1. Introduction
Modern radiotherapy has revolutionized the management of lung cancer, offering effective treatment options across all disease stages, either as standalone or in combination with other modalities [
1]. However, the therapeutic benefits of radiotherapy are often offset by its dose-limiting toxicities, particularly pneumonitis and fibrosis, resulting from collateral damage to surrounding healthy lung tissues [
2]. The ionizing radiation (IR) inflicts direct damage to alveolar epithelial cells—primarily type 1 pneumocytes—eliciting a persistent inflammatory response characterized by immune cell recruitment and cytokine release at the injury site. In many instances, this unresolved inflammation progresses into irreversible scarring, clinically manifested as radiation-induced pulmonary fibrosis (RPF) [
3].
RPF is a progressive and debilitating late toxicity that can occur after thoracic radiotherapy. It is driven by dysregulated immune signaling and chronic overexpression of profibrotic mediators such as transforming growth factor-β1 (TGF-β1), tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), and interleukin-6 (IL-6) [
4]. These factors trigger epithelial–mesenchymal transition (EMT), excessive extracellular matrix (ECM) deposition, fibroblast-to-myofibroblast differentiation, and progressive tissue stiffening [
5]. RPF remains a major clinical burden, as its incidence increases with dose-escalated radiotherapy and modern combined-modality treatment strategies. Importantly, no FDA-approved pharmacologic therapy currently exists for preventing or treating RPF, underscoring the need for novel therapeutic approaches.
Although RPF shares several pathological features with idiopathic pulmonary fibrosis (IPF)—including myofibroblast activation, ECM accumulation, and persistent inflammatory signaling—it differs fundamentally in its etiology, temporal dynamics, and transcriptomic architecture. RPF is initiated by ionizing-radiation–induced DNA damage and sterile inflammation, whereas IPF arises from repetitive microinjury and aberrant wound-repair processes of unknown origin. These distinctions highlight the importance of studying RPF as a biologically unique disease entity and justify the use of RPF-specific transcriptomic signatures for therapeutic discovery. Leveraging these disease-specific molecular profiles may enable more precise drug-repurposing strategies tailored to radiation-triggered fibrotic remodeling.
Despite advances in the understanding of pulmonary fibrosis, treatment options remain limited. To date, only two FDA-approved drugs-pirfenidone (Esbriet) and nintendanib (Ofev)-are available for fibrotic lung conditions [
6,
7,
8,
9]. Both agents act by inhibiting fibroblast proliferation and ECM production, and by modulating signaling cascades involved in fibrotic remodeling. However, their efficacy in RPF remains suboptimal, and their broad adoption is hampered by side effects and high costs.
Given the high attrition rates, extended timelines, and escalating costs associated with de novo drug discovery [
10], drug repurposing has emerged as a compelling strategy to expedite the development of new therapies for unmet medical needs [
11,
12]. This approach leverages large-scale transcriptomic databases, such as the Connectivity Map (CMap) and the Library of Integrated Network-Based Cellular Signatures (LINCS), which catalog drug-induced gene expression changes across diverse cell types and treatment conditions [
13,
14,
15].
Central to this strategy is the concept of “signature reversion,” wherein compounds that reverse disease-associated gene expression patterns are considered potential therapeutics [
13]. This methodology has been successfully applied in several disease domains, including cancer [
16,
17], muscle atrophy [
18], acute myeloid leukemia [
19], and neurodegenerative diseases such as Parkinson’s [
20].
In this study, we utilized REMEDY, a proprietary LINCS-based drug repurposing platform developed by Arontier Co. (Seoul, Republic of Korea), to identify candidate therapeutics for RPF. REMEDY integrates advanced pattern-matching algorithms and gene signature enrichment to systematically predict compounds that may reverse fibrotic transcriptomic profiles. Using differentially expressed genes (DEGs) from an established radiation-induced lung injury (RILI) model, our screening highlighted homoharringtonine (HHT), an FDA-approved alkaloid used in leukemia treatment, as a top-ranked candidate. HHT was prioritized not only based on its high negative connectivity score but also due to emerging evidence of its anti-fibrotic properties in other disease contexts. We proceeded to experimentally validate HHT in an in vitro model of RPF and investigated its mechanistic actions on key fibrosis-related signaling pathways. Although HHT has demonstrated anti-fibrotic activity in other organ systems, radiation-induced pulmonary fibrosis (RPF) arises from distinctly different initiating events—including DNA damage–driven epithelial injury, radiation-specific cytokine dynamics, and unique transcriptomic remodeling. Importantly, our study is the first to apply a LINCS/CMap-based reverse gene-signature approach to RPF-specific molecular profiles derived from an established radiation-induced lung injury model. By leveraging these radiation-context signatures rather than generic fibrosis datasets, we identified HHT as a top-ranked reverse-signature compound. This novelty lies in the disease context and the transcriptome-guided discovery framework rather than in proposing an entirely new molecular mechanism for HHT.
2. Materials and Methods
2.1. Reagent
HHT was purchased from Sigma-Aldrich (St. Louis, MO, USA). The compound was dissolved in dimethyl sulfoxide (DMSO), stored at 4 °C, and diluted to the required working concentrations immediately before use.
2.2. Cell Line and Radiation Treatment
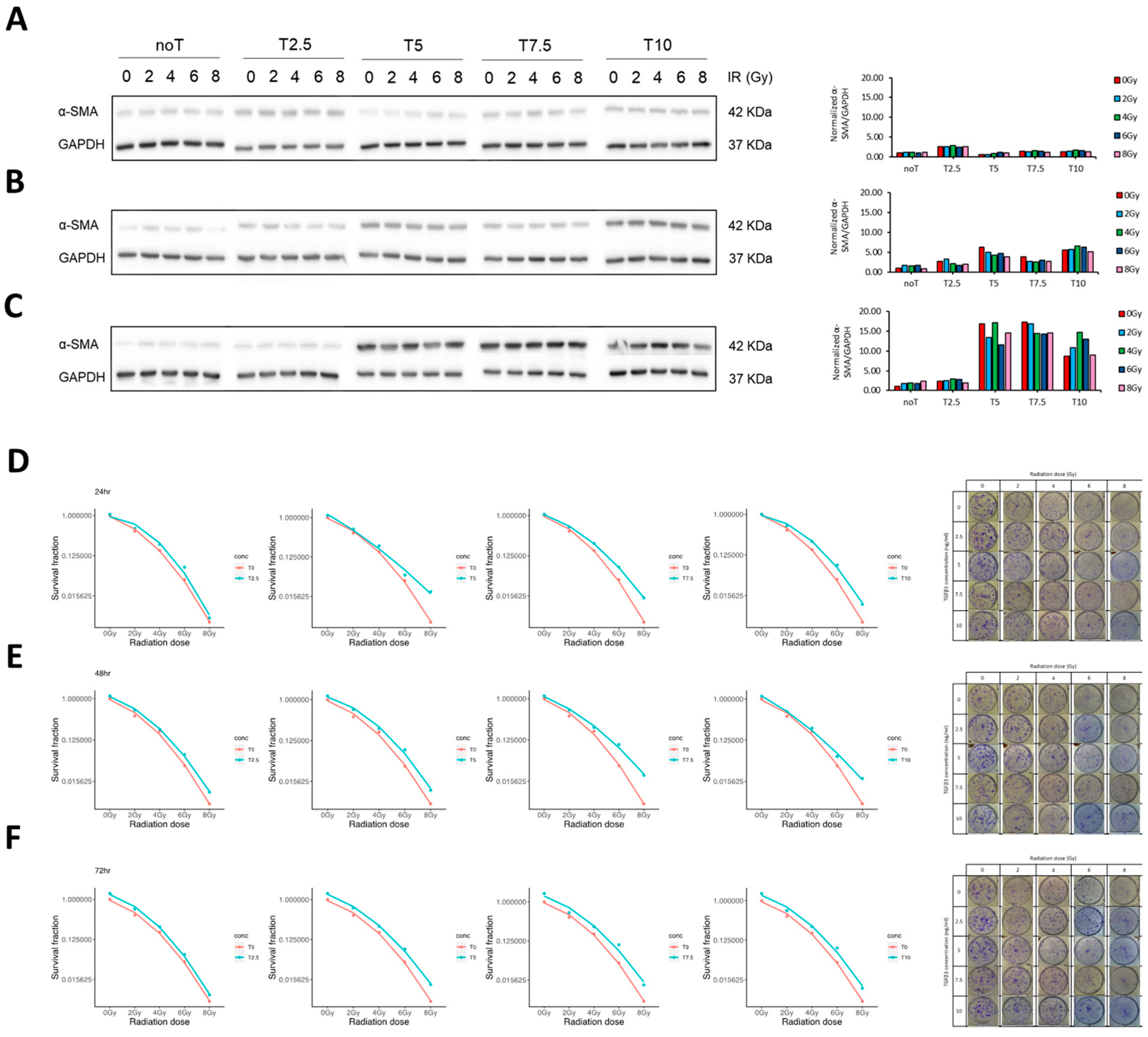
MRC-5, a lung fibroblast isolated from the lung tissue of a white male, 14-week-old embryo, were purchased from Korean Cell Line Bank (KCLB number: 10771). Cells were cultured in Minimal Essential Media (MEM; WELGENE, Gyeongsangbuk-do, Republic of Korea) supplied with 10% heated deactivated fetal bovine serum (FBS; WELGENE, Gyeongsangbuk-do, Republic of Korea) and maintained at 37 °C with 5% of CO2. For establishing the in vitro fibrotic model, cells were irradiated with 4 Gy (1.6 Gy/min, 300 kV, 7.86 mA) using X-RAD 320 (Precision, North Branford, CT, USA) and immediately supplied with approximately 5 ng/mL of recombinant human TGF-β1 (PeproTech, Cranbury, NJ, USA).
2.3. Differential Gene Expression (DEG) Collection
To identify drug candidates capable of reversing RILI-related gene signatures, previously published DEG datasets from a mouse model of radiation-induced lung injury were retrieved [
21]. These datasets included four experimental comparisons categorized by radiation dose (65 Gy and 75 Gy) and pathological phase (inflammatory vs. fibrotic); 65Gy_2W, 65Gy_6W, 75Gy_2W, and 75Gy_6W.
2.4. Drug/Target Prediction Using LINCS L1000 Database
Transcriptomic-based drug prediction was conducted using the REMEDY platform, developed by Arontier Inc. REMEDY operates on level 4 data from the LINCS L1000 molecular signature database, comprising over 1.2 million differential gene expression profiles normalized by z-score transformation. These datasets (Phase I: GSE92742; Phase II: GSE70138) were accessed via the Gene Expression Omnibus (GEO). Perturbagen information including Mode of Action (MoA) and clinical development phase was obtained from the Drug Repurposing Hub (
https://clue.io/repurposing accessed on 11 June 2024). Pattern-matching was performed using two algorithms: the original CMap Kolmogorov–Smirnov (KS)-based score and a modified KS-dependent score proprietary to Arontier Inc. Only reverse (negative) connectivity scores were considered, as all input signatures were disease-associated. Each compound’s frequency among the top 1000 ranked signatures was normalized and assessed for statistical significance using Fisher’s exact test. Candidates with fewer than four hits or a
p-value > 0.05 were excluded. Top-ranked perturbagens were selected for experimental validation.
2.5. Colony Formation Assay (CFA)
MRC5 cells were seeded in a designated plating cell number in 6-well plates overnight. Then, cells were treated with HHT and IR with 2, 4, 6, and 8 Gy using X-RAD 320 (Precision, North Branford, CT, USA). Cells were then re-cultured in MEM media supplied with 10% FBS at 37 °C with 5% (v/v) CO2 for nearly 2 weeks. Afterwards, cells were washed, fixed with cold methanol for 1 h, and then stained with 2% crystal violet. Colonies were quantified by visual observation of colony number counting. The final DMSO concentration in all treatments, including the 0 nm vehicle control, was ≤0.05% (v/v).
2.6. Scratch Wound Migration Assay
In order to conduct tip-scratch assay, MRC5 cells were seeded in 35 mm plates at 37 °C with 5% (v/v) CO2 for overnight so that cells were allowed to adhere and spread over the plate to create confluent (95–100%) monolayer cells. Then, cells were washed, media were changed with fresh one, and cells monolayer was carefully scratched from the middle using clean and sterilized pipette tip. Then, extend of cell migration was measured by periodically monitoring and measuring the cell-free area under microscope for 24 h. Vehicle exposure was standardized across groups (final DMSO ≤ 0.05%).
2.7. Cell Viability Assay
To investigate the cell viability following the treatment, cells were seeded in 96-microwell plates at 37 °C with 5% (v/v) CO2 overnight. Next day, cells were treated and incubated at 37 °C with 5% (v/v) CO2 for 24 h, 48 h, and 72 h. At each harvesting time, cells were treated with cell proliferation reagent WST-1 (Roche Hungary Ltd., Budapest, Hungary) in dark for 2 h. The plates were then read on a microplate spectrophotometer at 450 nm and the viability percentage was calculated by comparing the cell viability following the treated relative to the viability of treatment-free cells. To control for solvent effects, all HHT working dilutions were prepared in sterile water, with a final DMSO concentration ≤ 0.05% (v/v). The 0 nm group received the same vehicle solution without HHT.
2.8. Western Blotting
Cells were lysed in RIPA buffer (50 mMTris-HCl, pH 7.4; 1% Nonidet P-40; 0.25% sodium deoxycholate; 150 mm NaCl; 1 mm Na3VO4) containing protease inhibitors (2 mm phenylmethylsulfonyl fluoride, 100 μg/mL leupeptin, 10 μg/mL pepstatin, 1 μg/mL aprotinin, and 2 mm EDTA) and a phosphatase inhibitor cocktail (GenDEPOT, Baker, TX, USA). After incubation for 30 min, the lysates were centrifuged at 15,000 rpm for 20 min at 4 °C, and the supernatants were obtained for Western blotting. Evaluation of protein concentration was conducted using a BCA protein kit (Bio-Rad, Hercules, CA, USA). Extracted proteins (10 µg) were loaded and separated using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PADGE) for 2 h and transferred onto polyvinylidene fluoride membranes (GE Healthcare, Little Chalfont, UK) for 2 h. For membrane blocking, membranes were incubated with 5% skim milk for nearly 2 h at room temperature. Anti-α-SMA (1:3000, Abcam, Cambridge, UK), Anti-cyclin D1 (1:1000; Cell Signalling Technology, Danvers, MA, USA), anti-β catenin (total) (1:500; Cell Signalling Technology), anti-β catenin (active) (1:500; Cell Signalling Technology), anti-ROCK1 (1:000; Cell Signalling Technology), anti-GAPDH (1:2000; Cell Signalling Technology).
2.9. Collagen Staining
MRC-5 cells were seeded in 6-well plates and maintained in the abovementioned condition overnight. Then, cells were activated by IR with 4 Gy followed by addition of 5 ng/mL of recombinant human TGF-β1 and then incubated in the same condition for 72 h. Then, cells were washed by PBS, fixed with cold 75% ethanol for 1 h, washed again with PBS, and stained with Direct Red 80 dissolved in picric acid (w/v) for 1 h. Cells were washed with 0.01 N HCL to remove unbonded stain, and then photographed under microscope for image staining analysis using ImageJ v1.54d.
2.10. Molecular Docking
Receptor structures of α4β7 (PDB ID: 3V4V), α5β1 (PDB ID: 4WK0), and Frizzled (FzD; PDB ID: 5UWG) were retrieved from the Protein Data Bank (PDB). All receptor structures were prepared using AutoDock Tools 1.5.7 [
22] by removing crystallographic water molecules, adding polar hydrogens, computing Gasteiger partial charges, and converting the files into PDBQT format. The ligand HHT was obtained from PubChem, energy-minimized using the MMFF94 force field, and converted to PDBQT format using Open Babel [
23].
Molecular docking was performed using AutoDock Vina 1.2.0 [
24]. Grid boxes were positioned to cover the experimentally validated or predicted ligand-binding regions of each receptor, with grid centers defined as follows: α4β7 (x = −15.96 Å, y = 14.78 Å, z = 39.36 Å), α5β1 (x = 21.04 Å, y = 11.78 Å, z = −20.64 Å), and FzD (x = 4.00 Å, y = 109.00 Å, z = −9.00 Å) and dimensions size as follows: α4β7 (44 × 52 × 46 points), α5β1 (40 × 40 × 40 points), and FzD (51 × 51 × 51 points). A total of 10 binding poses were generated per docking run using an exhaustiveness of 16. Grid box dimensions were selected to ensure full coverage of the binding pocket while minimizing inclusion of irrelevant surface regions.
To validate the reliability of the docking protocol, each receptor’s co-crystallized ligand was redocked, yielding RMSD values ≤ 2.0 Å between native and redocked poses, confirming the robustness of the docking parameters. Binding significance was evaluated based on three criteria: (1) a predicted binding energy (ΔG) ≤ −6.0 kcal/mol or within 1.0 kcal/mol of the reference ligand for that receptor; (2) the presence of conserved interactions with key amino acids known from SAR or crystallographic studies; and (3) consistency of the docking pose with validated binding-site geometry.
Binding affinities, residue-level interaction patterns, and 2D/3D ligand contact maps were analyzed using Flare Viewer v10. All receptor–ligand structures, docking parameters, and grid definitions were documented to ensure methodological transparency and reproducibility.
2.11. Reproducibility and Replicates
All experiments were performed using three independent biological replicates, each derived from separately cultured cell populations prepared on different days. For viability assays (WST-1), colony-formation assays, and wound-healing assays, each biological replicate contained three technical replicates per experimental condition to ensure assay precision. Western blot analyses were performed using three biological replicates, and densitometric quantification reflects the mean values of these independent samples. Technical replicates were averaged before statistical testing so that biological variability served as the basis of statistical inference.
2.12. Statistical Analysis
Statistical analyses were performed using the rstatix package in the R programming language [
25]. Pairwise comparisons between individual treatment groups and the corresponding control were evaluated using two-tailed unpaired
t-tests, which were applied only when one experimental condition was compared directly with a single reference group (e.g., colony formation, migration assays, and most Western blot analyses). For experiments involving three or more treatment groups, such as the scratch-wound quantification and collagen deposition assays, data were analyzed using one-way ANOVA followed by Tukey’s post hoc test to correct for multiple comparisons. The replicate strategy followed the standardized framework described in the Reproducibility and Replicates section. Statistical significance was defined as
p < 0.05. All quantitative data are presented as mean ± standard error (SE) from three independent biological replicates.
4. Discussion
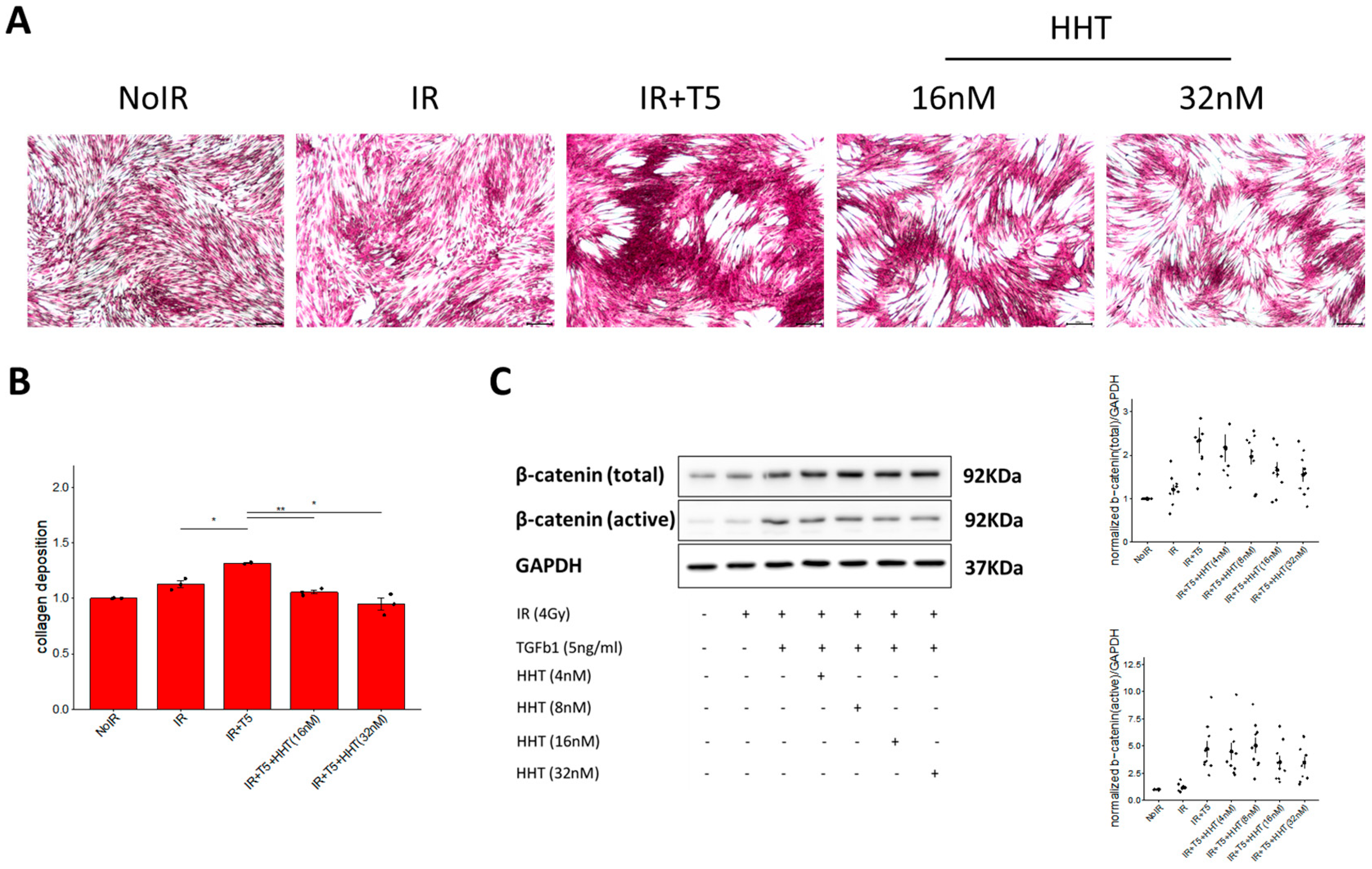
Genome-wide association studies (GWAS) have identified numerous genetic loci associated with disease susceptibility. However, listing these loci alone often fails to establish causality or provide mechanistic insight. To bridge this gap, perturbation-based approaches—where cellular systems are modulated and responses observed—are crucial for elucidating disease mechanisms and identifying therapeutic targets. The CMap, which links genes, drugs, and diseases via shared gene expression signatures, offers a powerful platform for understanding the mechanisms of small molecules. In this study, we employed REMEDY, a computational drug repurposing platform built on the LINCS dataset, to identify candidate therapeutics for RPF. Through this approach, we identified protein synthesis inhibitors—particularly HHT—as top-ranked candidates. Subsequent in vitro validation using irradiated, TGF-β1-activated fibroblasts confirmed the anti-fibrotic effects of HHT (
Figure 8).
While HHT’s anti-fibrotic activity has been described in arthrofibrosis, hepatic fibrosis, and cardiac fibrosis, these conditions do not share the same transcriptomic architecture as radiation-induced injury. Fibrosis biology exhibits a high degree of pathway convergence, particularly involving ROCK/RhoA and Wnt/β-catenin signaling. Therefore, overlap in mechanistic pathways is biologically expected and does not diminish the originality of the present work. The novelty of this study lies in demonstrating that RPF-specific molecular signatures can computationally predict HHT as a reverse-signature therapeutic candidate and in validating its effects within a radiation-dependent fibroblast activation model, a context that has never previously been explored.
Recent tools such as L1000 Fireworks Display (L1000FWD), the Drug Gene Interaction Database (DGIdb), and CMap have been increasingly utilized to predict drug–disease interactions by comparing disease-associated DEGs with drug-induced gene signatures [
14]. This strategy has demonstrated success across various pathologies, including cancer [
16,
17], muscle atrophy [
18], acute myelogenous leukemia [
19], and Parkinson’s disease [
20]. To our knowledge, the present study represents the first application of the LINCS/CMap platform to identify therapeutic candidates for RPF.
Establishing a relevant in vitro model that mimics the RPF microenvironment was a foundational step in this study. Previous studies typically relied on TGF-β1 alone to induce fibroblast activation and differentiation into myofibroblasts. However, this does not fully recapitulate the pathological conditions of radiation-induced lung injury (RILI). Here, we combined IR with TGF-β1 to establish a more physiologically relevant fibrotic model. Our results demonstrated that α-SMA—a hallmark of myofibroblast differentiation—was robustly upregulated following treatment with 5 ng/mL TGF-β1 at 72 h, even in the absence of IR. This finding partially contrasts prior reports where α-SMA induction was IR-dependent and further enhanced by TGF-β1 [
32]. Another important methodological consideration in establishing our in vitro RPF model was the selection of the radiation dose. We systematically evaluated a range of doses (0–8 Gy) to determine the optimal balance between fibroblast activation and cell viability. Our data showed that 4 Gy produced the most robust induction of α-SMA and stress-fiber remodeling, whereas lower doses such as 2 Gy induced only minimal activation despite better overall survival. In contrast, higher doses (6–8 Gy) markedly reduced clonogenic capacity and increased apoptosis, thereby limiting their suitability for downstream functional assays. Thus, 4 Gy represented an optimal “activation-without-cytotoxicity” window—sufficient to synergize with TGF-β1 and reproduce a reproducible fibrotic phenotype while maintaining viable cell populations for mechanistic and phenotypic analyses. This rationale supports the use of 4 Gy as the irradiation condition for the fibrotic model employed in this study.
It is important to note that previous studies employed higher IR doses (e.g., 6 Gy), which may have contributed to greater α-SMA expression. In our model, we used a 4 Gy dose to minimize radiation-induced apoptosis, which was observed to increase sharply at higher doses. We hypothesize that α-SMA induction in our model may be driven largely by TGF-β1, with IR contributing more to cell enlargement and migration than differentiation.
This also aligns with prior observations showing that low IR doses (e.g., 3 Gy) combined with TGF-β1 may not significantly increase α-SMA expression [
33]. Additionally, our model demonstrated IR- and TGF-β1-dependent enhancement of fibroblast migration, further validating its fidelity. The concurrence between our data and previous findings confirms that this two-hit model reliably reproduces key fibrogenic features and is suitable for testing anti-fibrotic agents.
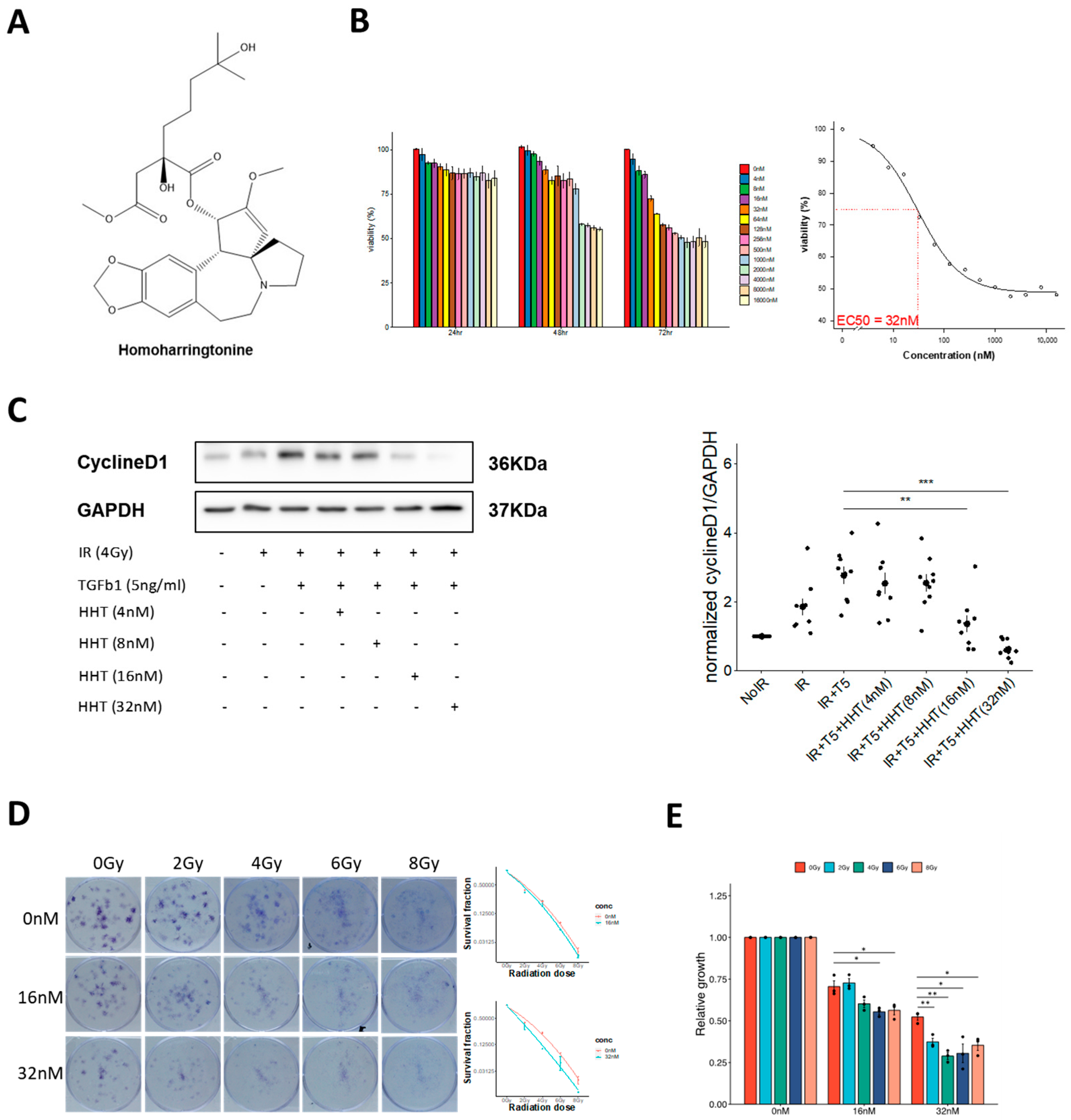
HHT is a well-characterized plant alkaloid and FDA-approved drug for the treatment of chronic myeloid leukemia. Recently, its therapeutic potential has been explored in various fibrotic conditions, including arthrofibrosis [
34], epidural fibrosis [
35], liver fibrosis [
17], and cardiac fibrosis [
36]. However, our study is the first to propose HHT as a candidate for treating RPF, specifically during the fibrotic phase. Prior research has reported effective anti-fibrotic activity at a concentration of ~34 nm [
36], which is consistent with the dose range employed in our experiments. Notably, HHT exhibited stronger anti-proliferative effects in irradiated (activated) fibroblasts compared to non-irradiated cells, suggesting preferential targeting of pathological phenotypes with reduced toxicity to normal tissue. Several molecular parallels exist between RPF and idiopathic pulmonary fibrosis (IPF), including epithelial–mesenchymal transition (EMT), excessive ECM deposition, and myofibroblast-driven lung tissue remodeling. The RhoA/ROCK pathway plays a central role in fibrogenesis by promoting fibroblast contractility, α-SMA expression, and collagen production. Dual inhibition of ROCK1 and ROCK2 has been shown to suppress COL1A1 and α-SMA expression and disrupt stress fiber formation in both animal models and MRC-5 cell lines [
37,
38]. Likewise, the canonical Wnt/β-catenin pathway is implicated in fibroblast activation and ECM production. Inhibition of β-catenin nuclear translocation—e.g., by bufotalin—has been shown to attenuate lung fibrosis in both bleomycin- and radiation-induced models [
39]. Given that pro-fibrotic remodeling is mediated by a limited set of conserved molecular pathways, most anti-fibrotic agents—including HHT—are naturally expected to converge on these canonical nodes. Accordingly, our goal was not to propose a novel molecular target for HHT, but to establish its relevance to radiation-triggered fibroblast activation using RPF-specific transcriptomic data. This distinction is essential, as the radiation-induced fibrotic environment differs markedly from idiopathic or post-surgical fibrosis, both in cytokine composition and in gene-expression patterning. By repositioning HHT to this radiation-specific disease context, our study fills a previously unaddressed therapeutic gap.
A limitation of the present study is the absence of established anti-fibrotic agents such as pirfenidone or nintedanib as positive controls. Although these agents are commonly used in preclinical fibrosis studies, including radiation-associated models, their effects in fibroblast-based assays are generally modest and often require prolonged exposure or more complex systems (e.g., co-culture or ECM-enriched conditions) to yield quantifiable changes. Because our primary aim was to conduct initial mechanistic and phenotypic validation of a transcriptome-identified hit (HHT) using a simplified IR/TGF-β1 activation model, we compared HHT only with the IR-treated negative control to isolate its direct influence on fibroblast activation without confounding pharmacologic background. Nonetheless, future work incorporating pirfenidone and nintedanib using extended culture periods and 3D/ECM-enhanced models is underway and will help benchmark the comparative potency and mechanistic distinctiveness of HHT.
An important consideration when repurposing HHT for a non-malignant indication such as RPF is its well-known myelosuppressive toxicity profile. In oncology, the dose-limiting toxicity of HHT is systemic myelosuppression, largely driven by its inhibition of global protein synthesis in rapidly proliferating hematopoietic cells. However, the concentrations applied in our in vitro experiments (10–50 nm) are substantially lower than those used in leukemia treatment, suggesting the possibility of a wider therapeutic window for applications that employ localized, low-dose, or short-exposure strategies. Moreover, several drug-delivery approaches—including localized pulmonary delivery, nanoformulation, liposomal encapsulation, and inhalation-based aerosolization—have been investigated in prior preclinical studies of other repurposed agents to limit systemic exposure while maximizing target-organ deposition. Although such delivery strategies were beyond the scope of the present study, they may theoretically reduce bone-marrow exposure and warrant systematic evaluation in future preclinical toxicology studies. Taken together, these considerations underscore the importance of carefully assessing HHT’s safety profile during its translational development while also highlighting feasible avenues to mitigate systemic toxicity as the compound advances toward in vivo investigation.
Despite the strengths of our computational–experimental workflow, several limitations should be acknowledged. First, the present study lacks in vivo validation, and future work using radiation-induced lung fibrosis models will be required to determine whether the transcriptome-guided effects of HHT translate into organism-level mitigation of fibrotic remodeling. Second, although we discuss potential strategies to limit systemic exposure, concerns regarding off-target or non-pulmonary toxicity remain and must be rigorously addressed in dedicated preclinical toxicology studies. Third, our computational predictions were derived exclusively from the LINCS L1000 dataset, which, despite its scale, captures only a subset of drug-induced transcriptional states. Additional cross-platform validation—incorporating alternative perturbational resources or full-length RNA-seq signatures—will therefore be important to strengthen the generalizability of our drug-repurposing results.
In addition, the docking analyses should be viewed as predictive and hypothesis-generating rather than definitive biochemical evidence of receptor–ligand binding. AutoDock Vina scoring functions approximate relative free energies and do not capture full receptor flexibility, solvent dynamics, or induced-fit conformational changes. Consequently, the predicted interactions between HHT and integrin or Frizzled receptors should be interpreted cautiously, and future biochemical binding and SAR studies will be required to validate these proposed interactions.
Collectively, these elements highlight that the originality of this study resides not in discovering a new biochemical action of HHT, but in demonstrating that a LINCS-based signature reversion strategy tailored to RPF can identify a viable therapeutic compound. This represents a conceptual advance in how radiation-related fibrotic disorders may be approached, enabling therapeutic discovery that is guided by disease-specific transcriptomic signatures rather than generalized fibrosis pathways.
Our data show that HHT significantly suppresses α-SMA, ROCK1, and β-catenin expression, consistent with inhibition of both the RhoA/ROCK and Wnt/β-catenin pathways. These results provide mechanistic support for HHT’s anti-fibrotic action and highlight its potential to interrupt multiple pro-fibrotic signaling cascades. Importantly, the anti-fibrotic effects observed with HHT cannot be explained solely by non-specific cytotoxicity arising from its canonical role as a protein synthesis inhibitor. First, the concentrations used in our in vitro assays (10–50 nm) fall within a range reported to exert minimal global cytotoxicity in fibroblast systems, yet these doses led to substantial reductions in α-SMA, ROCK1, and β-catenin expression. Second, inhibition of fibroblast migration occurred at concentrations that did not significantly affect cell viability, indicating a functional anti-fibrotic response independent of cell death. Third, irradiated fibroblasts displayed heightened sensitivity to HHT compared with non-irradiated cells, suggesting preferential targeting of activated or pathological phenotypes rather than generalized toxicity. Finally, molecular docking revealed strong binding affinities of HHT to α5β1, α4β7, and FzD receptors, supporting a mechanism that extends beyond translation inhibition alone. Together, these findings indicate that HHT mediates pathway-level modulation and phenotypic reprogramming rather than exerting merely translation-related cytotoxicity. Integrins, particularly α4- and α5-containing subtypes (e.g., α4β7, α5β1), are key regulators of fibrosis through their ability to activate RhoA/ROCK signaling and promote cytoskeletal reorganization and matrix stiffening [
40,
41,
42]. Prior studies have shown that HHT inhibits α5β1-mediated signaling in bladder cancer cells, either through direct receptor interaction or via suppression of protein translation [
43]. In our molecular docking simulations, HHT demonstrated strong binding affinities to α5β1, α4β7, and FzD receptors, further supporting its ability to modulate fibrosis-relevant pathways beyond translation inhibition.