Screening-Identified Oxazole-4-Carboxamide KB-2777 Exhibits In Vitro Anti-Coronavirus Activity
Abstract
1. Introduction
2. Materials and Methods
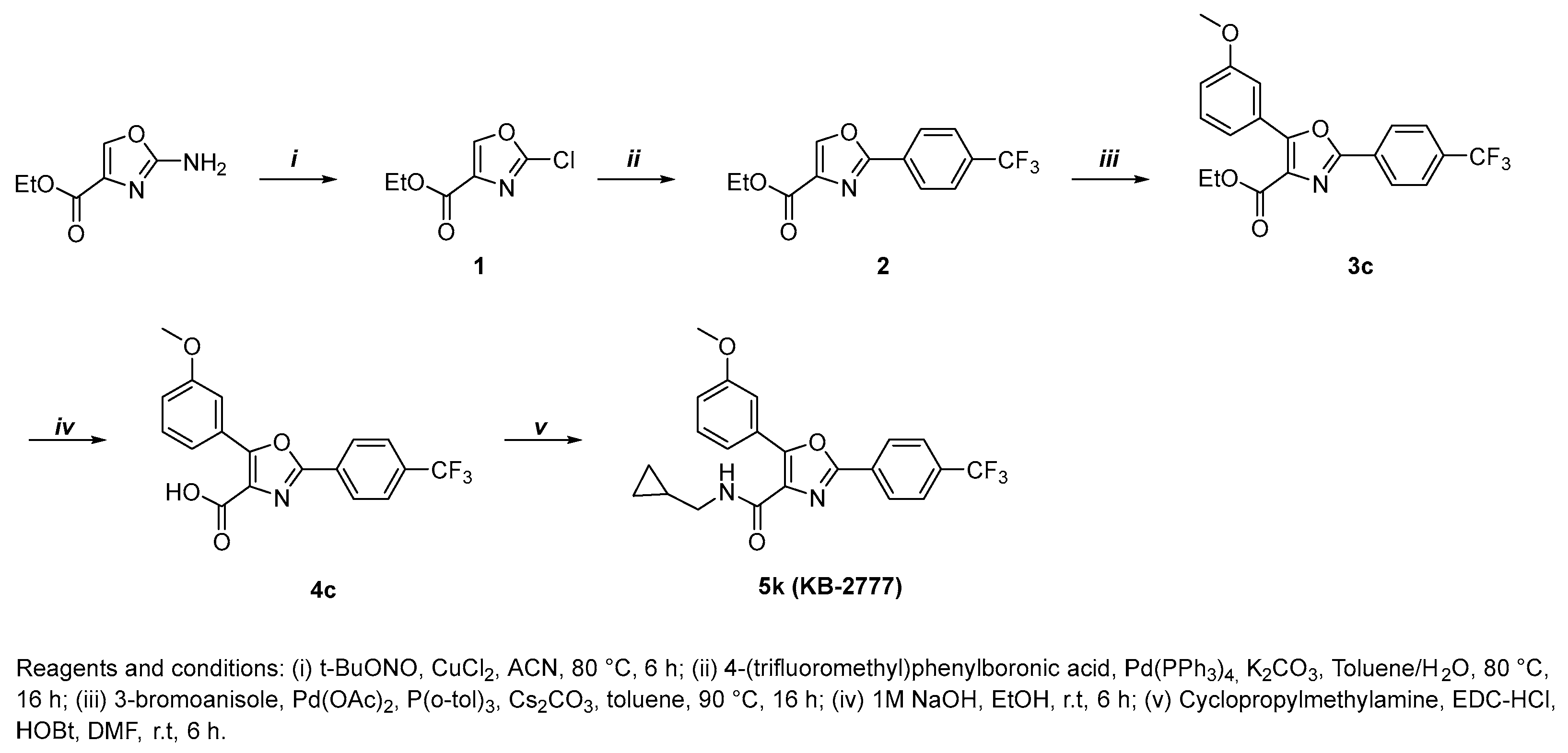
2.1. General Procedure for the Preparation of Compounds KB-2777
2.2. Compound KB-2777
2.2.1. Ethyl 2-Chlorooxazole-4-Carboxylate (1)
2.2.2. Ethyl 2-(4-(trifluoromethyl)phenyl)Oxazole-4-Carboxylate (2)
2.2.3. Ethyl 5-(3-methoxyphenyl)-2-(4-(trifluoromethyl)phenyl)Oxazole-4-Carboxylate (3c)
2.2.4. 5-(3-methoxyphenyl)-2-(4-(trifluoromethyl)phenyl)Oxazole-4-Carboxylic Acid (4c)
2.2.5. N-(cyclopropylmethyl)-5-(3-methoxyphenyl)-2-(4-(trifluoromethyl)phenyl)Oxazole-4-Carboxamide (5k, KB-2777)
2.3. Cytotoxicity (WST) and Antiviral Cytoprotection
2.4. Viral RNA Quantification
2.5. Immunofluorescence Assay (IFA)
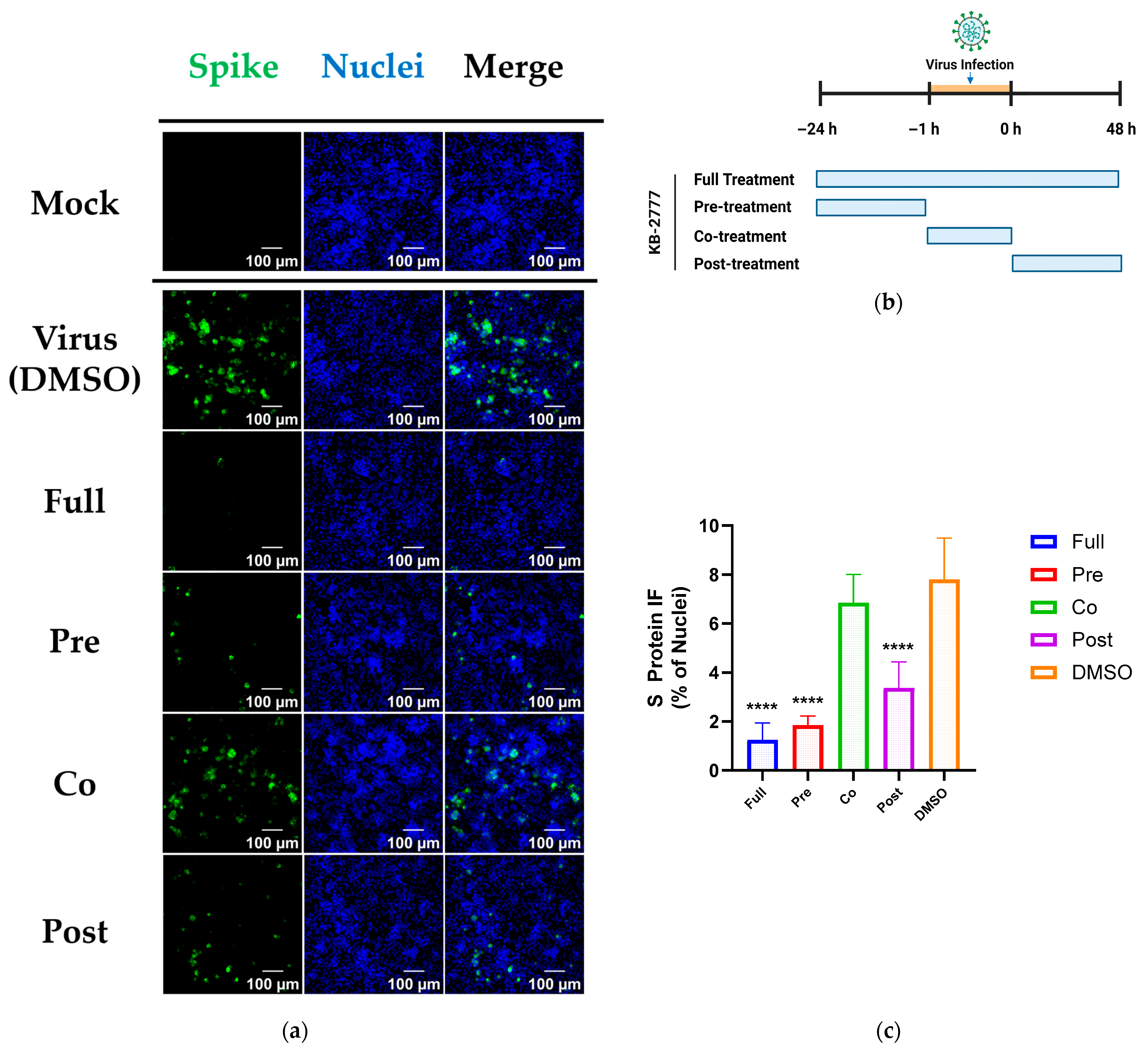
2.6. Time-of-Addition (TOA) Regimens
- Full: compound present from −24 h to 48 h relative to infection.
- Pre-treatment: −24 h to −1 h, wash at −1 h, then infection in drug-free medium.
- Co-treatment: −1 h to 0 h during adsorption only, wash at 0 h.
- Post-treatment: 0 h to 48 h after adsorption.
2.7. Host-Response Gene Expression Analysis (Two-Step RT-qPCR)
2.8. Combination (Drug–Drug Interaction) Studies
2.9. Propidium Iodide (PI) Live/Dead Imaging in HCoV-NL63/LLC-MK2 (48 h Post-Infection)
2.10. Data Processing and Statistics
3. Results
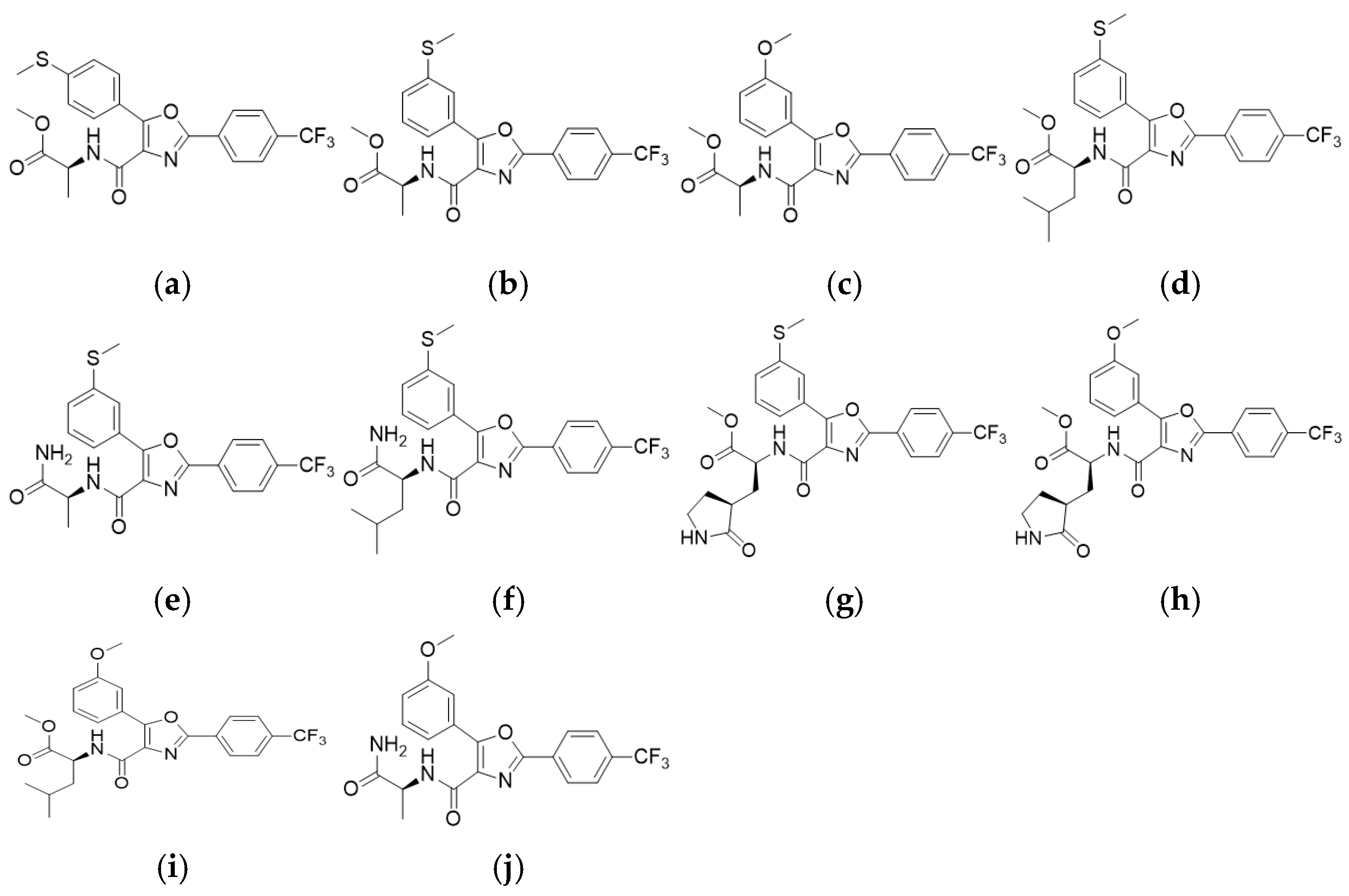
3.1. Primary Cytotoxicity and Antiviral Screening of Oxazole-4-Carboxamide Derivatives in HCoV-NL63–Infected LLC-MK2 Cells
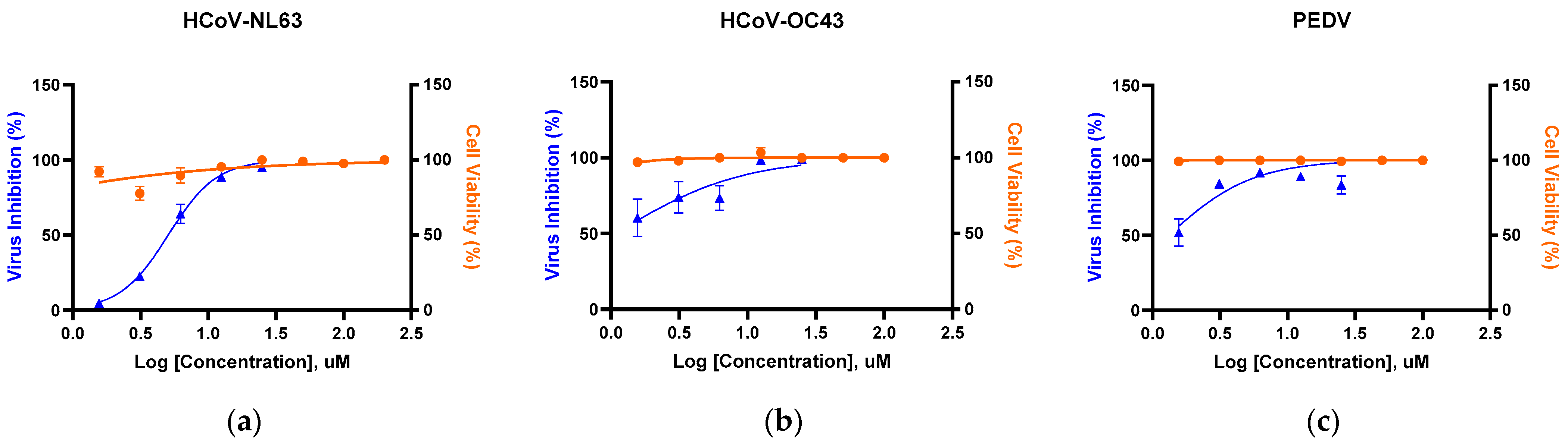
3.2. Antiviral Activity Across α/β-Coronaviruses by RT-qPCR
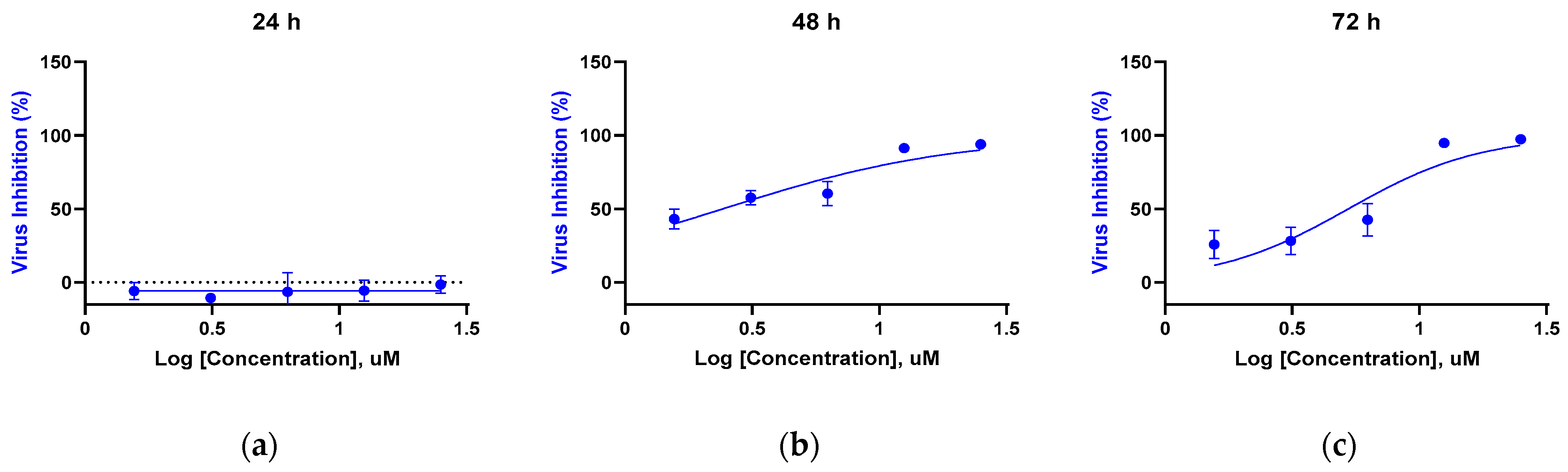
3.3. Time-Dependent Antiviral Activity Against HCoV-NL63 (RT-qPCR of Supernatants)
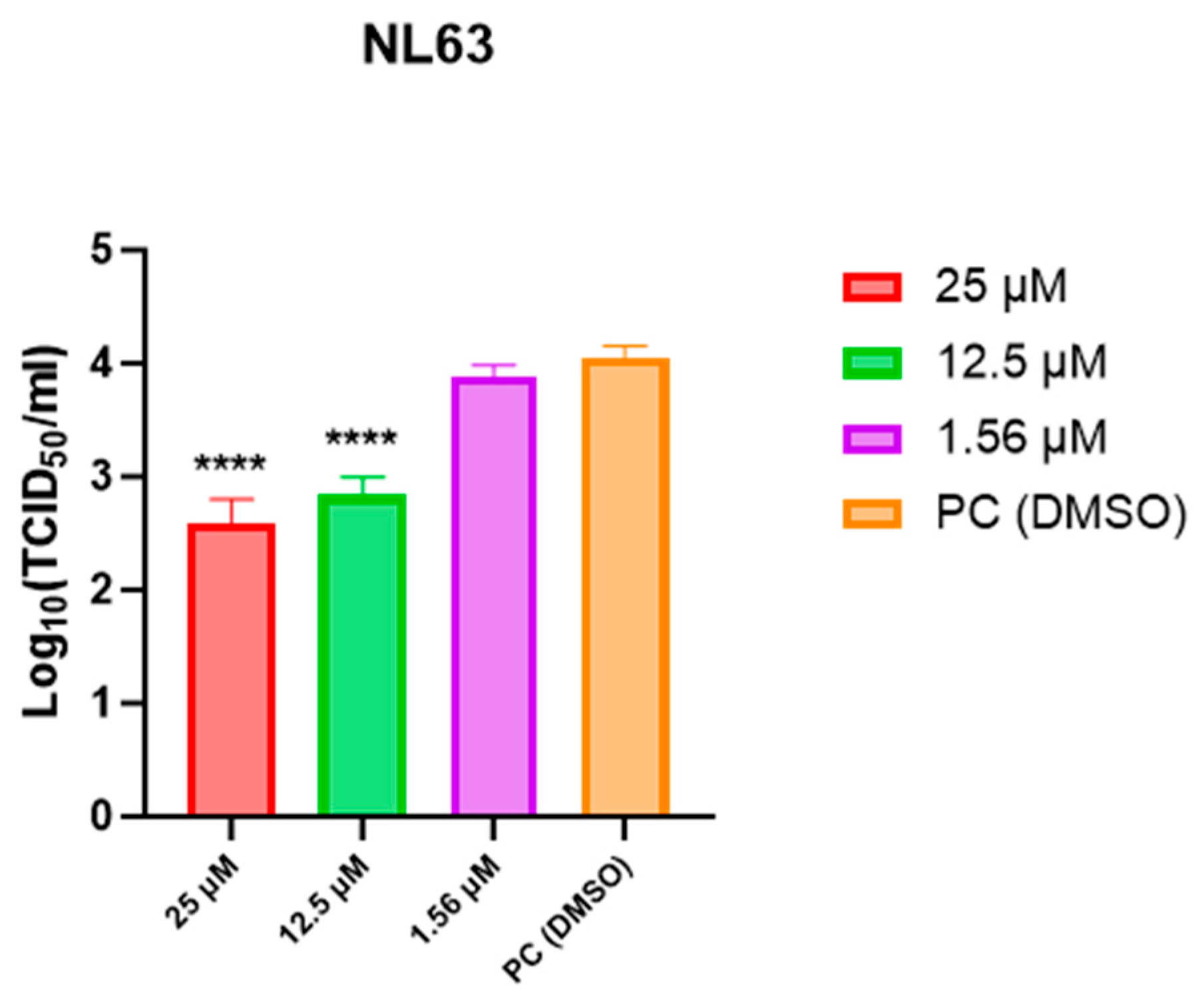
3.4. Infectious Titer Measurement at 48 h (TCID50)
3.5. Time-of-Addition Profile of KB-2777 Against HCoV-NL63 and HCoV-OC43
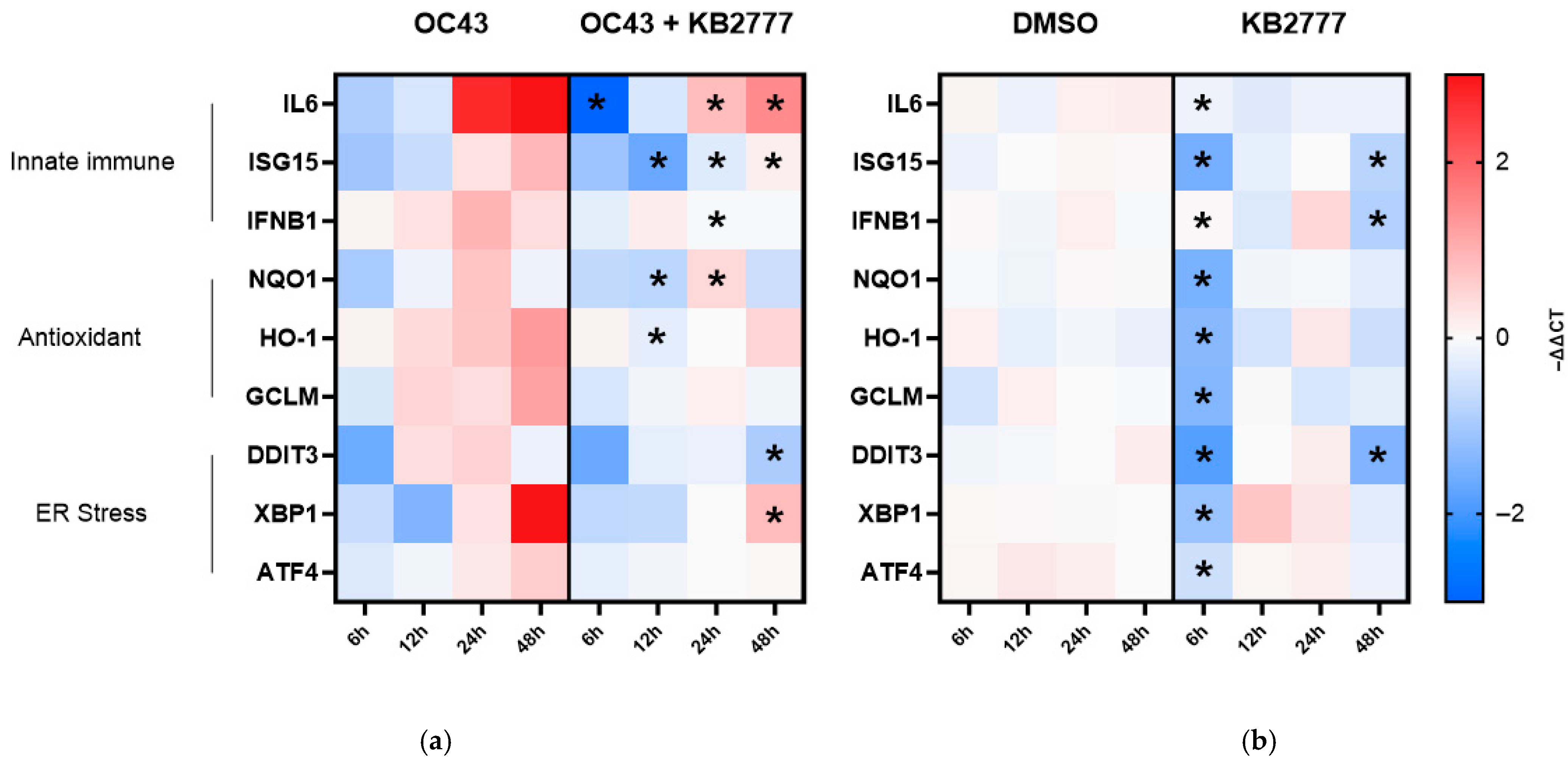
3.6. Targeted Host-Response Transcript Profiling
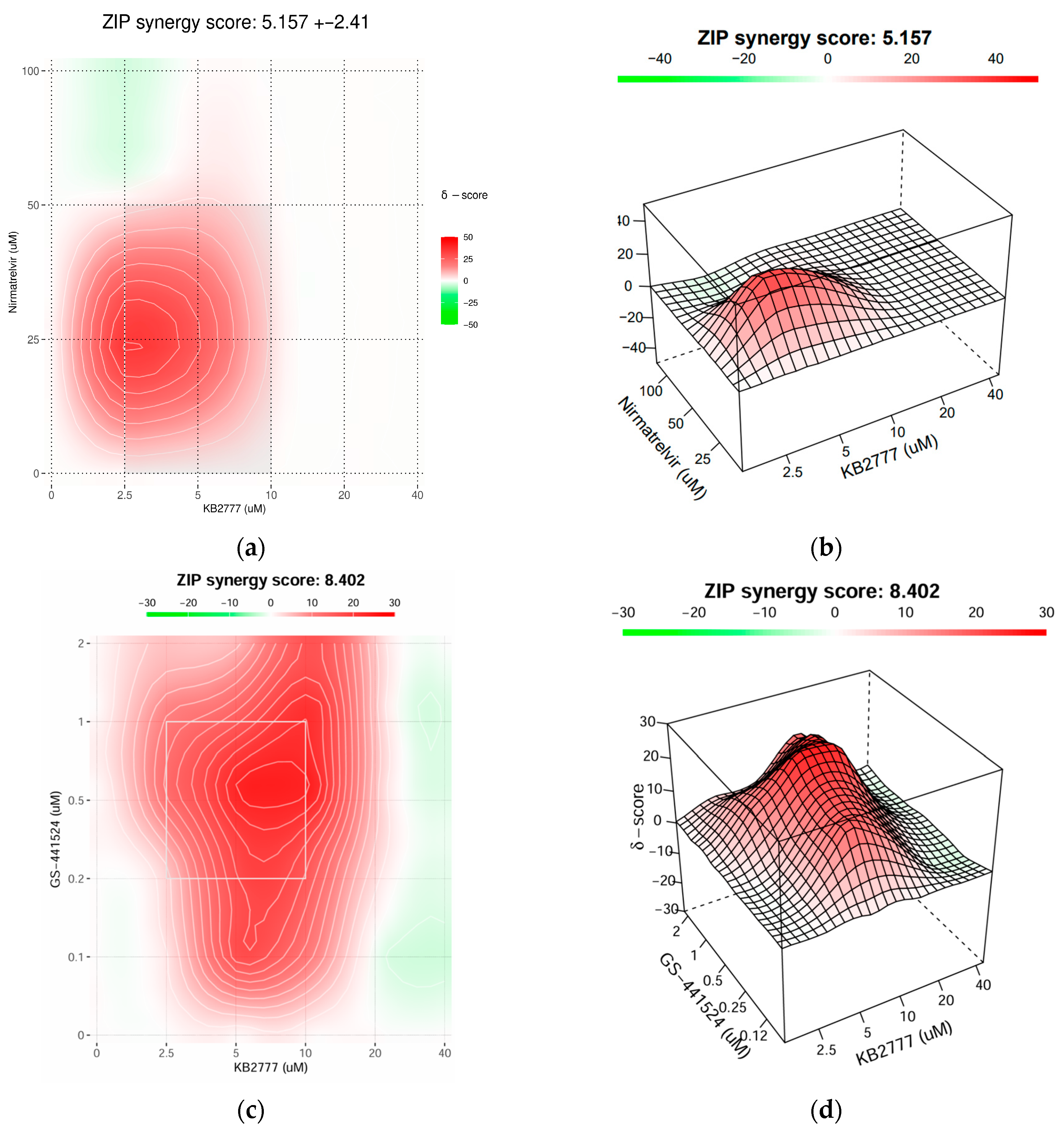
3.7. Combination Activity of KB-2777 with Approved DAAs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DAA | Direct-Acting Antiviral |
| TOA | Time-of-Addition |
| IFA | Immunofluorescence Assay |
| RT-qPCR | Reverse Transcription quantitative PCR |
| CPE | Cytopathic Effect |
| TCID50 | 50% Tissue Culture Infectious Dose |
| ER | Endoplasmic Reticulum |
| UPR | Unfolded Protein Response |
| NRF2 | Nuclear factor erythroid 2–related factor 2 |
| ZIP | Zero-Interaction Potency |
| HCoV-NL63 | Human coronavirus NL63 |
| PEDV | Porcine epidemic diarrhea virus |
| HCoV-OC43 | Human coronavirus OC43 |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
References
- Chen, R.; Gao, Y.; Liu, H.; Li, H.; Chen, W.; Ma, J. Advances in research on 3C-like protease (3CLpro) inhibitors against SARS-CoV-2 since 2020. RSC Med. Chem. 2023, 14, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Song, Y. Structure and function of SARS-CoV and SARS-CoV-2 main proteases and their inhibition: A comprehensive review. Eur. J. Med. Chem. 2023, 260, 115772. [Google Scholar] [CrossRef] [PubMed]
- Iketani, S.; Mohri, H.; Culbertson, B.; Hong, S.J.; Duan, Y.; Luck, M.I.; Annavajhala, M.K.; Guo, Y.; Sheng, Z.; Uhlemann, A.-C. Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir. Nature 2023, 613, 558–564. [Google Scholar] [CrossRef] [PubMed]
- Heilmann, E.; Costacurta, F.; Moghadasi, S.A.; Ye, C.; Pavan, M.; Bassani, D.; Volland, A.; Ascher, C.; Weiss, A.K.H.; Bante, D. SARS-CoV-2 3CLpro mutations selected in a VSV-based system confer resistance to nirmatrelvir, ensitrelvir, and GC376. Sci. Transl. Med. 2022, 15, eabq7360. [Google Scholar] [CrossRef]
- Zhang, L.; Xie, X.; Luo, H.; Qian, R.; Yang, Y.; Yu, H.; Huang, J.; Shi, P.-Y.; Hu, Q. Resistance mechanisms of SARS-CoV-2 3CLpro to the non-covalent inhibitor WU-04. Cell Discov. 2024, 10, 40. [Google Scholar] [CrossRef]
- Owen, D.R.; Allerton, C.M.; Anderson, A.S.; Aschenbrenner, L.; Avery, M.; Berritt, S.; Boras, B.; Cardin, R.D.; Carlo, A.; Coffman, K.J. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science 2021, 374, 1586–1593. [Google Scholar] [CrossRef]
- Gordon, C.J.; Tchesnokov, E.P.; Woolner, E.; Perry, J.K.; Feng, J.Y.; Porter, D.P.; Götte, M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020, 295, 6785–6797. [Google Scholar] [CrossRef]
- Choi, B.; Choudhary, M.C.; Regan, J.; Sparks, J.A.; Padera, R.F.; Qiu, X.; Solomon, I.H.; Kuo, H.-H.; Boucau, J.; Bowman, K. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N. Engl. J. Med. 2020, 383, 2291–2293. [Google Scholar] [CrossRef]
- Baum, A.; Fulton, B.O.; Wloga, E.; Copin, R.; Pascal, K.E.; Russo, V.; Giordano, S.; Lanza, K.; Negron, N.; Ni, M. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 2020, 369, 1014–1018. [Google Scholar] [CrossRef]
- Greaney, A.J.; Starr, T.N.; Barnes, C.O.; Weisblum, Y.; Schmidt, F.; Caskey, M.; Gaebler, C.; Cho, A.; Agudelo, M.; Finkin, S. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat. Commun. 2021, 12, 4196. [Google Scholar] [CrossRef]
- Pawlotsky, J.M. New hepatitis C therapies: The toolbox, strategies, and challenges. Gastroenterology 2014, 146, 1176–1192. [Google Scholar] [CrossRef] [PubMed]
- Havlir, D.V.; Tierney, C.; Friedland, G.H.; Pollard, R.B.; Smeaton, L.; Sommadossi, J.-P.; Fox, L.; Kessler, H.; Fife, K.H.; Richman, D.D. In vivo antagonism with zidovudine plus stavudine combination therapy. J. Infect. Dis. 2000, 182, 321–325. [Google Scholar] [CrossRef]
- Zhang, H.-Z.; Zhao, Z.-L.; Zhou, C.-H. Recent advance in oxazole-based medicinal chemistry. Eur. J. Med. Chem. 2018, 144, 444–492. [Google Scholar] [CrossRef]
- Patel, D.; Patel, K.; Patel, S.; Patel, B.; Patel, A. Review on therapeutic diversity of oxazole scaffold: An update. ChemistrySelect 2024, 9, e202403179. [Google Scholar] [CrossRef]
- Meanwell, N.A.; Belema, M. The discovery and development of daclatasvir: An inhibitor of the hepatitis C virus NS5A replication complex. In HCV: The Journey from Discovery to a Cure: Volume II.; Springer: Berlin/Heidelberg, Germany, 2019; pp. 27–55. [Google Scholar]
- Link, J.O.; Taylor, J.G.; Xu, L.; Mitchell, M.; Guo, H.; Liu, H.; Kato, D.; Kirschberg, T.; Sun, J.; Squires, N. Discovery of ledipasvir (GS-5885): A potent, once-daily oral NS5A inhibitor for the treatment of hepatitis C virus infection. J. Med. Chem. 2014, 57, 2033–2046. [Google Scholar] [CrossRef]
- Furuta, Y.; Komeno, T.; Nakamura, T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017, 93, 449–463. [Google Scholar] [CrossRef]
- Xue, F.; Luo, X.; Ye, C.; Ye, W.; Wang, Y. Inhibitory properties of 2-substituent-1H-benzimidazole-4-carboxamide derivatives against enteroviruses. Bioorg. Med. Chem. 2011, 19, 2641–2649. [Google Scholar] [CrossRef]
- Kim, Y.; Ma, C.; Park, S.; Shin, Y.; Lee, T.; Paek, J.; Hoon Kim, K.; Jang, G.; Cho, H.; Son, S. Rational Design, Synthesis and Evaluation of Oxazolo[4,5-c]-quinolinone Analogs as Novel Interleukin-33 Inhibitors. Chem. Asian J. 2021, 16, 3702–3712. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-J.; Song, D.-S.; Ha, G.-W.; Park, B.-K. Cloning and further sequence analysis of the spike gene of attenuated porcine epidemic diarrhea virus DR13. Virus Genes 2007, 35, 55–64. [Google Scholar] [CrossRef]
- Kärber, G. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Naunyn Schmiedebergs Arch. Exp. Pathol. Pharmakol. 1931, 162, 480–483. [Google Scholar] [CrossRef]
- Edwards, J.A.; Denis, F.; Talbot, P.J. Activation of glial cells by human coronavirus OC43 infection. J. Neuroimmunol. 2000, 108, 73–81. [Google Scholar] [CrossRef]
- Liu, S.Y.; Huang, M.; Fung, T.S.; Chen, R.A.; Liu, D.X. Characterization of the induction kinetics and antiviral functions of IRF1, ISG15 and ISG20 in cells infected with gammacoronavirus avian infectious bronchitis virus. Virology 2023, 582, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Oda, J.M.; den Hartigh, A.B.; Jackson, S.M.; Tronco, A.R.; Fink, S.L. The unfolded protein response components IRE1α and XBP1 promote human coronavirus infection. mBio 2023, 14, e00540-23. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Haas de Mello, A.; Morris, D.R.; Jones-Hall, Y.L.; Ivanciuc, T.; Sattler, R.A.; Paessler, S.; Menachery, V.D.; Garofalo, R.P.; Casola, A. SARS-CoV-2 inhibits NRF2-mediated antioxidant responses in airway epithelial cells and in the lung of a murine model of infection. Microbiol. Spectr. 2023, 11, e00378-23. [Google Scholar] [CrossRef]
- Gain, C.; Song, S.; Angtuaco, T.; Satta, S.; Kelesidis, T. The role of oxidative stress in the pathogenesis of infections with coronaviruses. Front. Microbiol. 2023, 13, 1111930. [Google Scholar] [CrossRef] [PubMed]
- Daskou, M.; Fotooh Abadi, L.; Gain, C.; Wong, M.; Sharma, E.; Kombe Kombe, A.J.; Nanduri, R.; Kelesidis, T. The role of the NRF2 pathway in the pathogenesis of viral respiratory infections. Pathogens 2024, 13, 39. [Google Scholar] [CrossRef]
- Liu, D.X.; Liang, J.Q.; Fung, T.S. Human Coronavirus-229E, -OC43, -NL63, and -HKU1 (Coronaviridae). In Encyclopedia of Virology, 4th ed.; Bamford, D.H., Zuckerman, M., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 428–440. [Google Scholar]
- Wang, X.; Tang, G.; Liu, Y.; Zhang, L.; Chen, B.; Han, Y.; Fu, Z.; Wang, L.; Hu, G.; Ma, Q. The role of IL-6 in coronavirus, especially in COVID-19. Front. Pharmacol. 2022, 13, 1033674. [Google Scholar] [CrossRef]
- Snijder, E.J.; Limpens, R.W.; de Wilde, A.H.; de Jong, A.W.; Zevenhoven-Dobbe, J.C.; Maier, H.J.; Faas, F.F.; Koster, A.J.; Bárcena, M. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 2020, 18, e3000715. [Google Scholar] [CrossRef]
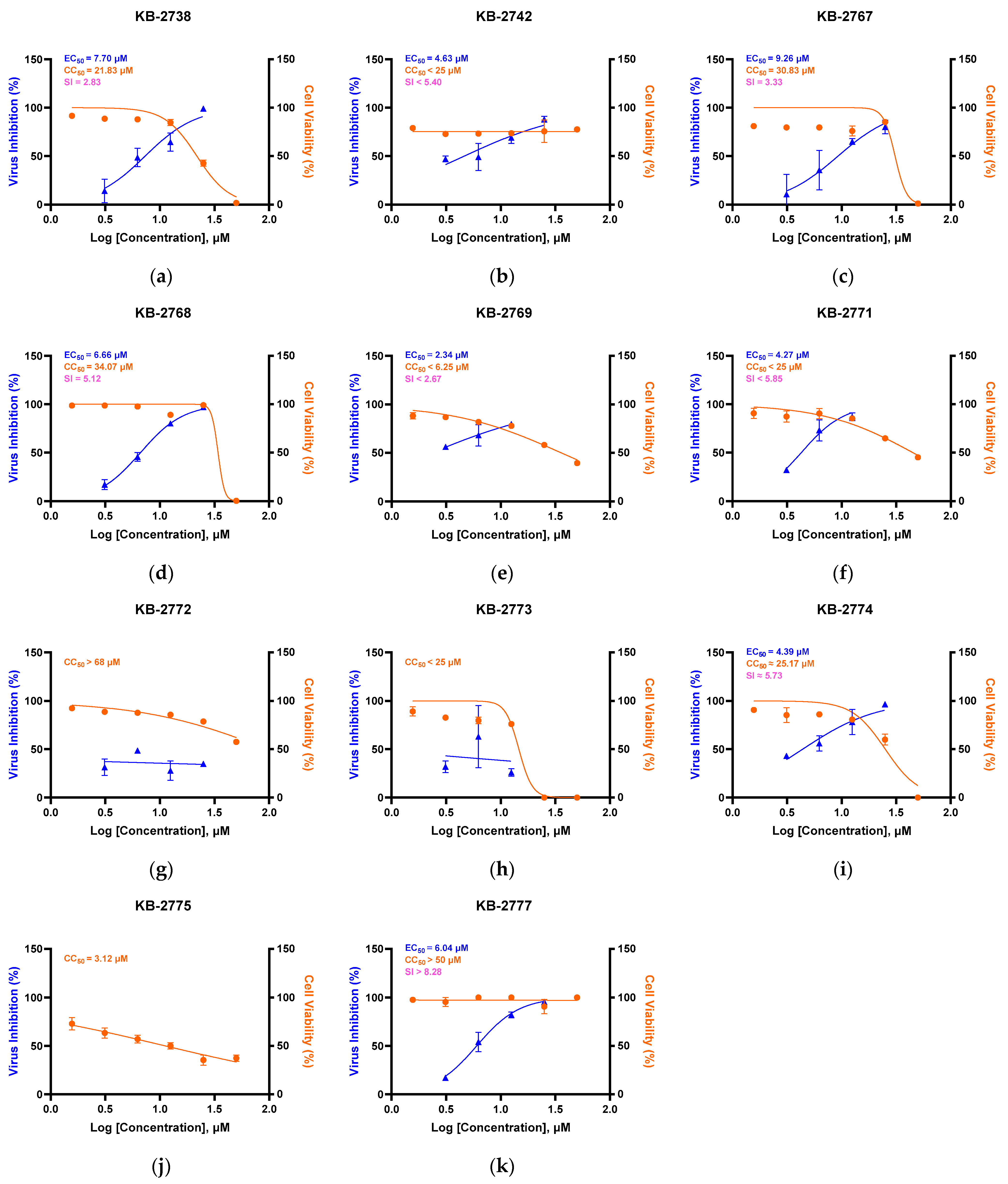
| Compound | EC50 (µM) | CC50 (µM) | SI (=CC50/EC50) |
|---|---|---|---|
| KB-2738 | 7.70 | 21.83 | 2.84 |
| KB-2742 | 4.63 | <25 | <5.40 |
| KB-2767 | 9.26 | 30.83 | 3.33 |
| KB-2768 | 6.66 | 34.07 | 5.12 |
| KB-2769 | 2.34 | <6.25 | <2.67 |
| KB-2771 | 4.27 | <25 | <5.85 |
| KB-2772 | - | >68 | - |
| KB-2773 | - | <25 | - |
| KB-2774 | 4.39 | 25.17 | 5.73 |
| KB-2775 | - | 3.12 | - |
| KB-2777 | 6.04 | >50 | >8.28 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, B.; Na, W.; Yeom, M.; Lim, J.-W.; Do, H.Q.; Jang, G.; Ban, M.-A.; Yang, J.-e.; Byun, Y.; Song, D. Screening-Identified Oxazole-4-Carboxamide KB-2777 Exhibits In Vitro Anti-Coronavirus Activity. Pharmaceutics 2025, 17, 1477. https://doi.org/10.3390/pharmaceutics17111477
Jung B, Na W, Yeom M, Lim J-W, Do HQ, Jang G, Ban M-A, Yang J-e, Byun Y, Song D. Screening-Identified Oxazole-4-Carboxamide KB-2777 Exhibits In Vitro Anti-Coronavirus Activity. Pharmaceutics. 2025; 17(11):1477. https://doi.org/10.3390/pharmaceutics17111477
Chicago/Turabian StyleJung, Bud, Woonsung Na, Minjoo Yeom, Jong-Woo Lim, Hai Quynh Do, Geonhee Jang, Min-A Ban, Ji-eun Yang, Youngjoo Byun, and Daesub Song. 2025. "Screening-Identified Oxazole-4-Carboxamide KB-2777 Exhibits In Vitro Anti-Coronavirus Activity" Pharmaceutics 17, no. 11: 1477. https://doi.org/10.3390/pharmaceutics17111477
APA StyleJung, B., Na, W., Yeom, M., Lim, J.-W., Do, H. Q., Jang, G., Ban, M.-A., Yang, J.-e., Byun, Y., & Song, D. (2025). Screening-Identified Oxazole-4-Carboxamide KB-2777 Exhibits In Vitro Anti-Coronavirus Activity. Pharmaceutics, 17(11), 1477. https://doi.org/10.3390/pharmaceutics17111477






