRef]
- Hendrickx, R.; Stichling, N.; Koelen, J.; Kuryk, L.; Lipiec, A.; Greber, U.F. Innate Immunity to Adenovirus. Hum. Gene Ther. 2014, 25, 265–284. [Google Scholar] [CrossRef]
- Haldrup, S.H.; Fabian-Jessing, B.K.; Jakobsen, T.S.; Lindholm, A.B.; Adsersen, R.L.; Aagaard, L.; Bek, T.; Askou, A.L.; Corydon, T.J. Subretinal AAV Delivery of RNAi-Therapeutics Targeting VEGFA Reduces Choroidal Neovascularization in a Large Animal Model. Mol. Ther. Methods Clin. Dev. 2024, 32, 101242. [Google Scholar] [CrossRef]
- Bie, Y.; Zhang, J.; Chen, J.; Zhang, Y.; Huang, M.; Zhang, L.; Zhou, X.; Qiu, Y. Design of Antiviral AGO2-Dependent Short Hairpin RNAs. Virol. Sin. 2024, 39, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Alsing, S.; Doktor, T.K.; Askou, A.L.; Jensen, E.G.; Ahmadov, U.; Kristensen, L.S.; Andresen, B.S.; Aagaard, L.; Corydon, T.J. VEGFA-Targeting MiR-AgshRNAs Combine Efficacy with Specificity and Safety for Retinal Gene Therapy. Mol. Ther. Nucleic Acids 2022, 28, 58–76. [Google Scholar] [CrossRef]
- Kaiser, P.K.; Symons, R.C.A.; Shah, S.M.; Quinlan, E.J.; Tabandeh, H.; Do, D.V.; Reisen, G.; Lockridge, J.A.; Short, B.; Guerciolini, R.; et al. RNAi-Based Treatment for Neovascular Age-Related Macular Degeneration by Sirna-027. Am. J. Ophthalmol. 2010, 150, 33–39.e2. [Google Scholar] [CrossRef]
- Zuckerman, J.E.; Gritli, I.; Tolcher, A.; Heidel, J.D.; Lim, D.; Morgan, R.; Chmielowski, B.; Ribas, A.; Davis, M.E.; Yen, Y. Correlating Animal and Human Phase Ia/Ib Clinical Data with CALAA-01, a Targeted, Polymer-Based Nanoparticle Containing SiRNA. Proc. Natl. Acad. Sci. USA 2014, 111, 11449–11454. [Google Scholar] [CrossRef]
- Davis, M.E.; Zuckerman, J.E.; Choi, C.H.J.; Seligson, D.; Tolcher, A.; Alabi, C.A.; Yen, Y.; Heidel, J.D.; Ribas, A. Evidence of RNAi in Humans from Systemically Administered SiRNA via Targeted Nanoparticles. Nature 2010, 464, 1067–1070. [Google Scholar] [CrossRef] [PubMed]
- Hattab, D.; Gazzali, A.M.; Bakhtiar, A. Clinical Advances of SiRNA-Based Nanotherapeutics for Cancer Treatment. Pharmaceutics 2021, 13, 1009. [Google Scholar] [CrossRef]
- Zhou, D.; Zhai, X.; Zhang, R. Ribonucleotide Reductase Regulatory Subunit M2 (RRM2) as a Potential Sero-Diagnostic Biomarker in Non-Small Cell Lung Cancer. PLoS ONE 2023, 18, e0291461. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Z.; Zhou, Z.; Chang, Y.; Liu, Y.; Shen, Y.; Li, Q.; Zhang, L. Ribonucleotide Reductase M2 (RRM2): Regulation, Function and Targeting Strategy in Human Cancer. Genes Dis. 2024, 11, 218–233. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Liu, B.; Liu, W.; Ma, Y.; Cao, Y.; Yan, S.; Zhang, P.; Zhou, L.; Zhan, Q.; et al. Targeting LncRNA16 by GalNAc-SiRNA Conjugates Facilitates Chemotherapeutic Sensibilization via the HBB/NDUFAF5/ROS Pathway. Sci. China Life Sci 2024, 67, 663–679. [Google Scholar] [CrossRef]
- Connerty, P.; Moles, E.; de Bock, C.E.; Jayatilleke, N.; Smith, J.L.; Meshinchi, S.; Mayoh, C.; Kavallaris, M.; Lock, R.B. Development of SiRNA-Loaded Lipid Nanoparticles Targeting Long Non-Coding RNA LINC01257 as a Novel and Safe Therapeutic Approach for t(8;21) Pediatric Acute Myeloid Leukemia. Pharmaceutics 2021, 13, 1681. [Google Scholar] [CrossRef]
- Miao, Z.; Li, J.; Wang, Y.; Shi, M.; Gu, X.; Zhang, X.; Wei, F.; Tang, X.; Zheng, L.; Xing, Y. Hsa_circ_0136666 Stimulates Gastric Cancer Progression and Tumor Immune Escape by Regulating the MiR-375/PRKDC Axis and PD-L1 Phosphorylation. Mol. Cancer 2023, 22, 205. [Google Scholar] [CrossRef]
- Dong, J.; Zheng, Z.; Zhou, M.; Wang, Y.; Chen, J.; Cen, J.; Cao, T.; Yang, T.; Xu, Y.; Shu, G.; et al. EGCG-LYS Fibrils-Mediated CircMAP2K2 Silencing Decreases the Proliferation and Metastasis Ability of Gastric Cancer Cells in Vitro and in Vivo. Adv. Sci. 2023, 10, 2304075. [Google Scholar] [CrossRef]
- You, S.; Luo, Z.; Cheng, N.; Wu, M.; Lai, Y.; Wang, F.; Zheng, X.; Wang, Y.; Liu, X.; Liu, J.; et al. Magnetically Responsive Nanoplatform Targeting CircRNA Circ_0058051 Inhibits Hepatocellular Carcinoma Progression. Drug Deliv. Transl. Res. 2023, 13, 782–794. [Google Scholar] [CrossRef] [PubMed]
- Rouget, C.; Papin, C.; Boureux, A.; Meunier, A.-C.; Franco, B.; Robine, N.; Lai, E.C.; Pelisson, A.; Simonelig, M. Maternal MRNA Deadenylation and Decay by the PiRNA Pathway in the Early Drosophila Embryo. Nature 2010, 467, 1128–1132. [Google Scholar] [CrossRef] [PubMed]
- Williams, Z.; Morozov, P.; Mihailovic, A.; Lin, C.; Puvvula, P.K.; Juranek, S.; Rosenwaks, Z.; Tuschl, T. Discovery and Characterization of PiRNAs in the Human Fetal Ovary. Cell Rep. 2015, 13, 854–863. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Munafò, M.; Ciabrelli, F.; Eastwood, E.L.; Fabry, M.H.; Kneuss, E.; Hannon, G.J. PiRNA-Guided Genome Defense: From Biogenesis to Silencing. Annu. Rev. Genet. 2018, 52, 131–157. [Google Scholar] [CrossRef]
- Jensen, S.; Brasset, E.; Parey, E.; Roest Crollius, H.; Sharakhov, I.V.; Vaury, C. Conserved Small Nucleotidic Elements at the Origin of Concerted PiRNA Biogenesis from Genes and LncRNAs. Cells 2020, 9, 1491. [Google Scholar] [CrossRef]
- Weigert, N.; Schweiger, A.-L.; Gross, J.; Matthes, M.; Corbacioglu, S.; Sommer, G.; Heise, T. Detection of a 7SL RNA-Derived Small Non-Coding RNA Using Molecular Beacons in Vitro and in Cells. Biol. Chem. 2023, 404, 1123–1136. [Google Scholar] [CrossRef]
- He, X.; Chen, X.; Zhang, X.; Duan, X.; Pan, T.; Hu, Q.; Zhang, Y.; Zhong, F.; Liu, J.; Zhang, H.; et al. An Lnc RNA (GAS5)/SnoRNA-Derived PiRNA Induces Activation of TRAIL Gene by Site-Specifically Recruiting MLL/COMPASS-like Complexes. Nucleic Acids Res. 2015, 43, 3712–3725. [Google Scholar] [CrossRef]
- Zhong, F.; Zhou, N.; Wu, K.; Guo, Y.; Tan, W.; Zhang, H.; Zhang, X.; Geng, G.; Pan, T.; Luo, H.; et al. A SnoRNA-Derived PiRNA Interacts with Human Interleukin-4 Pre-MRNA and Induces Its Decay in Nuclear Exosomes. Nucleic Acids Res. 2015, 43, 10474–10491. [Google Scholar] [CrossRef]
- Sun, Y.H.; Wang, R.H.; Du, K.; Zhu, J.; Zheng, J.; Xie, L.H.; Pereira, A.A.; Zhang, C.; Ricci, E.P.; Li, X.Z. Coupled Protein Synthesis and Ribosome-Guided PiRNA Processing on MRNAs. Nat. Commun. 2021, 12, 5970. [Google Scholar] [CrossRef]
- Sun, Y.H.; Lee, B.; Li, X.Z. The Birth of PiRNAs: How Mammalian PiRNAs Are Produced, Originated, and Evolved. Mamm. Genome 2022, 33, 293–311. [Google Scholar] [CrossRef] [PubMed]
- Rearick, D.; Prakash, A.; McSweeny, A.; Shepard, S.S.; Fedorova, L.; Fedorov, A. Critical Association of NcRNA with Introns. Nucleic Acids Res. 2011, 39, 2357–2366. [Google Scholar] [CrossRef]
- Fu, Q.; Wang, P.J. Mammalian PiRNAs: Biogenesis, Function, and Mysteries. Spermatogenesis 2014, 4, e27889. [Google Scholar] [CrossRef]
- Guo, B.; Li, D.; Du, L.; Zhu, X. PiRNAs: Biogenesis and Their Potential Roles in Cancer. Cancer Metastasis Rev. 2020, 39, 567–575. [Google Scholar] [CrossRef] [PubMed]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-Interacting RNAs: Small RNAs with Big Functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Hannon, G.J. One Loop to Rule Them All: The Ping-Pong Cycle and PiRNA-Guided Silencing. Trends Biochem. Sci. 2016, 41, 324–337. [Google Scholar] [CrossRef]
- Pippadpally, S.; Venkatesh, T. Deciphering PiRNA Biogenesis through Cytoplasmic Granules, Mitochondria and Exosomes. Arch. Biochem. Biophys. 2020, 695, 108597. [Google Scholar] [CrossRef]
- Perera, B.P.U.; Tsai, Z.T.-Y.; Colwell, M.L.; Jones, T.R.; Goodrich, J.M.; Wang, K.; Sartor, M.A.; Faulk, C.; Dolinoy, D.C. Somatic Expression of PiRNA and Associated Machinery in the Mouse Identifies Short, Tissue-Specific PiRNA. Epigenetics 2019, 14, 504–521. [Google Scholar] [CrossRef]
- Riquelme, I.; Pérez-Moreno, P.; Letelier, P.; Brebi, P.; Roa, J.C. The Emerging Role of PIWI-Interacting RNAs (PiRNAs) in Gastrointestinal Cancers: An Updated Perspective. Cancers 2021, 14, 202. [Google Scholar] [CrossRef]
- Patel, M.Z.; Jiang, Y.; Kakumani, P.K. Somatic PiRNA and PIWI-Mediated Post-Transcriptional Gene Regulation in Stem Cells and Disease. Front. Cell Dev. Biol. 2024, 12, 1495035. [Google Scholar] [CrossRef]
- Jia, D.-D.; Jiang, H.; Zhang, Y.-F.; Zhang, Y.; Qian, L.-L.; Zhang, Y.-F. The Regulatory Function of PiRNA/PIWI Complex in Cancer and Other Human Diseases: The Role of DNA Methylation. Int. J. Biol. Sci. 2022, 18, 3358–3373. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Xiong, Y.; Konno, Y.; Ihira, K.; Xu, D.; Kobayashi, N.; Yue, J.; Watari, H. Critical Roles of PIWIL1 in Human Tumors: Expression, Functions, Mechanisms, and Potential Clinical Implications. Front. Cell Dev. Biol. 2021, 9, 656993. [Google Scholar] [CrossRef]
- Wu, X.; Pan, Y.; Fang, Y.; Zhang, J.; Xie, M.; Yang, F.; Yu, T.; Ma, P.; Li, W.; Shu, Y. The Biogenesis and Functions of PiRNAs in Human Diseases. Mol. Ther. Nucleic Acids 2020, 21, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Mohn, F.; Handler, D.; Brennecke, J. PiRNA-Guided Slicing Specifies Transcripts for Zucchini-Dependent, Phased PiRNA Biogenesis. Science 2015, 348, 812–817. [Google Scholar] [CrossRef]
- Han, B.W.; Wang, W.; Li, C.; Weng, Z.; Zamore, P.D. PiRNA-Guided Transposon Cleavage Initiates Zucchini-Dependent, Phased PiRNA Production. Science 2015, 348, 817–821. [Google Scholar] [CrossRef]
- Aravin, A.A.; Naumova, N.M.; Tulin, A.V.; Vagin, V.V.; Rozovsky, Y.M.; Gvozdev, V.A. Double-Stranded RNA-Mediated Silencing of Genomic Tandem Repeats and Transposable Elements in the D. Melanogaster Germline. Curr. Biol. 2001, 11, 1017–1027. [Google Scholar] [CrossRef]
- Girard, A.; Sachidanandam, R.; Hannon, G.J.; Carmell, M.A. A Germline-Specific Class of Small RNAs Binds Mammalian Piwi Proteins. Nature 2006, 442, 199–202. [Google Scholar] [CrossRef]
- Wang, J.; Shi, Y.; Zhou, H.; Zhang, P.; Song, T.; Ying, Z.; Yu, H.; Li, Y.; Zhao, Y.; Zeng, X.; et al. PiRBase: Integrating PiRNA Annotation in All Aspects. Nucleic Acids Res. 2022, 50, D265–D272. [Google Scholar] [CrossRef]
- Guo, C.; Wang, X.; Ren, H. Databases and Computational Methods for the Identification of PiRNA-Related Molecules: A Survey. Comput. Struct. Biotechnol. J. 2024, 23, 813–833. [Google Scholar] [CrossRef]
- Martinez, V.D.; Vucic, E.A.; Thu, K.L.; Hubaux, R.; Enfield, K.S.S.; Pikor, L.A.; Becker-Santos, D.D.; Brown, C.J.; Lam, S.; Lam, W.L. Unique Somatic and Malignant Expression Patterns Implicate PIWI-Interacting RNAs in Cancer-Type Specific Biology. Sci. Rep. 2015, 5, 10423. [Google Scholar] [CrossRef]
- Lima, J.R.S.; Azevedo-Pinheiro, J.; Andrade, R.B.; Khayat, A.S.; de Assumpção, P.P.; Ribeiro-dos-Santos, Â.; Batista dos Santos, S.E.; Moreira, F.C. Identification and Characterization of Polymorphisms in PiRNA Regions. Curr. Issues Mol. Biol. 2022, 44, 942–951. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef] [PubMed]
- Gainetdinov, I.; Skvortsova, Y.; Kondratieva, S.; Funikov, S.; Azhikina, T. Two Modes of Targeting Transposable Elements by PiRNA Pathway in Human Testis. RNA 2017, 23, 1614–1625. [Google Scholar] [CrossRef] [PubMed]
- Dias Mirandela, M.; Zoch, A.; Leismann, J.; Webb, S.; Berrens, R.V.; Valsakumar, D.; Kabayama, Y.; Auchynnikava, T.; Schito, M.; Chowdhury, T.; et al. Two-Factor Authentication Underpins the Precision of the PiRNA Pathway. Nature 2024, 634, 979–985. [Google Scholar] [CrossRef]
- Zoch, A.; Konieczny, G.; Auchynnikava, T.; Stallmeyer, B.; Rotte, N.; Heep, M.; Berrens, R.V.; Schito, M.; Kabayama, Y.; Schöpp, T.; et al. C19ORF84 Connects PiRNA and DNA Methylation Machineries to Defend the Mammalian Germ Line. Mol. Cell 2024, 84, 1021–1035.e11. [Google Scholar] [CrossRef]
- Rajasethupathy, P.; Antonov, I.; Sheridan, R.; Frey, S.; Sander, C.; Tuschl, T.; Kandel, E.R. A Role for Neuronal PiRNAs in the Epigenetic Control of Memory-Related Synaptic Plasticity. Cell 2012, 149, 693–707. [Google Scholar] [CrossRef]
- Wu, D.; Fu, H.; Zhou, H.; Su, J.; Zhang, F.; Shen, J. Effects of Novel NcRNA Molecules, P15-piRNAs, on the Methylation of DNA and Histone H3 of the CDKN2B Promoter Region in U937 Cells. J. Cell. Biochem. 2015, 116, 2744–2754. [Google Scholar] [CrossRef]
- Sugimoto, K.; Kage, H.; Aki, N.; Sano, A.; Kitagawa, H.; Nagase, T.; Yatomi, Y.; Ohishi, N.; Takai, D. The Induction of H3K9 Methylation by PIWIL4 at the P16Ink4a Locus. Biochem. Biophys. Res. Commun. 2007, 359, 497–502. [Google Scholar] [CrossRef]
- Lu, Y.; Zhang, K.; Li, C.; Yao, Y.; Tao, D.; Liu, Y.; Zhang, S.; Ma, Y. Piwil2 Suppresses P53 by Inducing Phosphorylation of Signal Transducer and Activator of Transcription 3 in Tumor Cells. PLoS ONE 2012, 7, e30999. [Google Scholar] [CrossRef]
- Nagamori, I.; Kobayashi, H.; Nishimura, T.; Yamagishi, R.; Katahira, J.; Kuramochi-Miyagawa, S.; Kono, T.; Nakano, T. Relationship between PIWIL4-Mediated H3K4me2 Demethylation and PiRNA-Dependent DNA Methylation. Cell Rep. 2018, 25, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Goh, W.S.S.; Falciatori, I.; Tam, O.H.; Burgess, R.; Meikar, O.; Kotaja, N.; Hammell, M.; Hannon, G.J. PiRNA-Directed Cleavage of Meiotic Transcripts Regulates Spermatogenesis. Genes Dev. 2015, 29, 1032–1044. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Mai, D.; Zhang, B.; Jiang, X.; Zhang, J.; Bai, R.; Ye, Y.; Li, M.; Pan, L.; Su, J.; et al. PIWI-Interacting RNA-36712 Restrains Breast Cancer Progression and Chemoresistance by Interaction with SEPW1 Pseudogene SEPW1P RNA. Mol. Cancer 2019, 18, 9. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zheng, J.; Xue, Y.; Yu, H.; Gong, W.; Wang, P.; Li, Z.; Liu, Y. PIWIL3/OIP5-AS1/MiR-367-3p/CEBPA Feedback Loop Regulates the Biological Behavior of Glioma Cells. Theranostics 2018, 8, 1084–1105. [Google Scholar] [CrossRef]
- Peng, L.; Song, L.; Liu, C.; Lv, X.; Li, X.; Jie, J.; Zhao, D.; Li, D. PiR-55490 Inhibits the Growth of Lung Carcinoma by Suppressing MTOR Signaling. Tumor Biol. 2016, 37, 2749–2756. [Google Scholar] [CrossRef]
- Gou, L.-T.; Dai, P.; Yang, J.-H.; Xue, Y.; Hu, Y.-P.; Zhou, Y.; Kang, J.-Y.; Wang, X.; Li, H.; Hua, M.-M.; et al. Pachytene PiRNAs Instruct Massive MRNA Elimination during Late Spermiogenesis. Cell Res. 2014, 24, 680–700. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Patel, H.; Chen, J.; Wang, J.; Chen, Z.-S.; Wang, H. Epigenetic Modification of M6A Regulator Proteins in Cancer. Mol. Cancer 2023, 22, 102. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gao, L.; Cheng, L.; Lv, G.; Sun, B.; Wang, G.; Tang, Q. The Roles of N6-Methyladenosine and Its Target Regulatory Noncoding RNAs in Tumors: Classification, Mechanisms, and Potential Therapeutic Implications. Exp. Mol. Med. 2023, 55, 487–501. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef]
- Gao, X.-Q.; Zhang, Y.-H.; Liu, F.; Ponnusamy, M.; Zhao, X.-M.; Zhou, L.-Y.; Zhai, M.; Liu, C.-Y.; Li, X.-M.; Wang, M.; et al. The PiRNA CHAPIR Regulates Cardiac Hypertrophy by Controlling METTL3-Dependent N6-Methyladenosine Methylation of Parp10 MRNA. Nat. Cell Biol. 2020, 22, 1319–1331. [Google Scholar] [CrossRef]
- Han, H.; Fan, G.; Song, S.; Jiang, Y.; Qian, C.; Zhang, W.; Su, Q.; Xue, X.; Zhuang, W.; Li, B. PiRNA-30473 Contributes to Tumorigenesis and Poor Prognosis by Regulating M6A RNA Methylation in DLBCL. Blood 2021, 137, 1603–1614. [Google Scholar] [CrossRef] [PubMed]
- Mai, D.; Ding, P.; Tan, L.; Zhang, J.; Pan, Z.; Bai, R.; Li, C.; Li, M.; Zhou, Y.; Tan, W.; et al. PIWI-Interacting RNA-54265 Is Oncogenic and a Potential Therapeutic Target in Colorectal Adenocarcinoma. Theranostics 2018, 8, 5213–5230. [Google Scholar] [CrossRef]
- Yin, J.; Jiang, X.; Qi, W.; Ji, C.; Xie, X.; Zhang, D.; Cui, Z.; Wang, C.; Bai, Y.; Wang, J.; et al. PiR-823 Contributes to Colorectal Tumorigenesis by Enhancing the Transcriptional Activity of HSF1. Cancer Sci. 2017, 108, 1746–1756. [Google Scholar] [CrossRef] [PubMed]
- Das, B.; Jain, N.; Mallick, B. PiR-39980 Mediates Doxorubicin Resistance in Fibrosarcoma by Regulating Drug Accumulation and DNA Repair. Commun. Biol. 2021, 4, 1312. [Google Scholar] [CrossRef]
- Ou, B.; Liu, Y.; Gao, Z.; Xu, J.; Yan, Y.; Li, Y.; Zhang, J. Senescent Neutrophils-Derived Exosomal PiRNA-17560 Promotes Chemoresistance and EMT of Breast Cancer via FTO-Mediated M6A Demethylation. Cell Death Dis. 2022, 13, 905. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Yang, M.; Wei, Q.; Song, F.; Zhang, Y.; Wang, X.; Liu, B.; Li, J. Novel Evidence for Oncogenic PiRNA-823 as a Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. J. Cell. Mol. Med. 2020, 24, 9028–9040. [Google Scholar] [CrossRef]
- Zhang, W.; Zheng, Z.; Wang, K.; Mao, W.; Li, X.; Wang, G.; Zhang, Y.; Huang, J.; Zhang, N.; Wu, P.; et al. PiRNA-1742 Promotes Renal Cell Carcinoma Malignancy by Regulating USP8 Stability through Binding to HnRNPU and Thereby Inhibiting MUC12 Ubiquitination. Exp. Mol. Med. 2023, 55, 1258–1271. [Google Scholar] [CrossRef]
- Dong, Y.-Z.; Hu, T. Effects of MiR-143 Overexpression on Proliferation, Apoptosis, EGFR and Downstream Signaling Pathways in PC9/GR Cell Line. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 1709–1716. [Google Scholar] [CrossRef]
- Mi, T.; Tan, X.; Wang, Z.; Zhang, Z.; Jin, L.; Wang, J.; Li, M.; Wu, X.; He, D. Activation of the P53 Signaling Pathway by PiRNA-MW557525 Overexpression Induces a G0/G1 Phase Arrest Thus Inhibiting Neuroblastoma Growth. Eur. J. Med. Res. 2023, 28, 503. [Google Scholar] [CrossRef]
- Wu, L.; Huang, S.; Tian, W.; Liu, P.; Xie, Y.; Qiu, Y.; Li, X.; Tang, Y.; Zheng, S.; Sun, Y.; et al. PIWI-Interacting RNA-YBX1 Inhibits Proliferation and Metastasis by the MAPK Signaling Pathway via YBX1 in Triple-Negative Breast Cancer. Cell Death Discov. 2024, 10, 7. [Google Scholar] [CrossRef]
- Du, X.; Li, H.; Xie, X.; Shi, L.; Wu, F.; Li, G.; Lai, C.; Heng, B. PiRNA-31115 Promotes Cell Proliferation and Invasion via PI3K/AKT Pathway in Clear Cell Renal Carcinoma. Dis. Markers 2021, 2021, 6915329. [Google Scholar] [CrossRef]
- Fu, A.; Jacobs, D.I.; Hoffman, A.E.; Zheng, T.; Zhu, Y. PIWI-Interacting RNA 021285 Is Involved in Breast Tumorigenesis Possibly by Remodeling the Cancer Epigenome. Carcinogenesis 2015, 36, 1094–1102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Meng, X.; Pan, C.; Qu, F.; Gan, W.; Xiang, Z.; Han, X.; Li, D. PiR-31470 Epigenetically Suppresses the Expression of Glutathione S-Transferase Pi 1 in Prostate Cancer via DNA Methylation. Cell. Signal. 2020, 67, 109501. [Google Scholar] [CrossRef] [PubMed]
- Su, J.-F.; Zhao, F.; Gao, Z.-W.; Hou, Y.-J.; Li, Y.-Y.; Duan, L.-J.; Lun, S.-M.; Yang, H.-J.; Li, J.-K.; Dai, N.-T.; et al. PiR-823 Demonstrates Tumor Oncogenic Activity in Esophageal Squamous Cell Carcinoma through DNA Methylation Induction via DNA Methyltransferase 3B. Pathol. Res. Pract. 2020, 216, 152848. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Li, Y.; Lü, J.; Zhao, Q.; Guo, Y.; Lu, Z.; Ma, W.; Liu, P.; Pestell, R.G.; Liang, C.; et al. PiRNA-823 Is Involved in Cancer Stem Cell Regulation Through Altering DNA Methylation in Association with Luminal Breast Cancer. Front. Cell Dev. Biol. 2021, 9, 641052. [Google Scholar] [CrossRef]
- Merkerova, M.D.; Krejcik, Z. Transposable Elements and Piwi-interacting RNAs in Hemato-oncology with a Focus on Myelodysplastic Syndrome (Review). Int. J. Oncol. 2021, 59, 105. [Google Scholar] [CrossRef]
- Ernst, C.; Odom, D.T.; Kutter, C. The Emergence of PiRNAs against Transposon Invasion to Preserve Mammalian Genome Integrity. Nat. Commun. 2017, 8, 1411. [Google Scholar] [CrossRef]
- Tang, X.; Xie, X.; Wang, X.; Wang, Y.; Jiang, X.; Jiang, H. The Combination of PiR-823 and Eukaryotic Initiation Factor 3 B (EIF3B) Activates Hepatic Stellate Cells via Upregulating TGF-Β1 in Liver Fibrogenesis. Med. Sci. Monit. 2018, 24, 9151–9165. [Google Scholar] [CrossRef]
- Li, G.; Ni, A.; Tang, Y.; Li, S.; Meng, L. RNA Binding Proteins Involved in Regulation of Protein Synthesis to Initiate Biogenesis of Secondary Tumor in Hepatocellular Carcinoma in Mice. PeerJ 2020, 8, e8680. [Google Scholar] [CrossRef]
- Wu, Y.-J.; Wang, J.; Zhang, P.; Yuan, L.-X.; Ju, L.-L.; Wang, H.-X.; Chen, L.; Cao, Y.-L.; Cai, W.-H.; Ni, Y.; et al. PIWIL1 Interacting RNA PiR-017724 Inhibits Proliferation, Invasion, and Migration, and Inhibits the Development of HCC by Silencing PLIN3. Front. Oncol. 2023, 13, 1203821. [Google Scholar] [CrossRef]
- Ge, L.; Zhang, N.; Li, D.; Wu, Y.; Wang, H.; Wang, J. Circulating Exosomal Small RNAs Are Promising Non-invasive Diagnostic Biomarkers for Gastric Cancer. J. Cell Mol. Med. 2020, 24, 14502–14513. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liu, J.; Meng, A.; Zhang, L.; Wang, M.; Fan, H.; Peng, W.; Lu, J. Gastric Juice PiR-1245: A Promising Prognostic Biomarker for Gastric Cancer. J. Clin. Lab. Anal. 2020, 34, e23131. [Google Scholar] [CrossRef] [PubMed]
- Vinasco-Sandoval, T.; Moreira, F.C.; Vidal, A.F.; Pinto, P.; Ribeiro-dos-Santos, A.M.; Cruz, R.L.S.; Fonseca Cabral, G.; Anaissi, A.K.M.; Lopes, K.d.P.; Ribeiro-dos-Santos, A.; et al. Global Analyses of Expressed Piwi-Interacting RNAs in Gastric Cancer. Int. J. Mol. Sci. 2020, 21, 7656. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Wang, L.; Chen, X.; Lin, Y.; Zhang, S.; Fan, Z.; Peng, F. Impact of PIWIL1 Single Nucleotide Polymorphisms on Gastric Cancer Risk in a Chinese Population. Genet. Test. Mol. Biomark. 2023, 27, 185–192. [Google Scholar] [CrossRef]
- Sabbah, N.A.; Abdalla, W.M.; Mawla, W.A.; AbdAlMonem, N.; Gharib, A.F.; Abdul-Saboor, A.; Abdelazem, A.S.; Raafat, N. PiRNA-823 Is a Unique Potential Diagnostic Non-Invasive Biomarker in Colorectal Cancer Patients. Genes 2021, 12, 598. [Google Scholar] [CrossRef]
- Liu, Q.; Chen, Q.; Zhou, Z.; Tian, Z.; Zheng, X.; Wang, K. PiRNA-18 Inhibition Cell Proliferation, Migration and Invasion in Colorectal Cancer. Biochem. Genet. 2023, 61, 1881–1897. [Google Scholar] [CrossRef]
- Taghizadeh, M.; Jafari-Koshki, T.; Jafarlou, V.; Raeisi, M.; Alizadeh, L.; Roosta, Y.; Matin, S.; Jabari, R.; Sur, D.; Karimi, A. The Role of PiRNAs in Predicting and Prognosing in Cancer: A Focus on PiRNA-823 (a Systematic Review and Meta-Analysis). BMC Cancer 2024, 24, 484. [Google Scholar] [CrossRef]
- Li, S.; Kouznetsova, V.L.; Kesari, S.; Tsigelny, I.F. PiRNA in Machine-Learning-Based Diagnostics of Colorectal Cancer. Molecules 2024, 29, 4311. [Google Scholar] [CrossRef]
- Das, B.; Jain, N.; Mallick, B. PiR-39980 Promotes Cell Proliferation, Migration and Invasion, and Inhibits Apoptosis via Repression of SERPINB1 in Human Osteosarcoma. Biol. Cell 2020, 112, 73–91. [Google Scholar] [CrossRef]
- Liu, Y.; Dong, Y.; He, X.; Gong, A.; Gao, J.; Hao, X.; Wang, S.; Fan, Y.; Wang, Z.; Li, M.; et al. PiR-Hsa-211106 Inhibits the Progression of Lung Adenocarcinoma Through Pyruvate Carboxylase and Enhances Chemotherapy Sensitivity. Front. Oncol. 2021, 11, 651915. [Google Scholar] [CrossRef]
- Li, Y.; Dong, Y.; Zhao, S.; Gao, J.; Hao, X.; Wang, Z.; Li, M.; Wang, M.; Liu, Y.; Yu, X.; et al. Serum-Derived PiR-Hsa-164586 of Extracellular Vesicles as a Novel Biomarker for Early Diagnosis of Non-Small Cell Lung Cancer. Front. Oncol. 2022, 12, 850363. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Xu, L.; Wang, Y.; Chen, S.; Li, D.; Huo, X.; Li, R.; Zhu, X.; Chen, N.; Jin, Y.; et al. PiR-27222 Mediates PM2.5-Induced Lung Cancer by Resisting Cell PANoptosis through the WTAP/M6A Axis. Environ. Int. 2024, 190, 108928. [Google Scholar] [CrossRef]
- Xu, L.; Ma, W.; Huo, X.; Luo, J.; Li, R.; Zhu, X.; Kong, X.; Zhao, K.; Jin, Y.; Zhang, M.; et al. New Insights into the Function and Mechanisms of PiRNA PMLCPIR in Promoting PM2.5-Induced Lung Cancer. J. Adv. Res. 2024, in press. [Google Scholar] [CrossRef]
- Huang, S.; Chen, B.; Qiu, P.; Yan, Z.; Liang, Z.; Luo, K.; Huang, B.; Jiang, H. In Vitro Study of Piwi Interaction RNA-31106 Promoting Breast Carcinogenesis by Regulating METTL3-Mediated M6A RNA Methylation. Transl. Cancer Res. 2023, 12, 1588–1601. [Google Scholar] [CrossRef]
- Zhao, Q.; Qian, L.; Guo, Y.; Lü, J.; Li, D.; Xie, H.; Wang, Q.; Ma, W.; Liu, P.; Liu, Y.; et al. IL11 Signaling Mediates PiR-2158 Suppression of Cell Stemness and Angiogenesis in Breast Cancer. Theranostics 2023, 13, 2337–2349. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wang, J.; Sun, L.; Li, M.; He, X.; Jiang, J.; Zhou, Q. Piwi-Interacting RNA-651 Promotes Cell Proliferation and Migration and Inhibits Apoptosis in Breast Cancer by Facilitating DNMT1-Mediated PTEN Promoter Methylation. Cell Cycle 2021, 20, 1603–1616. [Google Scholar] [CrossRef] [PubMed]
- Ben, S.; Ding, Z.; Xin, J.; Li, F.; Cheng, Y.; Chen, S.; Fan, L.; Zhang, Q.; Li, S.; Du, M.; et al. PiRNA PROPER Suppresses DUSP1 Translation by Targeting N6-Methyladenosine-Mediated RNA Circularization to Promote Oncogenesis of Prostate Cancer. Adv. Sci. 2024, 11, 2402954. [Google Scholar] [CrossRef]
- Peng, Q.; Chen, Y.; Xie, T.; Pu, D.; Ho, V.W.-S.; Sun, J.; Liu, K.; Chan, R.C.-K.; Ding, X.; Teoh, J.Y.-C.; et al. PiRNA-4447944 Promotes Castration-Resistant Growth and Metastasis of Prostate Cancer by Inhibiting NEFH Expression through Forming the PiRNA-4447944-PIWIL2-NEFH Complex. Int. J. Biol. Sci. 2024, 20, 3638–3655. [Google Scholar] [CrossRef]
- Ding, L.; Wang, R.; Xu, W.; Shen, D.; Cheng, S.; Wang, H.; Lu, Z.; Zheng, Q.; Wang, L.; Xia, L.; et al. PIWI-Interacting RNA 57125 Restrains Clear Cell Renal Cell Carcinoma Metastasis by Downregulating CCL3 Expression. Cell Death Discov. 2021, 7, 333. [Google Scholar] [CrossRef]
- Hu, H.; Lu, J.; Xu, M.; Wang, J.; Zhang, Y.; Yang, S.; Wang, X.; Wang, M.; Xie, W.; Xu, W.; et al. PiR-Hsa-23533 Promotes Malignancy in Head and Neck Squamous Cell Carcinoma via USP7. Transl. Oncol. 2024, 45, 101990. [Google Scholar] [CrossRef]
- Yan, Y.; Tian, D.; Zhao, B.; Li, Z.; Huang, Z.; Li, K.; Chen, X.; Zhou, L.; Feng, Y.; Yang, Z. PiR-1919609 Is an Ideal Potential Target for Reversing Platinum Resistance in Ovarian Cancer. Technol. Cancer Res. Treat. 2024, 23, 15330338241249692. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, L.; Zu, W.; Jing, J.; Liu, G.; Sun, T.; Xie, Q. PIWI-Interacting RNA-17458 Is Oncogenic and a Potential Therapeutic Target in Cervical Cancer. J. Cancer 2023, 14, 1648–1659. [Google Scholar] [CrossRef]
- Zhong, Y.; Tian, Y.; Wang, Y.; Bai, J.; Long, Q.; Yan, L.; Gong, Z.; Gao, W.; Tang, Q. Small Extracellular Vesicle PiR-Hsa-30937 Derived from Pancreatic Neuroendocrine Neoplasms Upregulates CD276 in Macrophages to Promote Immune Evasion. Cancer Immunol. Res. 2024, 12, 840–853. [Google Scholar] [CrossRef]
- Nasseri, S.; Sharifi, M.; Mehrzad, V. Effects of Hsa-PiR-32877 Suppression with Antisense LNA GapmeRs on the Proliferation and Apoptosis of Human Acute Myeloid Leukemia Cells. Int. J. Mol. Cell Med. 2023, 12, 18–29. [Google Scholar] [CrossRef]
- Tosar, J.P. Letter to Editor Regarding “PiR-36249 and DHX36 Together Inhibit Testicular Cancer Cells Progression by Upregulating OAS2”. Noncoding RNA Res. 2023, 8, 589–590. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, N. PIWI Interacting RNA-13643 Contributes to Papillary Thyroid Cancer Development through Acting as a Novel Oncogene by Facilitating PRMT1 Mediated GLI1 Methylation. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2023, 1867, 130453. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, T.; Mallick, B. FDFT1 Repression by PiR-39980 Prevents Oncogenesis by Regulating Proliferation and Apoptosis through Hypoxia in Tongue Squamous Cell Carcinoma. Life Sci. 2023, 329, 121954. [Google Scholar] [CrossRef] [PubMed]
- Saha, B.; Chakravarty, S.; Ray, S.; Saha, H.; Das, K.; Ghosh, I.; Mallick, B.; Biswas, N.; Goswami, S. Correlating Tissue and Plasma-specific PiRNA Changes to Predict Their Possible Role in Pancreatic Malignancy and Chronic Inflammation. Biomed. Rep. 2024, 21, 186. [Google Scholar] [CrossRef]
- Xue, J.; Qin, S.; Ren, N.; Guo, B.; Shi, X.; Jia, E. Extracellular Vesicle Biomarkers in Circulation for the Diagnosis of Gastric Cancer: A Systematic Review and Meta-analysis. Oncol. Lett. 2023, 26, 423. [Google Scholar] [CrossRef]
- Rui, T.; Wang, K.; Xiang, A.; Guo, J.; Tang, N.; Jin, X.; Lin, Y.; Liu, J.; Zhang, X. Serum Exosome-Derived PiRNAs Could Be Promising Biomarkers for HCC Diagnosis. Int. J. Nanomed. 2023, 18, 1989–2001. [Google Scholar] [CrossRef]
- Li, J.; Wang, N.; Zhang, F.; Jin, S.; Dong, Y.; Dong, X.; Chen, Y.; Kong, X.; Tong, Y.; Mi, Q.; et al. PIWI-interacting RNAs Are Aberrantly Expressed and May Serve as Novel Biomarkers for Diagnosis of Lung Adenocarcinoma. Thorac. Cancer 2021, 12, 2468–2477. [Google Scholar] [CrossRef] [PubMed]
- Nayak, R.; Chattopadhyay, T.; Gupta, P.; Mallick, B. Integrative Analysis of Small Non-Coding RNAs Predicts a PiRNA/MiRNA-CCND1/BRAF/HRH1/ATXN3 Regulatory Circuit That Drives Oncogenesis in Glioblastoma. Mol. Omics 2023, 19, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Chiu, P.K.-F.; Wong, C.Y.-P.; Cheng, C.K.-L.; Teoh, J.Y.-C.; Ng, C.-F. Identification of PiRNA Targets in Urinary Extracellular Vesicles for the Diagnosis of Prostate Cancer. Diagnostics 2021, 11, 1828. [Google Scholar] [CrossRef] [PubMed]
- Chang, Z.; Ji, G.; Huang, R.; Chen, H.; Gao, Y.; Wang, W.; Sun, X.; Zhang, J.; Zheng, J.; Wei, Q. PIWI-Interacting RNAs PiR-13643 and PiR-21238 Are Promising Diagnostic Biomarkers of Papillary Thyroid Carcinoma. Aging 2020, 12, 9292–9310. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, H.; Ma, D.; Mu, Y.; Tan, X.; Hao, Q.; Feng, L.; Liang, J.; Xin, W.; Chen, Y.; et al. Serum PIWI-Interacting RNAs PiR-020619 and PiR-020450 Are Promising Novel Biomarkers for Early Detection of Colorectal Cancer. Cancer Epidemiol. Biomark. Prev. 2020, 29, 990–998. [Google Scholar] [CrossRef]
- Baba, A.B.; Rah, B.; Bhat, G.R.; Mushtaq, I.; Parveen, S.; Hassan, R.; Hameed Zargar, M.; Afroze, D. Transforming Growth Factor-Beta (TGF-β) Signaling in Cancer-A Betrayal Within. Front. Pharmacol. 2022, 13, 791272. [Google Scholar] [CrossRef]
- Öner, Ç.; Turgut Coşan, D.; Çolak, E. Estrogen and Androgen Hormone Levels Modulate the Expression of PIWI Interacting RNA in Prostate and Breast Cancer. PLoS ONE 2016, 11, e0159044. [Google Scholar] [CrossRef]
- Cheng, J.; Deng, H.; Xiao, B.; Zhou, H.; Zhou, F.; Shen, Z.; Guo, J. PiR-823, a Novel Non-Coding Small RNA, Demonstrates in Vitro and in Vivo Tumor Suppressive Activity in Human Gastric Cancer Cells. Cancer Lett. 2012, 315, 12–17. [Google Scholar] [CrossRef]
- Lumbreras, B.; Parker, L.A.; Caballero-Romeu, J.P.; Gómez-Pérez, L.; Puig-García, M.; López-Garrigós, M.; García, N.; Hernández-Aguado, I. Variables Associated with False-Positive PSA Results: A Cohort Study with Real-World Data. Cancers 2022, 15, 261. [Google Scholar] [CrossRef]
- Zhang, X.-O.; Dong, R.; Zhang, Y.; Zhang, J.-L.; Luo, Z.; Zhang, J.; Chen, L.-L.; Yang, L. Diverse Alternative Back-Splicing and Alternative Splicing Landscape of Circular RNAs. Genome Res. 2016, 26, 1277–1287. [Google Scholar] [CrossRef]
- Karousi, P.; Artemaki, P.I.; Sotiropoulou, C.D.; Christodoulou, S.; Scorilas, A.; Kontos, C.K. Identification of Two Novel Circular RNAs Deriving from BCL2L12 and Investigation of Their Potential Value as a Molecular Signature in Colorectal Cancer. Int. J. Mol. Sci. 2020, 21, 8867. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, X.-O.; Chen, T.; Xiang, J.-F.; Yin, Q.-F.; Xing, Y.-H.; Zhu, S.; Yang, L.; Chen, L.-L. Circular Intronic Long Noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef]
- Zhang, X.-O.; Wang, H.-B.; Zhang, Y.; Lu, X.; Chen, L.-L.; Yang, L. Complementary Sequence-Mediated Exon Circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs Are the Predominant Transcript Isoform from Hundreds of Human Genes in Diverse Cell Types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef]
- Huang, Y.; Zhu, Q. Mechanisms Regulating Abnormal Circular RNA Biogenesis in Cancer. Cancers 2021, 13, 4185. [Google Scholar] [CrossRef] [PubMed]
- Pervouchine, D.D. Circular Exonic RNAs: When RNA Structure Meets Topology. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2019, 1862, 194384. [Google Scholar] [CrossRef]
- Liang, D.; Tatomer, D.C.; Luo, Z.; Wu, H.; Yang, L.; Chen, L.-L.; Cherry, S.; Wilusz, J.E. The Output of Protein-Coding Genes Shifts to Circular RNAs When the Pre-MRNA Processing Machinery Is Limiting. Mol. Cell 2017, 68, 940–954.e3. [Google Scholar] [CrossRef]
- Suzuki, H.; Kameyama, T.; Ohe, K.; Tsukahara, T.; Mayeda, A. Nested Introns in an Intron: Evidence of Multi-step Splicing in a Large Intron of the Human Dystrophin Pre-mRNA. FEBS Lett. 2013, 587, 555–561. [Google Scholar] [CrossRef]
- Eger, N.; Schoppe, L.; Schuster, S.; Laufs, U.; Boeckel, J.-N. Circular RNA Splicing. In Circular RNAs: Biogenesis and Functions; Springer: Singapore, 2018; pp. 41–52. [Google Scholar]
- Zaphiropoulos, P.G. Circular RNAs from Transcripts of the Rat Cytochrome P450 2C24 Gene: Correlation with Exon Skipping. Proc. Natl. Acad. Sci. USA 1996, 93, 6536–6541. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs Are Abundant, Conserved, and Associated with ALU Repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Liang, D.; Wilusz, J.E. Short Intronic Repeat Sequences Facilitate Circular RNA Production. Genes Dev. 2014, 28, 2233–2247. [Google Scholar] [CrossRef] [PubMed]
- Xia, P.; Wang, S.; Ye, B.; Du, Y.; Li, C.; Xiong, Z.; Qu, Y.; Fan, Z. A Circular RNA Protects Dormant Hematopoietic Stem Cells from DNA Sensor CGAS-Mediated Exhaustion. Immunity 2018, 48, 688–701.e7. [Google Scholar] [CrossRef]
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G.; et al. Circular RNA Profiling Reveals an Abundant CircHIPK3 That Regulates Cell Growth by Sponging Multiple MiRNAs. Nat. Commun. 2016, 7, 11215. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Liao, X.; Gong, Y.; He, J.; Zhou, J.-K.; Tan, S.; Pu, W.; Huang, C.; Wei, Y.-Q.; Peng, Y. Circular RNA F-CircSR Derived from SLC34A2-ROS1 Fusion Gene Promotes Cell Migration in Non-Small Cell Lung Cancer. Mol. Cancer 2019, 18, 98. [Google Scholar] [CrossRef] [PubMed]
- Guarnerio, J.; Bezzi, M.; Jeong, J.C.; Paffenholz, S.V.; Berry, K.; Naldini, M.M.; Lo-Coco, F.; Tay, Y.; Beck, A.H.; Pandolfi, P.P. Oncogenic Role of Fusion-CircRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell 2016, 165, 289–302. [Google Scholar] [CrossRef]
- Rybak-Wolf, A.; Stottmeister, C.; Glažar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef]
- Wang, Z.-L.; Li, B.; Luo, Y.-X.; Lin, Q.; Liu, S.-R.; Zhang, X.-Q.; Zhou, H.; Yang, J.-H.; Qu, L.-H. Comprehensive Genomic Characterization of RNA-Binding Proteins across Human Cancers. Cell Rep. 2018, 22, 286–298. [Google Scholar] [CrossRef]
- Neelamraju, Y.; Hashemikhabir, S.; Janga, S.C. The Human RBPome: From Genes and Proteins to Human Disease. J. Proteom. 2015, 127, 61–70. [Google Scholar] [CrossRef]
- Gerstberger, S.; Hafner, M.; Tuschl, T. A Census of Human RNA-Binding Proteins. Nat. Rev. Genet. 2014, 15, 829–845. [Google Scholar] [CrossRef]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-Intron Circular RNAs Regulate Transcription in the Nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.T.; Orejuela, M.R.; Piechotta, M.; Levanon, E.Y.; Landthaler, M.; Dieterich, C.; et al. Analysis of Intron Sequences Reveals Hallmarks of Circular RNA Biogenesis in Animals. Cell Rep. 2015, 10, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, J.; Zheng, Y.; Zhang, J.; Chen, S.; Zhao, F. Comprehensive Identification of Internal Structure and Alternative Splicing Events in Circular RNAs. Nat. Commun. 2016, 7, 12060. [Google Scholar] [CrossRef] [PubMed]
- Katrekar, D.; Yen, J.; Xiang, Y.; Saha, A.; Meluzzi, D.; Savva, Y.; Mali, P. Efficient in Vitro and in Vivo RNA Editing via Recruitment of Endogenous ADARs Using Circular Guide RNAs. Nat. Biotechnol. 2022, 40, 938–945. [Google Scholar] [CrossRef]
- Liu, S.; Guo, X.Y.; Shang, Q.J.; Gao, P. The Biogenesis, Biological Functions and Modification of Circular RNAs. Exp. Mol. Pathol. 2023, 131, 104861. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Xu, J.; Wang, Y.; Min, Q.; Chen, X.; Zhang, W.; Chen, J.; Zhan, Q. Nuclear Genome-Derived Circular RNA CircPUM1 Localizes in Mitochondria and Regulates Oxidative Phosphorylation in Esophageal Squamous Cell Carcinoma. Signal Transduct. Target. Ther. 2022, 7, 40. [Google Scholar] [CrossRef]
- Li, S.; Li, X.; Xue, W.; Zhang, L.; Yang, L.-Z.; Cao, S.-M.; Lei, Y.-N.; Liu, C.-X.; Guo, S.-K.; Shan, L.; et al. Screening for Functional Circular RNAs Using the CRISPR–Cas13 System. Nat. Methods 2021, 18, 51–59. [Google Scholar] [CrossRef]
- Huang, C.; Liang, D.; Tatomer, D.C.; Wilusz, J.E. A Length-Dependent Evolutionarily Conserved Pathway Controls Nuclear Export of Circular RNAs. Genes Dev. 2018, 32, 639–644. [Google Scholar] [CrossRef]
- Zhou, C.; Molinie, B.; Daneshvar, K.; Pondick, J.V.; Wang, J.; Van Wittenberghe, N.; Xing, Y.; Giallourakis, C.C.; Mullen, A.C. Genome-Wide Maps of M6A CircRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns That Are Distinct from MRNAs. Cell Rep. 2017, 20, 2262–2276. [Google Scholar] [CrossRef]
- Liu, C.-X.; Li, X.; Nan, F.; Jiang, S.; Gao, X.; Guo, S.-K.; Xue, W.; Cui, Y.; Dong, K.; Ding, H.; et al. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell 2019, 177, 865–880.e21. [Google Scholar] [CrossRef]
- Chen, L.; Wang, Y.; Lin, J.; Song, Z.; Wang, Q.; Zhao, W.; Wang, Y.; Xiu, X.; Deng, Y.; Li, X.; et al. Exportin 4 Depletion Leads to Nuclear Accumulation of a Subset of Circular RNAs. Nat. Commun. 2022, 13, 5769. [Google Scholar] [CrossRef]
- Ngo, L.H.; Bert, A.G.; Dredge, B.K.; Williams, T.; Murphy, V.; Li, W.; Hamilton, W.B.; Carey, K.T.; Toubia, J.; Pillman, K.A.; et al. Nuclear Export of Circular RNA. Nature 2024, 627, 212–220. [Google Scholar] [CrossRef]
- Wang, M.; Yu, F.; Li, P.; Wang, K. Emerging Function and Clinical Significance of Exosomal CircRNAs in Cancer. Mol. Ther. Nucleic Acids 2020, 21, 367–383. [Google Scholar] [CrossRef]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA Delivery by Extracellular Vesicles in Mammalian Cells and Its Applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef]
- Chen, C.-K.; Cheng, R.; Demeter, J.; Chen, J.; Weingarten-Gabbay, S.; Jiang, L.; Snyder, M.P.; Weissman, J.S.; Segal, E.; Jackson, P.K.; et al. Structured Elements Drive Extensive Circular RNA Translation. Mol. Cell 2021, 81, 4300–4318.e13. [Google Scholar] [CrossRef] [PubMed]
- Welden, J.R.; Margvelani, G.; Arizaca Maquera, K.A.; Gudlavalleti, B.; Miranda Sardón, S.C.; Campos, A.R.; Robil, N.; Lee, D.C.; Hernandez, A.G.; Wang, W.-X.; et al. RNA Editing of Microtubule-Associated Protein Tau Circular RNAs Promotes Their Translation and Tau Tangle Formation. Nucleic Acids Res. 2022, 50, 12979–12996. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.-B. Dynamic M6A MRNA Methylation Directs Translational Control of Heat Shock Response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef] [PubMed]
- Fontemaggi, G.; Turco, C.; Esposito, G.; Di Agostino, S. New Molecular Mechanisms and Clinical Impact of CircRNAs in Human Cancer. Cancers 2021, 13, 3154. [Google Scholar] [CrossRef]
- Zeng, K.; Peng, J.; Xing, Y.; Zhang, L.; Zeng, P.; Li, W.; Zhang, W.; Pan, Z.; Zhou, C.; Lin, J. A Positive Feedback Circuit Driven by M6A-Modified Circular RNA Facilitates Colorectal Cancer Liver Metastasis. Mol. Cancer 2023, 22, 202. [Google Scholar] [CrossRef]
- Lu, J.; Ru, J.; Chen, Y.; Ling, Z.; Liu, H.; Ding, B.; Jiang, Y.; Ma, J.; Zhang, D.; Ge, J.; et al. N 6 -methyladenosine-modified CircSTX6 Promotes Hepatocellular Carcinoma Progression by Regulating the HNRNPD/ATF3 Axis and Encoding a 144 Amino Acid Polypeptide. Clin. Transl. Med. 2023, 13, e1451. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.-L.; Wang, Y.; et al. Extensive Translation of Circular RNAs Driven by N6-Methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA That Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e9. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Shin, M.-K.; Park, J.; Hwang, H.J.; Locker, N.; Ahn, J.; Kim, D.; Baek, D.; Park, Y.; Lee, Y.; et al. An Interaction between EIF4A3 and EIF3g Drives the Internal Initiation of Translation. Nucleic Acids Res. 2023, 51, 10950–10969. [Google Scholar] [CrossRef]
- Lin, H.-H.; Chang, C.-Y.; Huang, Y.-R.; Shen, C.-H.; Wu, Y.-C.; Chang, K.-L.; Lee, Y.-C.; Lin, Y.-C.; Ting, W.-C.; Chien, H.-J.; et al. Exon Junction Complex Mediates the Cap-Independent Translation of Circular RNA. Mol. Cancer Res. 2023, 21, 1220–1233. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, Z.; Zhang, M.; Zhou, H.; Wu, X.; Zhong, J.; Xiao, F.; Huang, N.; Yang, X.; Zeng, R.; et al. Rolling-Translated EGFR Variants Sustain EGFR Signaling and Promote Glioblastoma Tumorigenicity. Neuro-Oncology 2021, 23, 743–756. [Google Scholar] [CrossRef]
- Gao, X.; Xia, X.; Li, F.; Zhang, M.; Zhou, H.; Wu, X.; Zhong, J.; Zhao, Z.; Zhao, K.; Liu, D.; et al. Circular RNA-Encoded Oncogenic E-Cadherin Variant Promotes Glioblastoma Tumorigenicity through Activation of EGFR–STAT3 Signalling. Nat. Cell Biol. 2021, 23, 278–291. [Google Scholar] [CrossRef]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids Are Single-Stranded Covalently Closed Circular RNA Molecules Existing as Highly Base-Paired Rod-like Structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef]
- Nigro, J.M.; Cho, K.R.; Fearon, E.R.; Kern, S.E.; Ruppert, J.M.; Oliner, J.D.; Kinzler, K.W.; Vogelstein, B. Scrambled Exons. Cell 1991, 64, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Cocquerelle, C.; Daubersies, P.; Majérus, M.A.; Kerckaert, J.P.; Bailleul, B. Splicing with Inverted Order of Exons Occurs Proximal to Large Introns. EMBO J. 1992, 11, 1095–1098. [Google Scholar] [CrossRef]
- Capel, B.; Swain, A.; Nicolis, S.; Hacker, A.; Walter, M.; Koopman, P.; Goodfellow, P.; Lovell-Badge, R. Circular Transcripts of the Testis-Determining Gene Sry in Adult Mouse Testis. Cell 1993, 73, 1019–1030. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, X.; Liu, M.; Xu, F.; Zhang, Q.; Zhang, Y.; Weng, X.; Liu, S.; Du, Y.; Zhou, X. Direct Detection of CircRNA in Real Samples Using Reverse Transcription-Rolling Circle Amplification. Anal. Chim. Acta 2020, 1101, 169–175. [Google Scholar] [CrossRef]
- Goo, N.-I.; Kim, D.-E. Rolling Circle Amplification as Isothermal Gene Amplification in Molecular Diagnostics. Biochip. J. 2016, 10, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Dahl, M.; Daugaard, I.; Andersen, M.S.; Hansen, T.B.; Grønbæk, K.; Kjems, J.; Kristensen, L.S. Enzyme-Free Digital Counting of Endogenous Circular RNA Molecules in B-Cell Malignancies. Lab. Investig. 2018, 98, 1657–1669. [Google Scholar] [CrossRef]
- Li, S.; Teng, S.; Xu, J.; Su, G.; Zhang, Y.; Zhao, J.; Zhang, S.; Wang, H.; Qin, W.; Lu, Z.J.; et al. Microarray Is an Efficient Tool for CircRNA Profiling. Brief. Bioinform. 2019, 20, 1420–1433. [Google Scholar] [CrossRef]
- Chrzanowska, N.M.; Kowalewski, J.; Lewandowska, M.A. Use of Fluorescence In Situ Hybridization (FISH) in Diagnosis and Tailored Therapies in Solid Tumors. Molecules 2020, 25, 1864. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhong, Y.; Wang, X.; Shen, J.; An, W. Advances in Circular RNA and Its Applications. Int. J. Med. Sci. 2022, 19, 975–985. [Google Scholar] [CrossRef]
- Luo, J.; Xu, S.; Wang, J.; He, L.; Li, Z. Circular RNA CircWBSCR22 Facilitates Colorectal Cancer Metastasis by Enhancing CHD4’s Protein Stability. Int. J. Biol. Macromol. 2024, 282, 137135. [Google Scholar] [CrossRef]
- Wei, Z.; Zhang, C.; Song, Y.; Han, D.; Liu, J.; Song, X.; Chao, F.; Wang, S.; Xu, G.; Chen, G. CircUBE3A(2,3,4,5) Promotes Adenylate-Uridylate-Rich Binding Factor 1 Nuclear Translocation to Suppress Prostate Cancer Metastasis. Cancer Lett. 2024, 588, 216743. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Jiao, W.; Song, J.; Wang, J.; Chen, G.; Li, D.; Wang, X.; Bao, B.; Du, X.; Cheng, Y.; et al. Circ-HnRNPU Inhibits NONO-Mediated c-Myc Transactivation and MRNA Stabilization Essential for Glycosylation and Cancer Progression. J. Exp. Clin. Cancer Res. 2023, 42, 313. [Google Scholar] [CrossRef]
- Sun, Z.; Dang, P.; Guo, Y.; Liu, S.; Hu, S.; Sun, H.; Xu, Y.; Wang, W.; Chen, C.; Liu, J.; et al. Targeting CircAURKA Prevents Colorectal Cancer Progression via Enhancing CTNNB1 Protein Degradation. Oncogene 2024, 43, 3388–3401. [Google Scholar] [CrossRef]
- Abdelmohsen, K.; Panda, A.C.; Munk, R.; Grammatikakis, I.; Dudekula, D.B.; De, S.; Kim, J.; Noh, J.H.; Kim, K.M.; Martindale, J.L.; et al. Identification of HuR Target Circular RNAs Uncovers Suppression of PABPN1 Translation by CircPABPN1. RNA Biol. 2017, 14, 361–369. [Google Scholar] [CrossRef]
- Lv, Y.; Yuan, Z.; Chen, D.; Chen, Z.; Zhu, X.; Ying, X.; Huang, Y.; Ji, W.; Qi, D. Circular RNA LMBR1 Inhibits Bladder Cancer Progression by Enhancing Expression of the Protein ALDH1A3. Noncoding RNA Res. 2024, 9, 1235–1248. [Google Scholar] [CrossRef]
- Wang, C.; Liu, Y.; Zuo, Z.; Cui, D.; Xu, Y.; Li, L.; Jiang, Y. Dual Role of Exosomal CircCMTM3 Derived from GSCs in Impeding Degradation and Promoting Phosphorylation of STAT5A to Facilitate Vasculogenic Mimicry Formation in Glioblastoma. Theranostics 2024, 14, 5698–5724. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Li, Y.; Chang, W.; Feng, M.; Yang, Y.; Zhu, X.; Liu, Z.; Fu, Y. CircSEC24B Activates Autophagy and Induces Chemoresistance of Colorectal Cancer via OTUB1-Mediated Deubiquitination of SRPX2. Cell Death Dis. 2024, 15, 693. [Google Scholar] [CrossRef] [PubMed]
- Papachristou, D.J.; Sklirou, E.; Corradi, D.; Grassani, C.; Kontogeorgakos, V.; Rao, U.N.M. Immunohistochemical Analysis of the Endoribonucleases Drosha, Dicer and Ago2 in Smooth Muscle Tumours of Soft Tissues. Histopathology 2012, 60, E28–E36. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Okholm, T.L.H.; Venø, M.T.; Kjems, J. Circular RNAs Are Abundantly Expressed and Upregulated during Human Epidermal Stem Cell Differentiation. RNA Biol. 2018, 15, 280–291. [Google Scholar] [CrossRef]
- Hsiao, K.-Y.; Lin, Y.-C.; Gupta, S.K.; Chang, N.; Yen, L.; Sun, H.S.; Tsai, S.-J. Noncoding Effects of Circular RNA CCDC66 Promote Colon Cancer Growth and Metastasis. Cancer Res. 2017, 77, 2339–2350. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Yang, J.; Ran, F.; Shi, Y.; Yang, L.; Duan, Y.; Shi, Z.; Li, X.; Zhang, J.; Li, Z.; et al. CircBIRC6 Affects Prostate Cancer Progression by Regulating MiR-574-5p and DNAJB1. Cancer Biol. Ther. 2024, 25, 2399363. [Google Scholar] [CrossRef]
- Zhang, X.; Bian, Y.; Li, Q.; Yu, C.; Gao, Y.; Tian, B.; Xia, W.; Wang, W.; Xin, L.; Lin, H.; et al. EIF4A3-Mediated Oncogenic CircRNA Hsa_circ_0001165 Advances Esophageal Squamous Cell Carcinoma Progression through the MiR-381-3p/TNS3 Pathway. Cell Biol. Toxicol. 2024, 40, 84. [Google Scholar] [CrossRef]
- Zhang, Y.E.; Liang, Y.; Wu, Y.; Song, L.; Zhang, Z. CircTIAM1 Overexpression Promotes the Progression of Papillary Thyroid Cancer by Regulating the MiR-338-3p/LASP1 Axis. Oncol. Res. 2024, 32, 1747–1763. [Google Scholar] [CrossRef]
- Feng, Z.; Wu, J. Hsa_circ_0129047 Upregulates LYVE1 to Inhibit Hepatocellular Carcinoma Progression by Sponging MiR-492. Dis. Markers 2023, 2023, 6978234. [Google Scholar] [CrossRef]
- Su, X.; Hu, B.; Yi, J.; Zhao, Q.; Zhou, Y.; Zhu, X.; Wu, D.; Fan, Y.; Lin, J.; Cao, C.; et al. Crosstalk between CircBMI1 and MiR-338-5p/ID4 Inhibits Acute Myeloid Leukemia Progression. J. Leukoc. Biol. 2024, 116, 1080–1093. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Ding, X.; Fang, X.; Xu, J.; Chen, Y.; Qian, Y.; Zhang, J.; Yu, D.; Zhang, X.; Ma, X.; et al. Circ6834 Suppresses Non-Small Cell Lung Cancer Progression by Destabilizing ANHAK and Regulating MiR-873-5p/TXNIP Axis. Mol. Cancer 2024, 23, 128. [Google Scholar] [CrossRef]
- Gibb, E.A.; Brown, C.J.; Lam, W.L. The Functional Role of Long Non-Coding RNA in Human Carcinomas. Mol. Cancer 2011, 10, 38. [Google Scholar] [CrossRef]
- Entezari, M.; Ghanbarirad, M.; Taheriazam, A.; Sadrkhanloo, M.; Zabolian, A.; Goharrizi, M.A.S.B.; Hushmandi, K.; Aref, A.R.; Ashrafizadeh, M.; Zarrabi, A.; et al. Long Non-Coding RNAs and Exosomal LncRNAs: Potential Functions in Lung Cancer Progression, Drug Resistance and Tumor Microenvironment Remodeling. Biomed. Pharmacother. 2022, 150, 112963. [Google Scholar] [CrossRef]
- Kleaveland, B.; Shi, C.Y.; Stefano, J.; Bartel, D.P. A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain. Cell 2018, 174, 350–362.e17. [Google Scholar] [CrossRef]
- Silenzi, V.; D’Ambra, E.; Santini, T.; D’Uva, S.; Setti, A.; Salvi, N.; Nicoletti, C.; Scarfò, R.; Cordella, F.; Mongiardi, B.; et al. A Tripartite CircRNA/MRNA/MiRNA Interaction Regulates Glutamatergic Signaling in the Mouse Brain. Cell Rep. 2024, 43, 114766. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Zhao, G.; Yan, X.; Lv, Z.; Yin, H.; Zhang, S.; Song, W.; Li, X.; Li, L.; Du, Z.; et al. A Novel FLI1 Exonic Circular RNA Promotes Metastasis in Breast Cancer by Coordinately Regulating TET1 and DNMT1. Genome Biol. 2018, 19, 218. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. CircRNA Biogenesis Competes with Pre-MRNA Splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Barbagallo, D.; Caponnetto, A.; Cirnigliaro, M.; Brex, D.; Barbagallo, C.; D’Angeli, F.; Morrone, A.; Caltabiano, R.; Barbagallo, G.; Ragusa, M.; et al. CircSMARCA5 Inhibits Migration of Glioblastoma Multiforme Cells by Regulating a Molecular Axis Involving Splicing Factors SRSF1/SRSF3/PTB. Int. J. Mol. Sci. 2018, 19, 480. [Google Scholar] [CrossRef]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular Non-Coding RNA ANRIL Modulates Ribosomal RNA Maturation and Atherosclerosis in Humans. Nat. Commun. 2016, 7, 12429. [Google Scholar] [CrossRef]
- Yang, Q.; Du, W.W.; Wu, N.; Yang, W.; Awan, F.M.; Fang, L.; Ma, J.; Li, X.; Zeng, Y.; Yang, Z.; et al. A Circular RNA Promotes Tumorigenesis by Inducing C-Myc Nuclear Translocation. Cell Death Differ. 2017, 24, 1609–1620. [Google Scholar] [CrossRef] [PubMed]
- Conn, V.M.; Gabryelska, M.; Toubia, J.; Kirk, K.; Gantley, L.; Powell, J.A.; Cildir, G.; Marri, S.; Liu, R.; Stringer, B.W.; et al. Circular RNAs Drive Oncogenic Chromosomal Translocations within the MLL Recombinome in Leukemia. Cancer Cell 2023, 41, 1309–1326.e10. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.; Xia, W.; Chen, R.; Wang, S.; Xu, Y.; Ma, Z.; Xu, W.; Zhang, E.; Wang, J.; Fang, T.; et al. The Circular RNA CircPRKCI Promotes Tumor Growth in Lung Adenocarcinoma. Cancer Res. 2018, 78, 2839–2851. [Google Scholar] [CrossRef]
- Hanniford, D.; Ulloa-Morales, A.; Karz, A.; Berzoti-Coelho, M.G.; Moubarak, R.S.; Sánchez-Sendra, B.; Kloetgen, A.; Davalos, V.; Imig, J.; Wu, P.; et al. Epigenetic Silencing of CDR1as Drives IGF2BP3-Mediated Melanoma Invasion and Metastasis. Cancer Cell 2020, 37, 55–70.e15. [Google Scholar] [CrossRef]
- Ferreira, H.J.; Davalos, V.; de Moura, M.C.; Soler, M.; Perez-Salvia, M.; Bueno-Costa, A.; Setien, F.; Moran, S.; Villanueva, A.; Esteller, M. Circular RNA CpG Island Hypermethylation-Associated Silencing in Human Cancer. Oncotarget 2018, 9, 29208–29219. [Google Scholar] [CrossRef]
- Jakobsen, T.; Dahl, M.; Dimopoulos, K.; Grønbæk, K.; Kjems, J.; Kristensen, L.S. Genome-Wide Circular RNA Expression Patterns Reflect Resistance to Immunomodulatory Drugs in Multiple Myeloma Cells. Cancers 2021, 13, 365. [Google Scholar] [CrossRef]
- Bradley, R.K.; Anczuków, O. RNA Splicing Dysregulation and the Hallmarks of Cancer. Nat. Rev. Cancer 2023, 23, 135–155. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qian, J.; Gu, C.; Yang, Y. Alternative Splicing and Cancer: A Systematic Review. Signal Transduct. Target. Ther. 2021, 6, 78. [Google Scholar] [CrossRef]
- Kong, Y.; Luo, Y.; Zheng, S.; Yang, J.; Zhang, D.; Zhao, Y.; Zheng, H.; An, M.; Lin, Y.; Ai, L.; et al. Mutant KRAS Mediates CircARFGEF2 Biogenesis to Promote Lymphatic Metastasis of Pancreatic Ductal Adenocarcinoma. Cancer Res. 2023, 83, 3077–3094. [Google Scholar] [CrossRef]
- Aherrahrou, R.; Lue, D.; Civelek, M. Genetic Regulation of Circular RNA Expression in Human Aortic Smooth Muscle Cells and Vascular Traits. Hum. Genet. Genom. Adv. 2023, 4, 100164. [Google Scholar] [CrossRef]
- Kramer, M.C.; Liang, D.; Tatomer, D.C.; Gold, B.; March, Z.M.; Cherry, S.; Wilusz, J.E. Combinatorial Control of Drosophila Circular RNA Expression by Intronic Repeats, HnRNPs, and SR Proteins. Genes Dev. 2015, 29, 2168–2182. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, M.; Jin, M.; Jia, Q.; Wu, Y.; Hart, K.; Bargoma, E.; Pangallo, J.; Bradley, R.K.; Abdel-Wahab, O.; Jia, Z.; et al. RNA Splicing Factor Mutations Drive Aberrant Canonical and Cryptic Circular RNA Biogenesis in Leukemia. Blood 2023, 142, 1395. [Google Scholar] [CrossRef]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA Binding Protein Quaking Regulates Formation of CircRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef]
- Wang, M.; Hou, J.; Müller-McNicoll, M.; Chen, W.; Schuman, E.M. Long and Repeat-Rich Intronic Sequences Favor Circular RNA Formation under Conditions of Reduced Spliceosome Activity. iScience 2019, 20, 237–247. [Google Scholar] [CrossRef]
- Shu, X.; Yi, J.; Li, J.; Ying, Y.; Tang, Y.; Chen, Z.; Wang, J.; Zhang, F.; Lu, D.; Wu, Y.; et al. N6-Methyladenosine-Modified CircRPS6KC1 Regulated Cellular Senescence in Prostate Cancer via FOXM1/PCNA Axis. Cell. Signal. 2025, 125, 111510. [Google Scholar] [CrossRef]
- Pisignano, G.; Michael, D.C.; Visal, T.H.; Pirlog, R.; Ladomery, M.; Calin, G.A. Going Circular: History, Present, and Future of CircRNAs in Cancer. Oncogene 2023, 42, 2783–2800. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, J.; Yang, Y.; Liu, Z.; Sun, S.; Li, R.; Zhu, H.; Li, T.; Zheng, J.; Li, J.; et al. Circular RNAs in Human Diseases. MedComm 2024, 5, e699. [Google Scholar] [CrossRef]
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.-M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881.e13. [Google Scholar] [CrossRef]
- Gao, Y.; Shang, S.; Guo, S.; Li, X.; Zhou, H.; Liu, H.; Sun, Y.; Wang, J.; Wang, P.; Zhi, H.; et al. Lnc2Cancer 3.0: An Updated Resource for Experimentally Supported LncRNA/CircRNA Cancer Associations and Web Tools Based on RNA-Seq and ScRNA-Seq Data. Nucleic Acids Res. 2021, 49, D1251–D1258. [Google Scholar] [CrossRef]
- Meng, X.; Hu, D.; Zhang, P.; Chen, Q.; Chen, M. CircFunBase: A Database for Functional Circular RNAs. Database 2019, 2019, baz003. [Google Scholar] [CrossRef]
- Yang, J.-H.; Shao, P.; Zhou, H.; Chen, Y.-Q.; Qu, L.-H. DeepBase: A Database for Deeply Annotating and Mining Deep Sequencing Data. Nucleic Acids Res. 2010, 38, D123–D130. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wang, Q.; Shen, J.; Yang, B.B.; Ding, X. Circbank: A Comprehensive Database for CircRNA with Standard Nomenclature. RNA Biol. 2019, 16, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Qu, H. CircVAR Database: Genome-Wide Archive of Genetic Variants for Human Circular RNAs. BMC Genom. 2020, 21, 750. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Qiu, Z.; Chi-Shing Cho, W.; Liu, Z.; Chen, S.; Li, H.; Chen, K.; Li, Y.; Zuo, C.; Qiu, M. Synthetic CircRNA Therapeutics: Innovations, Strategies, and Future Horizons. MedComm 2024, 5, e720. [Google Scholar] [CrossRef]
- Chen, R.; Wang, S.K.; Belk, J.A.; Amaya, L.; Li, Z.; Cardenas, A.; Abe, B.T.; Chen, C.-K.; Wender, P.A.; Chang, H.Y. Engineering Circular RNA for Enhanced Protein Production. Nat. Biotechnol. 2023, 41, 262–272. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, S.; Wang, H.; Cao, J.; Huang, X.; Chen, Z.; Xu, P.; Sun, G.; Xu, J.; Lv, J.; et al. Circular RNA CircNRIP1 Acts as a MicroRNA-149-5p Sponge to Promote Gastric Cancer Progression via the AKT1/MTOR Pathway. Mol. Cancer 2019, 18, 20. [Google Scholar] [CrossRef]
- Sun, Y.-M.; Wang, W.-T.; Zeng, Z.-C.; Chen, T.-Q.; Han, C.; Pan, Q.; Huang, W.; Fang, K.; Sun, L.-Y.; Zhou, Y.-F.; et al. CircMYBL2, a CircRNA from MYBL2, Regulates FLT3 Translation by Recruiting PTBP1 to Promote FLT3-ITD AML Progression. Blood 2019, 134, 1533–1546. [Google Scholar] [CrossRef]
- Gu, Y.; Wang, Y.; He, L.; Zhang, J.; Zhu, X.; Liu, N.; Wang, J.; Lu, T.; He, L.; Tian, Y.; et al. Circular RNA CircIPO11 Drives Self-Renewal of Liver Cancer Initiating Cells via Hedgehog Signaling. Mol. Cancer 2021, 20, 132. [Google Scholar] [CrossRef]
- Xia, Q.; Ding, T.; Zhang, G.; Li, Z.; Zeng, L.; Zhu, Y.; Guo, J.; Hou, J.; Zhu, T.; Zheng, J.; et al. Circular RNA Expression Profiling Identifies Prostate Cancer- Specific CircRNAs in Prostate Cancer. Cell. Physiol. Biochem. 2018, 50, 1903–1915. [Google Scholar] [CrossRef]
- Enuka, Y.; Lauriola, M.; Feldman, M.E.; Sas-Chen, A.; Ulitsky, I.; Yarden, Y. Circular RNAs Are Long-Lived and Display Only Minimal Early Alterations in Response to a Growth Factor. Nucleic Acids Res. 2016, 44, 1370–1383. [Google Scholar] [CrossRef]
- Leung, K.T.; Cai, J.; Liu, Y.; Chan, K.Y.Y.; Shao, J.; Yang, H.; Hu, Q.; Xue, Y.; Wu, X.; Guo, X.; et al. Prognostic Implications of CD9 in Childhood Acute Lymphoblastic Leukemia: Insights from a Nationwide Multicenter Study in China. Leukemia 2024, 38, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.T.; Zhang, C.; Chan, K.Y.Y.; Li, K.; Cheung, J.T.K.; Ng, M.H.L.; Zhang, X.-B.; Sit, T.; Lee, W.Y.W.; Kang, W.; et al. CD9 Blockade Suppresses Disease Progression of High-Risk Pediatric B-Cell Precursor Acute Lymphoblastic Leukemia and Enhances Chemosensitivity. Leukemia 2020, 34, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Yuan, L.; Li, J.; Liu, Y.; Wang, H.; Ren, X. CircDENND4C, a Novel Serum Marker for Epithelial Ovarian Cancer, Acts as a Tumor Suppressor by Downregulating MiR-200b/c. Ann. Med. 2023, 55, 908–919. [Google Scholar] [CrossRef]
- Sun, X.-H.; Wang, Y.-T.; Li, G.-F.; Zhang, N.; Fan, L. Serum-Derived Three-CircRNA Signature as a Diagnostic Biomarker for Hepatocellular Carcinoma. Cancer Cell Int. 2020, 20, 226. [Google Scholar] [CrossRef]
- Omid-Shafaat, R.; Moayeri, H.; Rahimi, K.; Menbari, M.; Vahabzadeh, Z.; Hakhamaneshi, M.; Nouri, B.; Ghaderi, B.; Abdi, M. Serum Circ-FAF1/Circ-ELP3: A Novel Potential Biomarker for Breast Cancer Diagnosis. J. Clin. Lab. Anal. 2021, 35, e24008. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Dong, X.; Song, X. Tumor Educated Platelet: The Novel BioSource for Cancer Detection. Cancer Cell Int. 2023, 23, 91. [Google Scholar] [CrossRef]
- Sultana, Q.; Kar, J.; Verma, A.; Sanghvi, S.; Kaka, N.; Patel, N.; Sethi, Y.; Chopra, H.; Kamal, M.A.; Greig, N.H. A Comprehensive Review on Neuroendocrine Neoplasms: Presentation, Pathophysiology and Management. J. Clin. Med. 2023, 12, 5138. [Google Scholar] [CrossRef]
- Huang, D.; Zhu, X.; Ye, S.; Zhang, J.; Liao, J.; Zhang, N.; Zeng, X.; Wang, J.; Yang, B.; Zhang, Y.; et al. Tumour Circular RNAs Elicit Anti-Tumour Immunity by Encoding Cryptic Peptides. Nature 2024, 625, 593–602. [Google Scholar] [CrossRef]
- Naseer, Q.A.; Malik, A.; Zhang, F.; Chen, S. Exploring the Enigma: History, Present, and Future of Long Non-Coding RNAs in Cancer. Discov. Oncol. 2024, 15, 214. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Fernandes, J.C.R.; Acuña, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Noncoding RNA 2019, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.-C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Cao, J. The Functional Role of Long Non-Coding RNAs and Epigenetics. Biol. Proced. Online 2014, 16, 42. [Google Scholar] [CrossRef]
- Qian, Y.; Shi, L.; Luo, Z. Long Non-Coding RNAs in Cancer: Implications for Diagnosis, Prognosis, and Therapy. Front. Med. 2020, 7, 612393. [Google Scholar] [CrossRef] [PubMed]
- Karimi, B.; Dehghani Firoozabadi, A.; Peymani, M.; Ghaedi, K. Circulating Long Noncoding RNAs as Novel Bio-Tools: Focus on Autoimmune Diseases. Hum. Immunol. 2022, 83, 618–627. [Google Scholar] [CrossRef]
- Liang, J.; Xie, F.; Feng, J.; Huang, C.; Shen, J.; Han, Z.; Luo, W.; He, J.; Chen, H. Progress in the Application of Body Fluid and Tissue Level MRNAs-Non-Coding RNAs for the Early Diagnosis and Prognostic Evaluation of Systemic Lupus Erythematosus. Front. Immunol. 2022, 13, 1020891. [Google Scholar] [CrossRef]
- Li, M.; Zhao, Y.; Li, H.; Deng, X.; Sheng, M. Application Value of Circulating LncRNA in Diagnosis, Treatment, and Prognosis of Breast Cancer. Funct. Integr. Genom. 2023, 23, 61. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Fullwood, M.J. Roles, Functions, and Mechanisms of Long Non-Coding RNAs in Cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54. [Google Scholar] [CrossRef]
- Karimi, B.; Mokhtari, K.; Rozbahani, H.; Peymani, M.; Nabavi, N.; Entezari, M.; Rashidi, M.; Taheriazam, A.; Ghaedi, K.; Hashemi, M. Pathological Roles of MiRNAs and Pseudogene-Derived LncRNAs in Human Cancers, and Their Comparison as Prognosis/Diagnosis Biomarkers. Pathol. Res. Pract. 2024, 253, 155014. [Google Scholar] [CrossRef]
- Kritika, C. Transforming “Junk” DNA into Cancer Warriors: The Role of Pseudogenes in Hepatocellular Carcinoma. Cancer Diagn. Progn. 2024, 4, 214–222. [Google Scholar] [CrossRef]
- Sun, T.-T.; He, J.; Liang, Q.; Ren, L.-L.; Yan, T.-T.; Yu, T.-C.; Tang, J.-Y.; Bao, Y.-J.; Hu, Y.; Lin, Y.; et al. LncRNA GClnc1 Promotes Gastric Carcinogenesis and May Act as a Modular Scaffold of WDR5 and KAT2A Complexes to Specify the Histone Modification Pattern. Cancer Discov. 2016, 6, 784–801. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Lv, X.; Ru, Y.; Zhou, F.; Wang, N.; Xi, H.; Zhang, K.; Li, J.; Chang, R.; Xie, T.; et al. Circulating Exosomal Gastric Cancer–Associated Long Noncoding RNA1 as a Biomarker for Early Detection and Monitoring Progression of Gastric Cancer. JAMA Surg. 2020, 155, 572. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Lv, X.; Ru, Y.; Dong, J.; Chang, R.; Wu, D.; Chen, L.; Wang, X.; Guo, X. Circulating Exosomal Gastric Cancer-Associated Long Noncoding RNA1 as a Noninvasive Biomarker for Predicting Chemotherapy Response and Prognosis of Advanced Gastric Cancer: A Multi-Cohort, Multi-Phase Study. EBioMedicine 2022, 78, 103971. [Google Scholar] [CrossRef]
- Yang, J.; Qi, M.; Fei, X.; Wang, X.; Wang, K. LncRNA H19: A Novel Oncogene in Multiple Cancers. Int. J. Biol. Sci. 2021, 17, 3188–3208. [Google Scholar] [CrossRef]
- Alipoor, B.; Parvar, S.N.; Sabati, Z.; Ghaedi, H.; Ghasemi, H. An Updated Review of the H19 LncRNA in Human Cancer: Molecular Mechanism and Diagnostic and Therapeutic Importance. Mol. Biol. Rep. 2020, 47, 6357–6374. [Google Scholar] [CrossRef]
- Guo, G.; Kang, Q.; Chen, Q.; Chen, Z.; Wang, J.; Tan, L.; Chen, J.-L. High Expression of Long Non-coding RNA H19 Is Required for Efficient Tumorigenesis Induced by Bcr-Abl Oncogene. FEBS Lett. 2014, 588, 1780–1786. [Google Scholar] [CrossRef]
- Asadi, M.; Gholampour, M.A.; Kompani, F.; Alizadeh, S. Expression of Long Non-Coding RNA H19 in Acute Lymphoblastic Leukemia. Cell J. 2023, 25, 1. [Google Scholar] [CrossRef]
- Rojas, Á.; Gil-Gómez, A.; de la Cruz-Ojeda, P.; Muñoz-Hernández, R.; Sánchez-Torrijos, Y.; Gallego-Durán, R.; Millán, R.; Rico, M.C.; Montero-Vallejo, R.; Gato-Zambrano, S.; et al. Long Non-coding RNA H19 as a Biomarker for Hepatocellular Carcinoma. Liver Int. 2022, 42, 1410–1422. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhou, J.; Zhang, W.; Lin, J.; Ma, J.; Wen, X.; Yuan, Q.; Li, X.; Xu, Z.; Qian, J. H19 Overexpression Promotes Leukemogenesis and Predicts Unfavorable Prognosis in Acute Myeloid Leukemia. Clin. Epigenet. 2018, 10, 47. [Google Scholar] [CrossRef]
- May, A.M.; Frey, A.-V.; Bogatyreva, L.; Benkisser-Petersen, M.; Hauschke, D.; Lübbert, M.; Wäsch, R.; Werner, M.; Hasskarl, J.; Lassmann, S. ID2 and ID3 Protein Expression Mirrors Granulopoietic Maturation and Discriminates between Acute Leukemia Subtypes. Histochem. Cell Biol. 2014, 141, 431–440. [Google Scholar] [CrossRef]
- Zhao, T.-F.; Jia, H.-Z.; Zhang, Z.-Z.; Zhao, X.-S.; Zou, Y.-F.; Zhang, W.; Wan, J.; Chen, X.-F. LncRNA H19 Regulates ID2 Expression through Competitive Binding to Hsa-MiR-19a/b in Acute Myelocytic Leukemia. Mol. Med. Rep. 2017, 16, 3687–3693. [Google Scholar] [CrossRef] [PubMed]
- Nokkeaw, A.; Thamjamrassri, P.; Chantaravisoot, N.; Tangkijvanich, P.; Ariyachet, C. Long Non-Coding RNA H19 Promotes Proliferation in Hepatocellular Carcinoma Cells via H19/MiR-107/CDK6 Axis. Oncol. Res. 2023, 31, 989–1005. [Google Scholar] [CrossRef]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A Long Noncoding RNA Maintains Active Chromatin to Coordinate Homeotic Gene Expression. Nature 2011, 472, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ma, Q.; Wu, X.; Chen, P. LncRNA HOTTIP Promotes Ovarian Cancer Cell Invasion And Metastasis By Stabilizing Hif-1α In The Anoxic Cellular Microenvironment. Acta Endocrinol. (Buchar.) 2022, 18, 263–270. [Google Scholar] [CrossRef]
- Feng, H.; Zhao, F.; Luo, J.; Xu, S.; Liang, Z.; Xu, W.; Bao, Y.; Qin, G. Long Non-Coding RNA HOTTIP Exerts an Oncogenic Function by Regulating HOXA13 in Nasopharyngeal Carcinoma. Mol. Biol. Rep. 2023, 50, 6807–6818. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Hu, Z.; Fu, Y.; Wang, J. Long Non-Coding RNA HOTTIP Promotes Renal Cell Carcinoma Progression through the Regulation of the MiR-506 Pathway. Aging 2024, 16, 10832–10840. [Google Scholar] [CrossRef]
- Quagliata, L.; Matter, M.S.; Piscuoglio, S.; Arabi, L.; Ruiz, C.; Procino, A.; Kovac, M.; Moretti, F.; Makowska, Z.; Boldanova, T.; et al. Long Noncoding RNA HOTTIP/HOXA13 Expression Is Associated with Disease Progression and Predicts Outcome in Hepatocellular Carcinoma Patients. Hepatology 2014, 59, 911–923. [Google Scholar] [CrossRef]
- Liu, T.; Wang, H.; Yu, H.; Bi, M.; Yan, Z.; Hong, S.; Li, S. The Long Non-Coding RNA HOTTIP Is Highly Expressed in Colorectal Cancer and Enhances Cell Proliferation and Invasion. Mol. Ther. Nucleic Acids 2020, 19, 612–618. [Google Scholar] [CrossRef]
- Wei, H.; Xu, Z.; Chen, L.; Wei, Q.; Huang, Z.; Liu, G.; Li, W.; Wang, J.; Tang, Q.; Pu, J. Long Non-Coding RNA PAARH Promotes Hepatocellular Carcinoma Progression and Angiogenesis via Upregulating HOTTIP and Activating HIF-1α/VEGF Signaling. Cell Death Dis. 2022, 13, 102. [Google Scholar] [CrossRef]
- Jin, J.; Byun, J.-K.; Choi, Y.-K.; Park, K.-G. Targeting Glutamine Metabolism as a Therapeutic Strategy for Cancer. Exp. Mol. Med. 2023, 55, 706–715. [Google Scholar] [CrossRef]
- Kim, S.S.; Baek, G.O.; Son, J.A.; Ahn, H.R.; Yoon, M.K.; Cho, H.J.; Yoon, J.H.; Nam, S.W.; Cheong, J.Y.; Eun, J.W. Early Detection of Hepatocellular Carcinoma via Liquid Biopsy: Panel of Small Extracellular Vesicle-derived Long Noncoding RNAs Identified as Markers. Mol. Oncol. 2021, 15, 2715–2731. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.; Jiang, Y.; Wang, N.; Su, H.; Han, X. Long Noncoding RNAs MALAT1 and HOTTIP Act as Serum Biomarkers for Hepatocellular Carcinoma. Cancer Control 2024, 31, 10732748241284821. [Google Scholar] [CrossRef]
- Ali Akbar-Esfahani, S.; Karimipoor, M.; Bahreini, F.; Soltania, A.R.; Aletaha, N.; Mahdavinezhad, A. Diagnostic Value of Plasma Long Non-Coding RNA HOTTIP as a Non-Invasive Biomarker for Colorectal Cancer (A Case- Control Study). Int. J. Mol. Cell. Med. 2019, 8, 240–247. [Google Scholar] [CrossRef]
- Chen, X.; Liu, Y.; Zhang, Q.; Liu, B.; Cheng, Y.; Zhang, Y.; Sun, Y.; Liu, J.; Gen, H. Exosomal Long Non-Coding RNA HOTTIP Increases Resistance of Colorectal Cancer Cells to Mitomycin via Impairing MiR-214-Mediated Degradation of KPNA3. Front. Cell Dev. Biol. 2021, 8, 582723. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Xu, Q.; Sun, L.; Wen, J.; Fang, X.; Xing, C.; Yuan, Y. Four Novel Polymorphisms in Long Non-Coding RNA HOTTIP Are Associated with the Risk and Prognosis of Colorectal Cancer. Biosci. Rep. 2019, 39, BSR20180573. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Guan, M.-X.; Zhou, T.; Cai, X.; Shan, G. Emerging Functions of Mitochondria-Encoded Noncoding RNAs. Trends Genet. 2023, 39, 125–139. [Google Scholar] [CrossRef]
- Piergentili, R.; Sechi, S. Non-Coding RNAs of Mitochondrial Origin: Roles in Cell Division and Implications in Cancer. Int. J. Mol. Sci. 2024, 25, 7498. [Google Scholar] [CrossRef]
- Burzio, V.A.; Villota, C.; Villegas, J.; Landerer, E.; Boccardo, E.; Villa, L.L.; Martínez, R.; Lopez, C.; Gaete, F.; Toro, V.; et al. Expression of a Family of Noncoding Mitochondrial RNAs Distinguishes Normal from Cancer Cells. Proc. Natl. Acad. Sci. USA 2009, 106, 9430–9434. [Google Scholar] [CrossRef]
- Villegas, J.; Burzio, V.; Villota, C.; Landerer, E.; Martinez, R.; Santander, M.; Martinez, R.; Pinto, R.; Vera, M.I.; Boccardo, E.; et al. Expression of a Novel Non-Coding Mitochondrial RNA in Human Proliferating Cells. Nucleic Acids Res. 2007, 35, 7336–7347. [Google Scholar] [CrossRef]
- Owens, G.K.; Kumar, M.S.; Wamhoff, B.R. Molecular Regulation of Vascular Smooth Muscle Cell Differentiation in Development and Disease. Physiol. Rev. 2004, 84, 767–801. [Google Scholar] [CrossRef]
- Kong, P.; Wang, X.; Gao, Y.K.; Zhang, D.D.; Huang, X.F.; Song, Y.; Zhang, W.D.; Guo, R.J.; Li, H.; Han, M. RGS5 Maintaining Vascular Homeostasis Is Altered by the Tumor Microenvironment. Biol. Direct 2023, 18, 78. [Google Scholar] [CrossRef]
- Zhang, X.; Yan, X.; Cao, J.; Yang, Z.; Cao, X.; Zhang, Y.; Liang, L.; Zheng, M.; Liu, X.; Zhang, J.; et al. SM22α+ Vascular Mural Cells Are Essential for Vessel Stability in Tumors and Undergo Phenotype Transition Regulated by Notch Signaling. J. Exp. Clin. Cancer Res. 2020, 39, 1–14. [Google Scholar] [CrossRef]
- Bongolo, C.C.; Thokerunga, E.; Fidele, N.B.; Souraka, T.D.M.; Kisembo, P.; Rugera, S.P.; Worley, P.F.; Tu, J.C. Upregulation of the Long Non-Coding RNA, LIPCAR Promotes Proliferation, Migration, and Metastasis of Hepatocellular Carcinoma. Cancer Biomark. 2022, 35, 245–256. [Google Scholar] [CrossRef]
- Wang, H.; Song, T.; Zhao, Y.; Zhao, J.; Wang, X.; Fu, X. Long Non-Coding RNA LICPAR Regulates Atrial Fibrosis via TGF-β/Smad Pathway in Atrial Fibrillation. Tissue Cell 2020, 67, 101440. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Liu, S.; Zhou, L.; Li, X.; Meng, Y.; Li, Y.; Li, L.; Jiao, B.; Bai, L.; Yu, Y.; et al. Aberrant Shuttling of Long Noncoding RNAs during the Mitochondria-Nuclear Crosstalk in Hepatocellular Carcinoma Cells. Am. J. Cancer Res. 2019, 9, 1008. [Google Scholar]
- Xu, Q.; Liao, Z.; Gong, Z.; Liu, X.; Yang, Y.; Wang, Z.; Yang, W.; Hou, L.; Yang, J.; Song, J.; et al. Down-Regulation of EVA1A by MiR-103a-3p Promotes Hepatocellular Carcinoma Cells Proliferation and Migration. Cell Mol. Biol. Lett. 2022, 27, 93. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhu, X. MiR-103a-3p Contributes to the Progression of Colorectal Cancer by Regulating GREM2 Expression. Yonsei Med. J. 2022, 63, 520–529. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Huhe, M.; Lou, J. MicroRNA-103a-3p Promotes Cell Proliferation and Invasion in Non-Small-Cell Lung Cancer Cells through Akt Pathway by Targeting PTEN. BioMed Res. Int. 2021, 2021, 7590976. [Google Scholar] [CrossRef]
- Fang, P.; Jiang, Q.; Liu, S.; Gu, J.; Hu, K.; Wang, Z. Circ_0002099 Is a Novel Molecular Therapeutic Target for Bladder Cancer. Drug Dev. Res. 2022, 83, 1890–1905. [Google Scholar] [CrossRef]
- Huang, J.; Lin, F.; Xu, C.; Xu, Y. LINC00662 Facilitates Osteosarcoma Progression via Sponging MiR-103a-3p and Regulating SIK2 Expression. J. Tissue Eng. Regen. Med. 2021, 15, 1082–1091. [Google Scholar] [CrossRef]
- Chen, Z.; He, Q.; Lu, T.; Wu, J.; Shi, G.; He, L.; Zong, H.; Liu, B.; Zhu, P. McPGK1-Dependent Mitochondrial Import of PGK1 Promotes Metabolic Reprogramming and Self-Renewal of Liver TICs. Nat. Commun. 2023, 14, 1121. [Google Scholar] [CrossRef]
- Li, J.; Bai, R.; Yang, W.; Miao, H.; Li, Y.; Dai, H.; Li, L.; Zhao, Y.; Song, X. The Mitochondrial-derived LncRNA MDL1 Mediates a Mitochondria-to-nucleus Retrograde Regulation by Inhibiting the Nuclear Translocation of P53. MedComm—Oncology 2022, 1, e15. [Google Scholar] [CrossRef]
- Fischer, M. Census and Evaluation of P53 Target Genes. Oncogene 2017, 36, 3943–3956. [Google Scholar] [CrossRef] [PubMed]
- Garrido, P.; Casas-Benito, A.; Larrayoz, I.M.; Narro-Íñiguez, J.; Rubio-Mediavilla, S.; Zozaya, E.; Martín-Carnicero, A.; Martínez, A. Expression of Mitochondrial Long Non-Coding RNAs, MDL1 and MDL1AS, Are Good Prognostic and/or Diagnostic Biomarkers for Several Cancers, Including Colorectal Cancer. Cancers 2024, 16, 960. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Sun, H.; Wang, C.; Liu, W.; Liu, M.; Zhu, Y.; Xu, W.; Jin, H.; Li, J. Mitochondrial Genome-Derived CircRNA Mc-COX2 Functions as an Oncogene in Chronic Lymphocytic Leukemia. Mol. Ther. Nucleic Acids 2020, 20, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, X.; Li, J.; Hu, S.; Deng, Y.; Yin, H.; Bao, X.; Zhang, Q.C.; Wang, G.; Wang, B.; et al. Identification of MeacciRNAs and Their Roles in the Mitochondrial Entry of Proteins. Sci. China Life Sci. 2020, 63, 1429–1449. [Google Scholar] [CrossRef]
- Sainero-Alcolado, L.; Liaño-Pons, J.; Ruiz-Pérez, M.V.; Arsenian-Henriksson, M. Targeting Mitochondrial Metabolism for Precision Medicine in Cancer. Cell Death Differ. 2022, 29, 1304–1317. [Google Scholar] [CrossRef]
- Lin, Y.; Yang, B.; Huang, Y.; Zhang, Y.; Jiang, Y.; Ma, L.; Shen, Y.-Q. Mitochondrial DNA-Targeted Therapy: A Novel Approach to Combat Cancer. Cell Insight 2023, 2, 100113. [Google Scholar] [CrossRef]
- Bonekamp, N.A.; Peter, B.; Hillen, H.S.; Felser, A.; Bergbrede, T.; Choidas, A.; Horn, M.; Unger, A.; Di Lucrezia, R.; Atanassov, I.; et al. Small-Molecule Inhibitors of Human Mitochondrial DNA Transcription. Nature 2020, 588, 712–716. [Google Scholar] [CrossRef]
- Araya, M.; Sepúlveda, F.; Villegas, J.; Alarcón, L.; Burzio, L.O.; Burzio, V.A.; Borgna, V. Knockdown of Antisense Noncoding Mitochondrial RNA Reduces Tumorigenicity of Patient-Derived Clear Cell Renal Carcinoma Cells in an Orthotopic Xenograft Mouse Model. Cancers 2024, 16, 830. [Google Scholar] [CrossRef]
- Fitzpatrick, C.; Bendek, M.F.; Briones, M.; Farfán, N.; Silva, V.A.; Nardocci, G.; Montecino, M.; Boland, A.; Deleuze, J.F.; Villegas, J.; et al. Mitochondrial NcRNA Targeting Induces Cell Cycle Arrest and Tumor Growth Inhibition of MDA-MB-231 Breast Cancer Cells through Reduction of Key Cell Cycle Progression Factors. Cell Death Dis. 2019, 10, 423. [Google Scholar] [CrossRef] [PubMed]
- Bendek, M.F.; Fitzpatrick, C.; Jeldes, E.; Boland, A.; Deleuze, J.F.; Farfán, N.; Villegas, J.; Nardocci, G.; Montecino, M.; Burzio, L.O.; et al. Inverse Modulation of Aurora Kinase A and Topoisomerase IIα in Normal and Tumor Breast Cells upon Knockdown of Mitochondrial ASncmtRNA. Noncoding RNA 2023, 9, 59. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).