Serendipitous Hinge Modulation Hypothetically Reprograms Caerin 1.1-LC Antibacterial Mechanism and Gram-Negative Selectivity
Abstract
1. Introduction
2. Materials and Methods
2.1. Collections of Skin Secretions from Litoria Caerulea
2.2. Molecular Cloning
2.3. Peptide Synthesis, Purification and Identification
2.4. Antimicrobial Assay
2.5. Time-Killing Assay
2.6. Salt and Serum Sensitivity Assay
2.7. Haemolysis Assay
2.8. MTT Assay
2.9. In Vivo Toxicity and Antimicrobial Activity
2.10. LPS Neutralisation Assay
2.11. LPS-Binding Assay
2.12. NPN Outer Membrane Permeability Assay
2.13. ONPG Inner Membrane Permeability Assay
2.14. Bacterial Membrane Permeability Assay
2.15. Localisation of FITC-Labelled Peptide on Bacteria
2.16. Membrane Depolarisation Assay
2.17. Intracellular ATP Assay
2.18. Statistical Analysis
3. Results
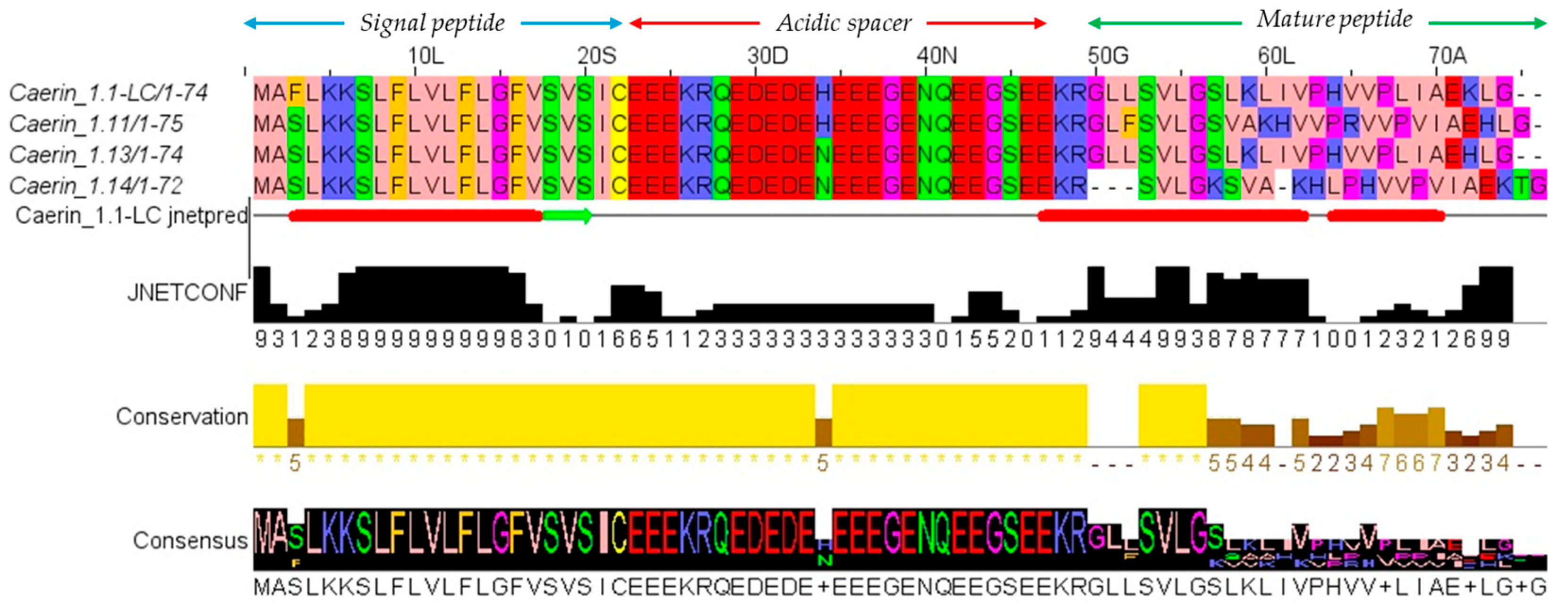
3.1. Molecular Cloning of Caerin 1.1-LC Precursor-Encoding cDNA
3.2. Peptide Design, Characterisation and Structure Analysis
3.3. Antimicrobial Activity
3.4. Haemolysis, Therapeutic Index and Cytotoxicity
3.5. Salt Sensitivity of Caerin 1.1-LC and Its Analogues
3.6. In Vivo Antimicrobial Activity on Larvae Model
3.7. LPS Neutralisation Activity
3.8. Time-Killing Kinetics Studies
3.9. LPS-Binding Affinity
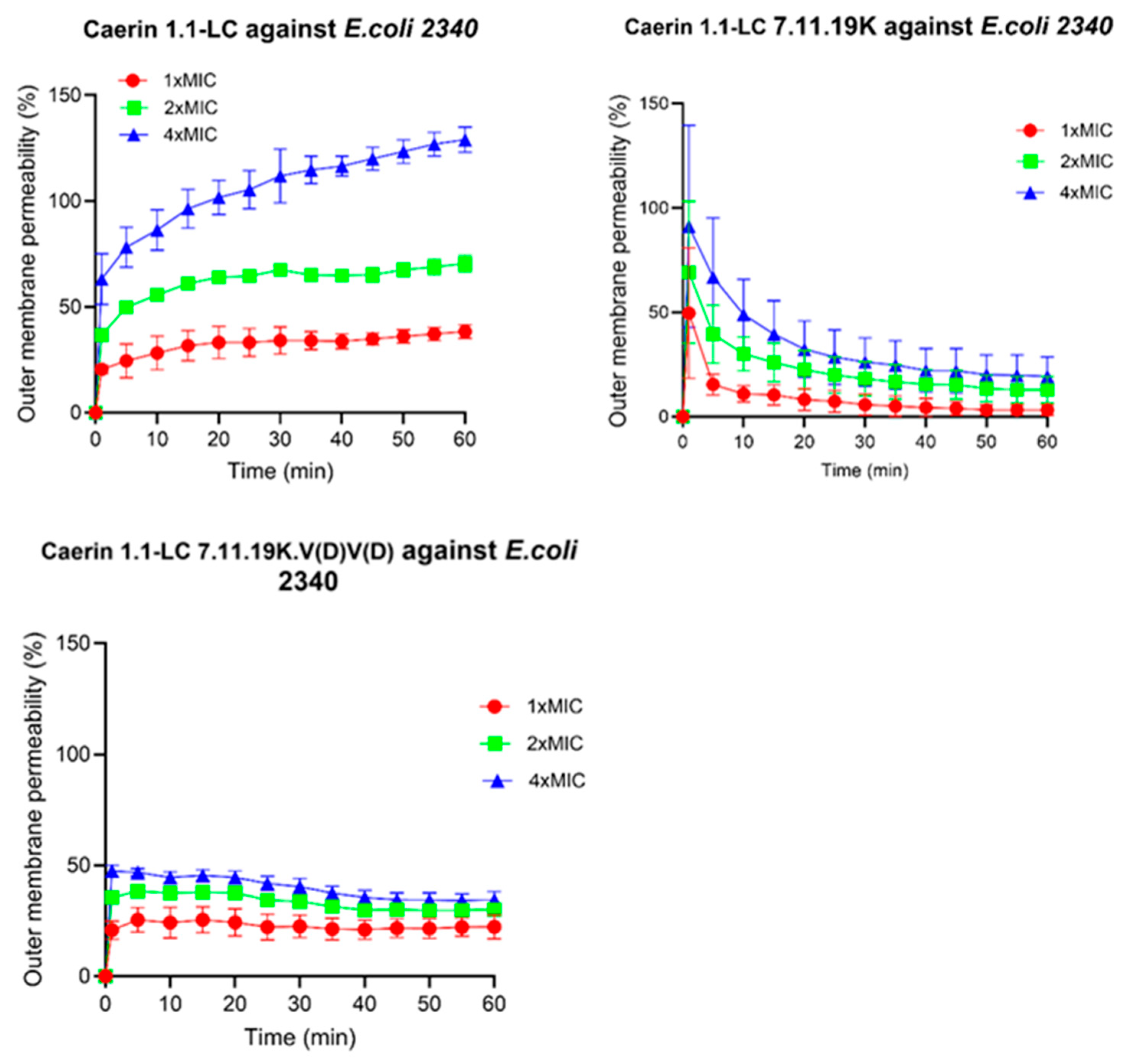
3.10. Outer Membrane Permeabilisation
3.11. Intracellular Membrane Permeability
3.12. Further Validation of Membrane Integrity
3.13. Localisation of FITC-Labelled Peptides
3.14. Membrane Depolarisation Activity
3.15. Disruption of the Intracellular ATP Level
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Haney, E.F.; Mansour, S.C.; Hancock, R.E.W. Antimicrobial Peptides: An Introduction. In Methods in Molecular Biology (Clifton N.J.); Humana Press: New York, NY, USA, 2017; Volume 1548, pp. 3–22. [Google Scholar] [CrossRef]
- Shinu, P.; Al Mouslem, A.K.; Nair, A.B.; Venugopala, K.N.; Attimarad, M.; Singh, V.A.; Nagaraja, S.; Alotaibi, G.; Deb, P.K. Progress Report: Antimicrobial Drug Discovery in the Resistance Era. Pharmaceuticals 2022, 15, 413. [Google Scholar] [CrossRef] [PubMed]
- Jackson, N.; Czaplewski, L.; Piddock, L.J.V. Discovery and Development of New Antibacterial Drugs: Learning from Experience? J. Antimicrob. Chemother. 2018, 73, 1452–1459. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Separovic, F.; O’Brien-Simpson, N.M.; Wade, J.D. Chemically Modified and Conjugated Antimicrobial Peptides against Superbugs. Chem. Soc. Rev. 2021, 50, 4932–4973. [Google Scholar] [CrossRef]
- Hutchings, M.; Truman, A.; Wilkinson, B. Antibiotics: Past, Present and Future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef]
- Mwangi, J.; Hao, X.; Lai, R.; Zhang, Z.Y. Antimicrobial Peptides: New Hope in the War against Multidrug Resistance. Zool. Res. 2019, 40, 488–505. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Garg, A.; Srivastava, A.; Arora, P.K. The Role of Antimicrobial Peptides in Overcoming Antibiotic Resistance. Microbe 2025, 7, 100337. [Google Scholar] [CrossRef]
- Sarkar, T.; Chetia, M.; Chatterjee, S. Antimicrobial Peptides and Proteins: From Nature’s Reservoir to the Laboratory and Beyond. Front. Chem. 2021, 9, 691532. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Björn, C.; Ekblom, J. Antimicrobial Peptides as Therapeutic Agents: Opportunities and Challenges. Crit. Rev. Biotechnol. 2020, 40, 978–992. [Google Scholar] [CrossRef]
- Tan, P.; Fu, H.; Ma, X. Design, Optimization, and Nanotechnology of Antimicrobial Peptides: From Exploration to Applications. Nano Today 2021, 39, 101229. [Google Scholar] [CrossRef]
- Xiao, L.; Yang, X.; Li, J.; Zhang, P.; Tang, S.; Cao, D.; Chen, S.; Li, H.; Zhang, W.; Chen, G.; et al. Caerin 1 Peptides, the Potential Jack-of-All-Trades for the Multiple Antibiotic-Resistant Bacterial Infection Treatment and Cancer Immunotherapy. Biomed. Res. Int. 2022, 2022, 7841219. [Google Scholar] [CrossRef]
- Pukala, T.L.; Brinkworth, C.S.; Carver, J.A.; Bowie, J.H. Investigating the Importance of the Flexible Hinge in Caerin 1.1: Solution Structures and Activity of Two Synthetically Modified Caerin Peptides. Biochemistry 2004, 43, 937–944. [Google Scholar] [CrossRef]
- Vermeer, L.S.; Lan, Y.; Abbate, V.; Ruh, E.; Bui, T.T.; Wilkinson, L.J.; Kanno, T.; Jumagulova, E.; Kozlowska, J.; Patel, J.; et al. Conformational Flexibility Determines Selectivity and Antibacterial, Antiplasmodial, and Anticancer Potency of Cationic α-Helical Peptides*. J. Biol. Chem. 2012, 287, 34120–34133. [Google Scholar] [CrossRef]
- Lü, Y.; Bai, J.; Tan, D.; Chen, T.; Shan, A. The Effects of Hinge Structure on the Biological Activity of Antimicrobial Peptides and Its Application in Molecular Design: A Review. Shengwu Gongcheng Xuebao/Chin. J. Biotechnol. 2021, 37, 3142–3150. [Google Scholar] [CrossRef]
- Chia, B.C.S.; Carver, J.A.; Mulhern, T.D.; Bowie, J.H. Maculatin 1.1, an Anti-Microbial Peptide from the Australian Tree Frog, Litoria Genimaculata. Solution Structure and Biological Activity. Eur. J. Biochem. 2000, 267, 1894–1908. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.L.; Kim, Y.; Park, Y.; Jae, I.K.; Park, I.S.; Hahm, K.S.; Song, Y.S. The Role of the Central L- or D-Pro Residue on Structure and Mode of Action of a Cell-Selective α-Helical IsCT-Derived Antimicrobial Peptide. Biochem. Biophys. Res. Commun. 2005, 334, 1329–1335. [Google Scholar] [CrossRef]
- Timmons, P.B.; O’Flynn, D.; Conlon, J.M.; Hewage, C.M. Insights into Conformation and Membrane Interactions of the Acyclic and Dicarba-Bridged Brevinin-1BYa Antimicrobial Peptides. Eur. Biophys. J. 2019, 48, 701–710. [Google Scholar] [CrossRef]
- Chen, T.; Scott, C.; Tang, L.; Zhou, M.; Shaw, C. The Structural Organization of Aurein Precursor CDNAs from the Skin Secretion of the Australian Green and Golden Bell Frog, Litoria Aurea. Regul. Pept. 2005, 128, 75–83. [Google Scholar] [CrossRef]
- Li, L.; Wu, Q.; Wang, X.; Lu, H.; Xi, X.; Zhou, M.; Watson, C.J.; Chen, T.; Wang, L. Discovery of Novel Caeridins from the Skin Secretion of the Australian White’s Tree Frog, Litoria Caerulea. Int. J. Genom. 2018, 2018, 8158453. [Google Scholar] [CrossRef]
- Shi, J.; Chen, C.; Wang, D.; Wang, Z.; Liu, Y. The Antimicrobial Peptide LI14 Combats Multidrug-Resistant Bacterial Infections. Commun. Biol. 2022, 5, 926. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Sun, R.; Yao, A.; Zhou, M.; Chen, X.; Ma, C.; Wang, T.; Jiang, Y.; Chen, T.; Shaw, C.; et al. A Promising Antibiotic Candidate, Brevinin-1 Analogue 5R, against Drug-Resistant Bacteria, with Insights into Its Membrane-Targeting Mechanism. Comput. Struct. Biotechnol. J. 2023, 21, 5719–5737. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.; Chai, J.; Deng, Z.; Ye, T.; Chen, W.; Li, D.; Chen, X.; Chen, M.; Xu, X. Functional Characterization of a Novel Lipopolysaccharide-Binding Antimicrobial and Anti-Inflammatory Peptide in Vitro and in Vivo. J. Med. Chem. 2018, 61, 10709–10723. [Google Scholar] [CrossRef]
- Wang, J.; Hu, J.; Pu, W.; Chen, X.; Ma, C.; Jiang, Y.; Wang, T.; Chen, T.; Shaw, C.; Zhou, M.; et al. Discovery, Development and Optimisation of a Novel Frog Antimicrobial Peptide with Combined Mode of Action against Drug-Resistant Bacteria. Comput. Struct. Biotechnol. J. 2024, 23, 3391–3406. [Google Scholar] [CrossRef]
- Yang, Z.; He, S.; Wang, J.; Yang, Y.; Zhang, L.; Li, Y.; Shan, A. Rational Design of Short Peptide Variants by Using Kunitzin-RE, an Amphibian-Derived Bioactivity Peptide, for Acquired Potent Broad-Spectrum Antimicrobial and Improved Therapeutic Potential of Commensalism Coinfection of Pathogens. J. Med. Chem. 2019, 62, 4586–4605. [Google Scholar] [CrossRef]
- Meier, S.; Ridgway, Z.M.; Picciano, A.L.; Caputo, G.A. Impacts of Hydrophobic Mismatch on Antimicrobial Peptide Efficacy and Bilayer Permeabilization. Antibiotics 2023, 12, 1624. [Google Scholar] [CrossRef] [PubMed]
- Hao, G.; Shi, Y.-H.; Tang, Y.-L.; Le, G.-W. The Membrane Action Mechanism of Analogs of the Antimicrobial Peptide Buforin 2. Peptides 2009, 30, 1421–1427. [Google Scholar] [CrossRef] [PubMed]
- Ki, M.R.; Kim, S.H.; Park, T.I.; Pack, S.P. Self-Entrapment of Antimicrobial Peptides in Silica Particles for Stable and Effective Antimicrobial Peptide Delivery System. Int. J. Mol. Sci. 2023, 24, 16423. [Google Scholar] [CrossRef] [PubMed]
- Noda, T.; Kim, J.; Huang, W.-P.; Baba, M.; Tokunaga, C.; Ohsumi, Y.; Klionsky, D.J. Apg9p/Cvt7p Is an Integral Membrane Protein Required for Transport Vesicle Formation in the Cvt and Autophagy Pathways. J. Cell Biol. 2000, 148, 465–480. [Google Scholar] [CrossRef]
- Song, R.; Han, X.; Jie, H.; Zhang, X.; Li, S.; Sun, E. Effects and Mechanisms of Celastrol on the Formation of Neutrophil Extracellular Traps (NETs). Ann. Transl. Med. 2023, 11, 16. [Google Scholar] [CrossRef]
- Belokoneva, O.S.; Satake, H.; Mal’tseva, E.L.; Pal’mina, N.P.; Villegas, E.; Nakajima, T.; Corzo, G. Pore Formation of Phospholipid Membranes by the Action of Two Hemolytic Arachnid Peptides of Different Size. Biochim. Biophys. Acta (BBA)-Biomembr. 2004, 1664, 182–188. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lai, Z.; Tan, P.; Zhu, Y.; Shao, C.; Shan, A.; Li, L. Highly Stabilized α-Helical Coiled Coils Kill Gram-Negative Bacteria by Multicomplementary Mechanisms under Acidic Condition. ACS Appl. Mater. Interfaces 2019, 11, 22113–22128. [Google Scholar] [CrossRef]
- Wong, H.; Bowie, J.H.; Carver, J.A. The Solution Structure and Activity of Caerin 1.1, an Antimicrobial Peptide from the Australian Green Tree Frog, Litoria Splendida. Eur. J. Biochem. 1997, 247, 545–557. [Google Scholar] [CrossRef]
- Yang, S.T.; Jeon, J.H.; Kim, Y.; Shin, S.Y.; Hahm, K.S.; Kim, J. Il Possible Role of a PXXP Central Hinge in the Antibacterial Activity and Membrane Interaction of PMAP-23, a Member of Cathelicidin Family. Biochemistry 2006, 45, 1775–1784. [Google Scholar] [CrossRef]
- Mishra, A.K.; Choi, J.; Moon, E.; Baek, K.H. Tryptophan-Rich and Proline-Rich Antimicrobial Peptides. Molecules 2018, 23, 815. [Google Scholar] [CrossRef] [PubMed]
- Cui, Q.; Xu, Q.; Liu, L.; Guan, L.; Jiang, X.; Inam, M.; Kong, L.; Ma, H.X. Preparation, Characterization and Pharmacokinetic Study of N-Terminal PEGylated D-Form Antimicrobial Peptide OM19r-8. J. Pharm. Sci. 2021, 110, 1111–1119. [Google Scholar] [CrossRef] [PubMed]
- Oliva, R.; Chino, M.; Pane, K.; Pistorio, V.; De Santis, A.; Pizzo, E.; D’Errico, G.; Pavone, V.; Lombardi, A.; Del Vecchio, P.; et al. Exploring the Role of Unnatural Amino Acids in Antimicrobial Peptides. Sci. Rep. 2018, 8, 8888. [Google Scholar] [CrossRef] [PubMed]
- Frimpong, A.O.; Xu, X.; Jia, X.; Zhang, Y. Divalent Cation Induced Re-Entrant Condensation Behavior for Lipopolysaccharides. J. Chem. Phys. 2022, 157, 154902. [Google Scholar] [CrossRef]
- Hongming, H.; Huang, Y.; Shunhang, F.; Xiaoyan, Z.; Chu, C.; Hongyu, Y.; Rui, L.; Mengxin, L.; Juan, L.; Yancheng, W.; et al. High Salt Condition Alters LPS Synthesis and Induces the Emergence of Drug Resistance Mutations in Helicobacter Pylori. Antimicrob. Agents Chemother. 2024, 68, e00587-24. [Google Scholar] [CrossRef]
- Wei, L.; Yang, J.; He, X.; Mo, G.; Hong, J.; Yan, X.; Lin, D.; Lai, R. Structure and Function of a Potent Lipopolysaccharide-Binding Antimicrobial and Anti-Inflammatory Peptide. J. Med. Chem. 2013, 56, 3546–3556. [Google Scholar] [CrossRef]
- Tian, M.; Wang, K.; Liang, Y.; Chai, J.; Wu, J.; Zhang, H.; Huang, X.; Chen, X.; Xu, X. The First Brevinin-1 Antimicrobial Peptide with LPS-Neutralizing and Anti-Inflammatory Activities in Vitro and in Vivo. Front. Microbiol. 2023, 14, 1102576. [Google Scholar] [CrossRef]
- Rice, A.; Zourou, A.C.; Cotten, M.L.; Pastor, R.W. A Unified Model of Transient Poration Induced by Antimicrobial Peptides. Proc. Natl. Acad. Sci. USA 2025, 122, e2510294122. [Google Scholar] [CrossRef]
- Qiu, J.; Wu, D.; Xu, C.; Zhuang, Y.; Lu, Y.; Sun, A.; Lu, X.; Han, J.; Ni, L. Current Advances and Emerging Prospects of Specifically Targeted Antimicrobial Peptides: A Comprehensive Review. Int. J. Biol. Macromol. 2025, 322, 147037. [Google Scholar] [CrossRef] [PubMed]










| Peptide Name | Peptide Sequence |
|---|---|
| Caerin 1.1-LC | GLLSVLGSLKLIVPHVVPLIAEKL-NH2 |
| Caerin 1.1-LC-11K | GLLSVLGSLKKIVPHVVPLIAEKL-NH2 |
| Caerin 1.1-LC-11K.22K | GLLSVLGSLKKIVPHVVPLIAKKL-NH2 |
| Caerin 1.1-LC-11K.22K.12W | GLLSVLGSLKKWVPHVVPLIAKKL-NH2 |
| Caerin 1.1-LC-11K.22K.19W | GLLSVLGSLKKIVPHVVPWIAKKL-NH2 |
| Caerin 1.1-LC 7.11.19K | GLLSVLKSLKKIVPHVVPKIAEKL-NH2 |
| Caerin 1.1-LC 2P-2A | GLLSVLGSLKLIVAHVVALIAEKL-NH2 |
| Caerin 1.1-LC PGGGP | GLLSVLGSLKLIVPGGGPLIAEKL-NH2 |
| Caerin 1.1-LC PRVVP | GLLSVLGSLKLIVPRVVPLIAEKL-NH2 |
| Caerin 1.1-LC PKVVP | GLLSVLGSLKLIVPKVVPLIAEKL-NH2 |
| Caerin 1.1-LC PHLLP | GLLSVLGSLKLIVPHLLPLIAEKL-NH2 |
| Caerin 1.1-LC PHIIP | GLLSVLGSLKLIVPHIIPLIAEKL-NH2 |
| Caerin 1.1-LC PHWWP | GLLSVLGSLKLIVPHWWPLIAEKL-NH2 |
| Caerin 1.1-LC 11K.19K.PKAVP | GLLSVLGSLKKIVPKAVPKIAEKL-NH2 |
| Caerin 1.1-LC 7.11.19K.V(D)V(D) | GLLSVLKSLKKIVPHvvPKIAEKL-NH2 |
| Caerin 1.1-LC PKAL(D)P | GLLSVLGSLKLIVPKAlPKIAEKL-NH2 |
| Peptide Name | Length (AA) | Hydrophobicity * | Net Charge at pH 7 |
|---|---|---|---|
| Caerin 1.1-LC | 24 | 0.815 | +2 |
| Caerin 1.1-LC-11K | 24 | 0.703 | +3 |
| Caerin 1.1-LC-11K.22K | 24 | 0.688 | +5 |
| Caerin 1.1-LC-11K.12K.12W | 24 | 0.707 | +5 |
| Caerin 1.1-LC-11K.12K.19W | 24 | 0.711 | +5 |
| Caerin 1.1-LC 7.11.19K | 24 | 0.550 | +5 |
| Caerin 1.1-LC 2P-2A | 24 | 0.781 | +2 |
| Caerin 1.1-LC PGGGP | 24 | 0.708 | +2 |
| Caerin 1.1-LC PRVVP | 24 | 0.768 | +3 |
| Caerin 1.1-LC PKVVP | 24 | 0.768 | +3 |
| Caerin 1.1-LC PHLLP | 24 | 0.855 | +2 |
| Caerin 1.1-LC PHIIP | 24 | 0.863 | +2 |
| Caerin 1.1-LC PHWWP | 24 | 0.901 | +2 |
| Caerin 1.1-LC 11K.19K.PKAVP | 24 | 0.506 | +5 |
| Caerin 1.1-LC 7.11.19K.V(D)V(D) | 24 | 0.550 | +5 |
| Caerin 1.1-LC PKAL(D)P | 24 | 0.526 | +6 |
| Peptide | Gram-Negative Bacteria Strains | Gram-Positive Bacteria Strains | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| E. coli (ATCC 8739) | E. coli (ATCC BAA-2340) | E. coli (NCTC 13846) | K. pneumoniae (ATCC 43816) | K. pneumoniae (ATCC BAA-2342) | P. aeruginosa (ATCC 9027) | A. baumannii (ATCC BAA-747) | S. aureus (ATCC 6538) | E. faecalis (NCTC 12697) | MRSA (NCTC 12493) | |
| Caerin 1.1-LC | 16/64 | 8/16 | 4/32 | 16/64 | 16/64 | 64/128 | 16/128 | 64/128 | >128 | 32/128 |
| Caerin 1.1-LC 11K | 8/16 | 2/8 | 4/8 | 16/32 | 8/16 | 16/32 | 32/32 | 8/32 | 128/128 | 8/8 |
| Caerin 1.1-LC 11K.22K | 1/2 | 2/2 | 4/8 | 8/16 | 4/4 | 4/8 | 4/16 | 4/16 | 64/64 | 4/8 |
| Caerin 1.1-LC 11K.12K.12W | 2/2 | 2/4 | 2/4 | 2/4 | 8/8 | 2/4 | 4/4 | 1/2 | 32/32 | 1/16 |
| Caerin 1.1-LC 11K.12K.19W | 2/2 | 2/2 | 2/4 | 4/4 | 4/4 | 4/4 | 4/4 | 2/2 | 16/32 | 2/2 |
| Caerin 1.1-LC 7.11.19K | 2/2 | 2/2 | 4/4 | 4/4 | 4/4 | 4/4 | 4/8 | 4/4 | 128/128 | 8/8 |
| Caerin 1.1-LC 2P-2A | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 |
| Caerin 1.1-LC PGGGP | 128/128 | 128/>128 | 128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 |
| Caerin 1.1-LC PRVVP | 2/8 | 2/4 | 4/16 | 8/16 | 8/32 | 8/8 | 32/32 | 8/16 | >128 | 4/8 |
| Caerin 1.1-LC PKVVP | 4/8 | 2/4 | 2/4 | 16/32 | 8/32 | 8/32 | 32/32 | 16/32 | >128 | 8/8 |
| Caerin 1.1-LC PHLLP | 4/8 | 4/4 | 4/4 | 8/8 | 8/16 | 32/64 | 4/8 | 4/4 | 32/32 | 4/4 |
| Caerin 1.1-LC PHIIP | 4/8 | 4/8 | 4/8 | 16/16 | 8/16 | 32/64 | 8/8 | 4/8 | >128 | 4/4 |
| Caerin 1.1-LC PHWWP | 32/64 | 16/32 | 64/128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 |
| Caerin 1.1-LC 11K.19K.PKAVP | 8/8 | 8/8 | 2/2 | 64/64 | >128 | 32/64 | 128/128 | >128 | >128 | 64/64 |
| Caerin 1.1-LC 7.11.19K.V(D)V(D) | 2/4 | 4/4 | 8/8 | 64/128 | 16/32 | 4/4 | 32/128 | 128/128 | >128 | 64/64 |
| Caerin 1.1-LC PKAL(D)P | 2/2 | 2/2 | 8/8 | 4/4 | 4/4 | 4/4 | 32/32 | 32/32 | >128 | 4/8 |
| Peptides | HC10 | GM MIC * Gram-Negative | GM MIC * Gram-Positive | TI * Gram-Negative | TI * Gram-Positive |
|---|---|---|---|---|---|
| Caerin 1.1-LC | 37.5 | 80.63 | 14.25 | 0.47 | 2.63 |
| Caerin 1.1-LC 11K | 35.8 | 8.8 | 20.1 | 4.06 | 1.78 |
| Caerin 1.1-LC 11K.22K | 10.3 | 10.08 | 3.17 | 1.02 | 3.25 |
| Caerin 1.1-LC 11K.12K.12W | 8.5 | 3.17 | 2 | 2.68 | 4.25 |
| Caerin 1.1-LC 11K.12K.19W | 10.5 | 4 | 2.97 | 2.62 | 3.53 |
| Caerin 1.1-LC 7.11.19K | 46.5 | 16 | 3.28 | 2.91 | 14.18 |
| Caerin 1.1-LC 2P-2A | 256 | 256 | 256 | 1 | 1 |
| Caerin 1.1-LC PGGGP | 256 | 190.21 | 256 | 1.34 | 1 |
| Caerin 1.1-LC PRVVP | 21.2 | 20.16 | 5.94 | 1.05 | 3.57 |
| Caerin 1.1-LC PKVVP | 25.8 | 32 | 12.14 | 0.81 | 2.12 |
| Caerin 1.1-LC PHLLP | 1.6 | 8 | 6.56 | 0.2 | 0.244 |
| Caerin 1.1-LC PHIIP | 2.4 | 16 | 8 | 0.15 | 0.3 |
| Caerin 1.1-LC PHWWP | 256 | 105 | 256 | 2.44 | 1 |
| Caerin 1.1-LC 11K.19K.PKAVP | 256 | 26.08 | 161.5 | 9.82 | 1.59 |
| Caerin 1.1-LC 7.11.19K.V(D)V(D) | 256 | 9.75 | 128 | 26.26 | 2 |
| Caerin 1.1-LC PKAL(D)P | 43.4 | 4.87 | 64 | 8.91 | 0.68 |
| Cell Line | IC50 | |
|---|---|---|
| Caerin 1.1-LC | Caerin 1.1-LC 7.11.19K.V(D)V(D) | |
| MRC-5 | 14.8 | NI * |
| HEK 293 | 10.8 | 129.6 |
| HMEC-1 | 79.3 | NI |
| HaCaT | 61.2 | NI |
| RAW 264.7 | 6.0 | NI |
| Peptide | MIC (μM) | |||||||
|---|---|---|---|---|---|---|---|---|
| Na+ | K+ | NH4+ | Mg2+ | Ca2+ | Fe3+ | FBS | Control | |
| Caerin 1.1-LC | 16 | 16 | 16 | 32 | 64 | 16 | 32 | 16 |
| Caerin 1.1-LC 11K.22K | 8 | 8 | 8 | 16 | 32 | 8 | 16 | 4 |
| Caerin 1.1-LC 11K.12K.12W | 4 | 4 | 4 | 8 | 32 | 4 | 8 | 4 |
| Caerin 1.1-LC 11K.12K.19W | 4 | 4 | 4 | 4 | 8 | 4 | 8 | 4 |
| Caerin 1.1-LC 7.11.19K | 2 | 2 | 2 | 4 | 64 | 2 | 16 | 2 |
| Caerin 1.1-LC PKAL(D)P | 4 | 2 | 2 | 16 | 128 | 2 | 16 | 2 |
| Caerin 1.1-LC 7.11.19K.V(D)V(D) | 16 | 4 | 4 | 32 | 128 | 4 | 16 | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Z.; Zhao, R.; Zhang, Y.; Ma, X.; Jiang, Y.; Wang, T.; Chen, X.; Ma, C.; Chen, T.; Shaw, C.; et al. Serendipitous Hinge Modulation Hypothetically Reprograms Caerin 1.1-LC Antibacterial Mechanism and Gram-Negative Selectivity. Pharmaceutics 2025, 17, 1500. https://doi.org/10.3390/pharmaceutics17111500
Sun Z, Zhao R, Zhang Y, Ma X, Jiang Y, Wang T, Chen X, Ma C, Chen T, Shaw C, et al. Serendipitous Hinge Modulation Hypothetically Reprograms Caerin 1.1-LC Antibacterial Mechanism and Gram-Negative Selectivity. Pharmaceutics. 2025; 17(11):1500. https://doi.org/10.3390/pharmaceutics17111500
Chicago/Turabian StyleSun, Zhengze, Ruixin Zhao, Yueao Zhang, Xiaonan Ma, Yangyang Jiang, Tao Wang, Xiaoling Chen, Chengbang Ma, Tianbao Chen, Chris Shaw, and et al. 2025. "Serendipitous Hinge Modulation Hypothetically Reprograms Caerin 1.1-LC Antibacterial Mechanism and Gram-Negative Selectivity" Pharmaceutics 17, no. 11: 1500. https://doi.org/10.3390/pharmaceutics17111500
APA StyleSun, Z., Zhao, R., Zhang, Y., Ma, X., Jiang, Y., Wang, T., Chen, X., Ma, C., Chen, T., Shaw, C., Zhou, M., & Wang, L. (2025). Serendipitous Hinge Modulation Hypothetically Reprograms Caerin 1.1-LC Antibacterial Mechanism and Gram-Negative Selectivity. Pharmaceutics, 17(11), 1500. https://doi.org/10.3390/pharmaceutics17111500








