Pharmacogenomic Drug–Target Network Analysis Reveals Similarity Profiles Among FDA–Approved Cancer Drugs
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Resources and Integration
2.2. Statistical Analysis: Robust Calculation of Gene–Drug Correlations
2.3. Calculation of a Similarity Score Based on the Number of Shared Gene Targets
2.4. B-Index Calculation and Clustering Analysis
2.5. Drug Structural Analysis
2.6. Gene–Drug Bipartite Network Modeling
3. Results
3.1. Construction of a Drug-Gene Target Bipartite Network
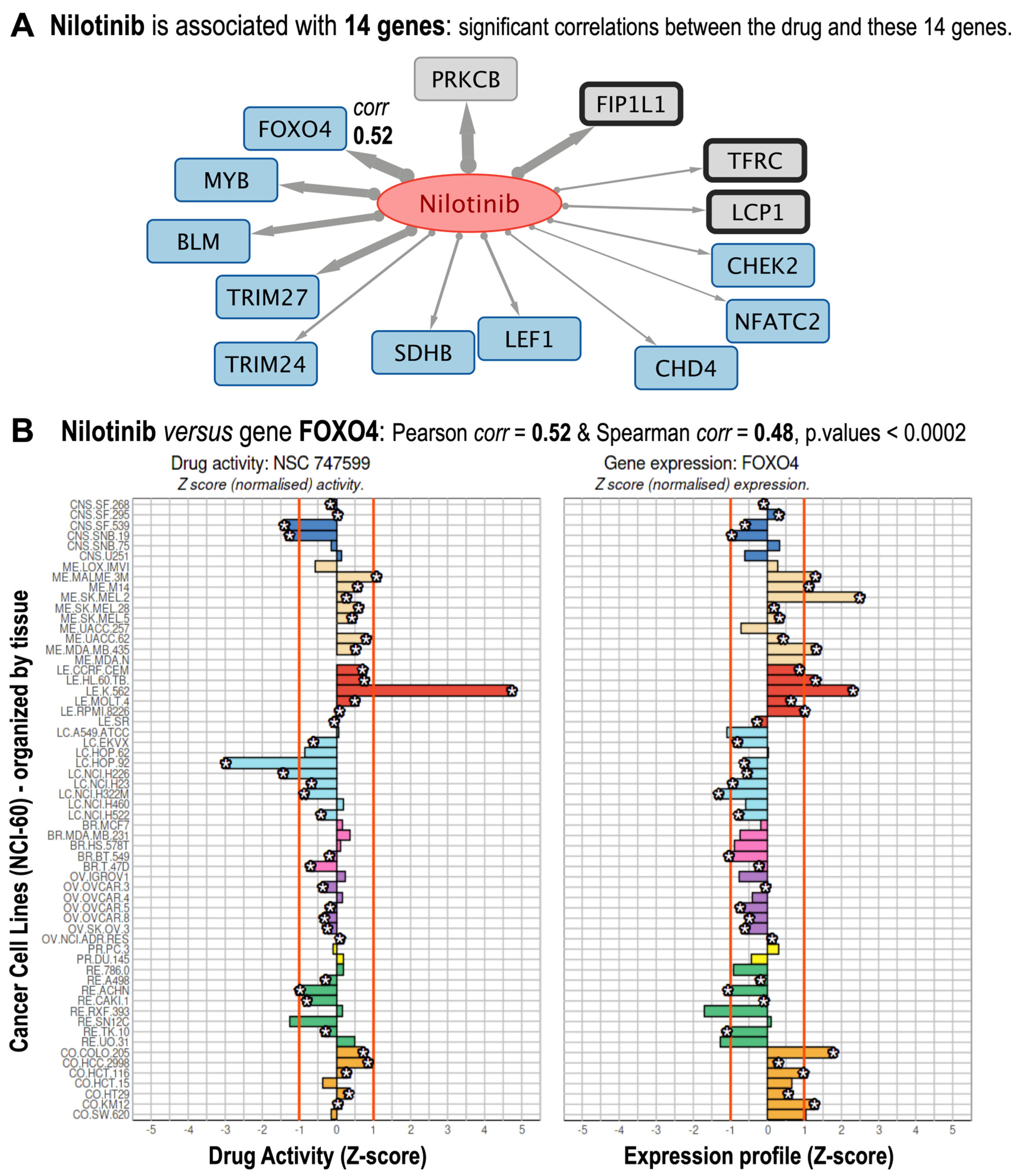
3.2. Identification of Known and Putative FDA-Approved Cancer Drug–Gene Interactions
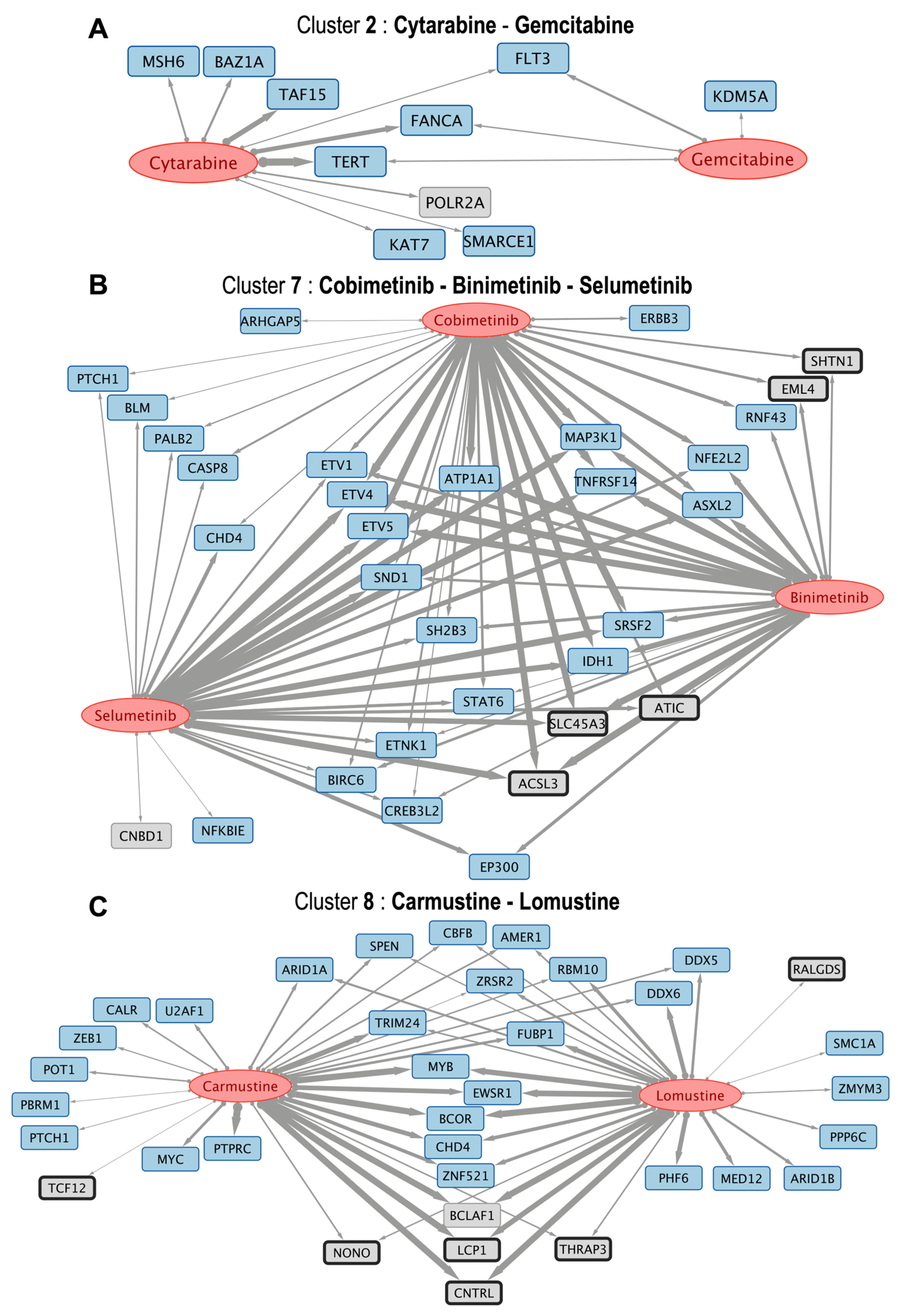
3.3. Pairwise Drug-to-Drug Clustering Based on the Analysis of the Common Gene Interactions
3.4. Determination of Drug-to-Drug Structural Similarities
3.5. Global Drug-to-Drug Comparison Using Common Targets and Structural Similarity
3.6. Agreement Between Drugs Structural Similarity and Drugs Common Gene Targets
4. Discussion
4.1. Drug–Gene Bipartite Network: Drug Activity and Gene Expression Analysis
4.2. Development of a New Index for the Association Between Drugs
4.3. Identification of Known and Novel Interactions Between FDA-Approved Drugs and Cancer Genes
4.4. Structural Similarity Between Drugs and Shared Gene-Network: The Case of the ERBB Family
4.5. Strengths and Limitations of the Proposed Approach, and Comparison with Similar Studies
4.6. Future Perspectives
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barreto, R.B.; Izidoro, A.M.; Miranda, M.H.F. New Oncologic Drugs from 2008 to 2023—Differences in Approval and Access between the United States, Europe and Brazil. Curr. Oncol. 2024, 31, 4443–4454. [Google Scholar] [CrossRef] [PubMed]
- De Las Rivas, J.; Alonso-López, D.; Arroyo, M.M. Human Interactomics: Comparative Analysis of Different Protein Interaction Resources and Construction of a Cancer Protein-Drug Bipartite Network. Adv. Protein Chem. Struct. Biol. 2018, 111, 263–282. [Google Scholar] [CrossRef]
- Shankavaram, U.T.; Varma, S.; Kane, D.; Sunshine, M.; Chary, K.K.; Reinhold, W.C.; Pommier, Y.; Weinstein, J.N. CellMiner: A Relational Database and Query Tool for the NCI-60 Cancer Cell Lines. BMC Genom. 2009, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- Sondka, Z.; Bamford, S.; Cole, C.G.; Ward, S.A.; Dunham, I.; Forbes, S.A. The COSMIC Cancer Gene Census: Describing Genetic Dysfunction across All Human Cancers. Nat. Rev. Cancer 2018, 18, 696–705. [Google Scholar] [CrossRef]
- Reinhold, W.C.; Sunshine, M.; Liu, H.; Varma, S.; Kohn, K.W.; Morris, J.; Doroshow, J.; Pommier, Y. CellMiner: A Web-Based Suite of Genomic and Pharmacologic Tools to Explore Transcript and Drug Patterns in the NCI-60 Cell Line Set. Cancer Res. 2012, 72, 3499–3511. [Google Scholar] [CrossRef]
- Todeschini, R.; Consonni, V.; Xiang, H.; Holliday, J.; Buscema, M.; Willett, P. Similarity Coefficients for Binary Chemoinformatics Data: Overview and Extended Comparison Using Simulated and Real Data Sets. J. Chem. Inf. Model. 2012, 52, 2884–2901. [Google Scholar] [CrossRef]
- Backman, T.W.H.; Cao, Y.; Girke, T. ChemMine Tools: An Online Service for Analyzing and Clustering Small Molecules. Nucleic Acids Res. 2011, 39, W486–W491. [Google Scholar] [CrossRef]
- Wang, Y.; Backman, T.W.H.; Horan, K.; Girke, T. fmcsR: Mismatch Tolerant Maximum Common Substructure Searching in R. Bioinformatics 2013, 29, 2792–2794. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, S.; Li, F.; Zhou, Y.; Zhang, Y.; Wang, Z.; Zhang, R.; Zhu, J.; Ren, Y.; Tan, Y.; et al. Therapeutic Target Database 2020: Enriched Resource for Facilitating Research and Early Development of Targeted Therapeutics. Nucleic Acids Res. 2020, 48, D1031–D1041. [Google Scholar] [CrossRef] [PubMed]
- Reinhold, W.C.; Sunshine, M.; Varma, S.; Doroshow, J.H.; Pommier, Y. Using CellMiner 1.6 for Systems Pharmacology and Genomic Analysis of the NCI-60. Clin. Cancer Res. 2015, 21, 3841–3852. [Google Scholar] [CrossRef]
- Gmeiner, W.H.; Reinhold, W.C.; Pommier, Y. Genome-Wide mRNA and microRNA Profiling of the NCI 60 Cell-Line Screen and Comparison of FdUMP[10] with Fluorouracil, Floxuridine, and Topoisomerase 1 Poisons. Mol. Cancer Ther. 2010, 9, 3105–3114. [Google Scholar] [CrossRef]
- Arroyo, M.M.; Berral-González, A.; Bueno-Fortes, S.; Alonso-López, D.; Rivas, J.D.L. Mining Drug-Target Associations in Cancer: Analysis of Gene Expression and Drug Activity Correlations. Biomolecules 2020, 10, 667. [Google Scholar] [CrossRef] [PubMed]
- Jaccard, P. The Distribution of the Flora in the Alpine Zone. New Phytol. 1912, 11, 37–50. [Google Scholar] [CrossRef]
- Russell, P.F.; Rao, T.R. On Habitat and Association of Species of Anopheline Larvae in South-eastern Madras. J. Malar. Inst. India 1940, 3, 153–178. [Google Scholar]
- Tanimoto, T.T. An Elementary Mathematical Theory of Classification and Prediction; International Business Machines Corporation: New York, NY, USA, 1958; 11p. [Google Scholar]
- Dice, L.R. Measures of the Amount of Ecologic Association between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Sorensen, T. A Method of Establishing Groups of Equal Amplitude in Plant Sociology Based on Similarity of Species and Its Application to Analyses of the Vegetation on Danish Commons. K. Dan. Vidensk. Selsk. 1948, 5, 1–34. [Google Scholar]
- Patterson, D.E.; Cramer, R.D.; Ferguson, A.M.; Clark, R.D.; Weinberger, L.E. Neighborhood Behavior: A Useful Concept for Validation of “Molecular Diversity” Descriptors. J. Med. Chem. 1996, 39, 3049–3059. [Google Scholar] [CrossRef]
- Garland, P.; Apperley, J. Nilotinib: Evaluation and Analysis of Its Role in Chronic Myeloid Leukemia. Future Oncol. 2011, 7, 201–218. [Google Scholar] [CrossRef]
- Yang, H.; Zhao, R.; Yang, H.-Y.; Lee, M.-H. Constitutively Active FOXO4 Inhibits Akt Activity, Regulates p27 Kip1 Stability, and Suppresses HER2-Mediated Tumorigenicity. Oncogene 2005, 24, 1924–1935. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Targeting BCR-Abl in the Treatment of Philadelphia-Chromosome Positive Chronic Myelogenous Leukemia. Pharmacol. Res. 2022, 178, 106156. [Google Scholar] [CrossRef]
- Yu, H.A.; Pao, W. Targeted Therapies: Afatinib—New Therapy Option for EGFR-Mutant Lung Cancer. Nat. Rev. Clin. Oncol. 2013, 10, 551–552. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Santos, A.; von Mering, C.; Jensen, L.J.; Bork, P.; Kuhn, M. STITCH 5: Augmenting Protein-Chemical Interaction Networks with Tissue and Affinity Data. Nucleic Acids Res. 2016, 44, D380–D384. [Google Scholar] [CrossRef]
- Wang, J.; Xiao, B.; Kimura, E.; Mongan, M.; Hsu, W.W.; Medvedovic, M.; Puga, A.; Xia, Y. Crosstalk of MAP3K1 and EGFR signaling mediates gene-environment interactions that block developmental tissue closure. J. Biol. Chem. 2024, 300, 107486. [Google Scholar] [CrossRef]
- Dratwa, M.; Wysoczańska, B.; Łacina, P.; Kubik, T.; Bogunia-Kubik, K. TERT-Regulation and Roles in Cancer Formation. Front. Immunol. 2020, 11, 589929. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhang, Y.; Jian, Y.; Gu, L.; Zhang, D.; Zhou, H.; Wang, Y.; Xu, Z.-X. The Regulations of Telomerase Reverse Transcriptase (TERT) in Cancer. Cell Death Dis. 2024, 15, 90. [Google Scholar] [CrossRef]
- Oh, S.; Shin, S.; Janknecht, R. ETV1, 4 and 5: An oncogenic subfamily of ETS transcription factors. Biochim. Biophys. Acta. 2012, 1826, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, S.; Li, C.; Xiao, Z.; Hu, J.; Zhao, C. TNF Family-Based Signature Predicts Prognosis, Tumor Microenvironment, and Molecular Subtypes in Bladder Carcinoma. Front. Cell Dev. Biol. 2021, 9, 800967. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Zheng, H.; Madan-Lala, R.; Dai, W.; Gimbrone, N.T.; Chen, Z.; Kinose, F.; Blackstone, S.A.; Smalley, K.S.M.; Cress, W.D.; et al. MEK Inhibition Modulates Cytokine Response to Mediate Therapeutic Efficacy in Lung Cancer. Cancer Res. 2019, 79, 5812–5825. [Google Scholar] [CrossRef]
- Yu, Z.; Zhu, J.; Wang, H.; Li, H.; Jin, X. Function of BCLAF1 in Human Disease. Oncol. Lett. 2022, 23, 58. [Google Scholar] [CrossRef]
- Mennel, H.D.; Szymás, J. Chemotherapy of Brain Tumors—Experimental Results. In Advances in Neurosurgery; Springer: Berlin/Heidelberg, Germany, 1978; pp. 289–291. ISBN 9783540089643. [Google Scholar]
- Howe, G.A.; Xiao, B.; Zhao, H.; Al-Zahrani, K.N.; Hasim, M.S.; Villeneuve, J.; Sekhon, H.S.; Goss, G.D.; Sabourin, L.A.; Dimitroulakos, J.; et al. Focal Adhesion Kinase Inhibitors in Combination with Erlotinib Demonstrate Enhanced Anti-Tumor Activity in Non-Small Cell Lung Cancer. PLoS ONE 2016, 11, e0150567. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Y.; Fan, C.; Wang, L.; Li, A.; Zhou, H.; Cai, L.; Miao, Y.; Li, Q.; Qiu, X.; et al. Lasp1 Promotes Malignant Phenotype of Non-Small-Cell Lung Cancer via Inducing Phosphorylation of FAK-AKT Pathway. Oncotarget 2017, 8, 75102–75113. [Google Scholar] [CrossRef] [PubMed]
- Gaviraghi, M.; Rabellino, A.; Andolfo, A.; Brand, M.; Brombin, C.; Bagnato, P.; De Feudis, G.; Raimondi, A.; Locatelli, A.; Tosoni, D.; et al. Direct Stimulation of ERBB2 Highlights a Novel Cytostatic Signaling Pathway Driven by the Receptor Thr Phosphorylation. Sci. Rep. 2020, 10, 16906. [Google Scholar] [CrossRef]
- Daks, A.A.; Fedorova, O.A.; Shuvalov, O.Y.; Parfenev, S.E.; Barlev, N.A. The Role of ERBB2/HER2 Tyrosine Kinase Receptor in the Regulation of Cell Death. Biochemistry (Mosc). 2020, 85, 1277–1287. [Google Scholar] [CrossRef]
- Caputo, R.; Buono, G.; Di Lauro, V.; Cianniello, D.; Von Arx, C.; Pensabene, M.; Pagliuca, M.; Pacilio, C.; Di Rella, F.; Verrazzo, A.; et al. Neratinib as Adjuvant Therapy in Patients with HER2 Positive Breast Cancer: Expert Opinion. Future Oncol. 2023, 19, 1695–1708. [Google Scholar] [CrossRef]
- Ye, W.; Chen, C.; Gao, Y.; Zheng, Z.-S.; Xu, Y.; Yun, M.; Weng, H.-W.; Xie, D.; Ye, S.; Zhang, J.-X. Overexpression of SLC34A2 Is an Independent Prognostic Indicator in Bladder Cancer and Its Depletion Suppresses Tumor Growth via Decreasing c-Myc Expression and Transcriptional Activity. Cell Death Dis. 2017, 8, e2581. [Google Scholar] [CrossRef]
- Lai, W.-Y.; Chuang, T.-P.; Borenäs, M.; Palmer, R.; Hallberg, B. Abstract 3544: ALK Signaling Activity Stabilizes SLC3A2 Protein Levels in Neuroblastoma Tumorigenesis. Cancer Res. 2023, 83, 3544. [Google Scholar] [CrossRef]
- Harada, Y.; Sato, A.; Nakamura, H.; Kai, K.; Kitamura, S.; Nakamura, T.; Kurihara, Y.; Ikeda, S.; Sueoka, E.; Kimura, S.; et al. Anti-Cancer Effect of Afatinib, Dual Inhibitor of HER2 and EGFR, on Novel Mutation HER2 E401G in Models of Patient-Derived Cancer. BMC Cancer 2023, 23, 77. [Google Scholar] [CrossRef]
- Li, R.; Sant, S.; Brown, E.; Caramia, F.; Nikolic, B.; Clarke, K.; Byrne, A.; Lara Gonzalez, L.E.; Savas, P.; Luen, S.J.; et al. Tucatinib Promotes Immune Activation and Synergizes with Programmed Cell Death-1 and Programmed Cell Death-Ligand 1 Inhibition in HER2-Positive Breast Cancer. J. Natl. Cancer Inst. 2023, 115, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Frenel, J.-S.; Zeghondy, J.; Guérin-Charbonnel, C.; Mailliez, A.; Volant, E.; Poumeaud, F.; Patsouris, A.; Arnedos, M.; Bailleux, C.; Cabal, J.; et al. Tucatinib Combination Treatment After Trastuzumab-Deruxtecan in Patients with ERBB2-Positive Metastatic Breast Cancer. JAMA Netw. Open 2024, 7, e244435. [Google Scholar] [CrossRef]
- Song, P.; Mansur, A.; Sorace, A. Abstract 3990: Imaging Molecular Alterations during Tucatinib Response in Preclinical Models of HER2+ Breast Cancer. Cancer Res. 2023, 83, 3990. [Google Scholar] [CrossRef]
- Paull, K.D.; Shoemaker, R.H.; Hodes, L.; Monks, A.; Scudiero, D.A.; Rubinstein, L.; Plowman, J.; Boyd, M.R. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: Development of mean graph and COMPARE algorithm. J. Natl. Cancer Inst. 1989, 81, 1088–1092. [Google Scholar] [CrossRef]
- Covell, D.G. Integrating constitutive gene expression and chemoactivity: Mining the NCI60 anticancer screen. PLoS ONE 2012, 7, e44631. [Google Scholar] [CrossRef] [PubMed]
- Reinhold, W.C.; Wilson, K.; Elloumi, F.; Bradwell, K.R.; Ceribelli, M.; Varma, S.; Wang, Y.; Duveau, D.; Menon, N.; Trepel, J.; et al. CellMinerCDB: NCATS Is a Web-Based Portal Integrating Public Cancer Cell Line Databases for Pharmacogenomic Explorations. Cancer Res. 2023, 83, 1941–1952. [Google Scholar] [CrossRef] [PubMed]
- Xia, F.; Allen, J.; Balaprakash, P.; Brettin, T.; Garcia-Cardona, C.; Clyde, A.; Cohn, J.; Doroshow, J.; Duan, X.; Dubinkina, V.; et al. A cross-study analysis of drug response prediction in cancer cell lines. Brief Bioinform. 2022, 23, bbab356. [Google Scholar] [CrossRef]
- Tao, W.; Liu, Y.; Lin, X.; Song, B.; Zeng, X. Prediction of multi-relational drug-gene interaction via Dynamic hyperGraph Contrastive Learning. Brief Bioinform. 2023, 24, bbad371. [Google Scholar] [CrossRef]
- Ariey-Bonnet, J.; Carrasco, K.; Le Grand, M.; Hoffer, L.; Betzi, S.; Feracci, M.; Tsvetkov, P.; Devred, F.; Collette, Y.; Morelli, X.; et al. In silico molecular target prediction unveils mebendazole as a potent MAPK14 inhibitor. Mol. Oncol. 2020, 14, 3083–3099. [Google Scholar] [CrossRef]
- Guo, Q.; Hernandez-Hernandez, S.; Ballester, P.J. UMAP-based clustering split for rigorous evaluation of AI models for virtual screening on cancer cell lines. J. Cheminform. 2025, 17, 94. [Google Scholar] [CrossRef]
- Cannon, M.; Stevenson, J.; Stahl, K.; Basu, R.; Coffman, A.; Kiwala, S.; McMichael, J.F.; Kuzma, K.; Morrissey, D.; Cotto, K.; et al. DGIdb 5.0: Rebuilding the drug-gene interaction database for precision medicine and drug discovery platforms. Nucleic Acids Res. 2024, 52, D1227–D1235. [Google Scholar] [CrossRef]
- Hartung, M.; Anastasi, E.; Mamdouh, Z.M.; Nogales, C.; Schmidt, H.H.H.W.; Baumbach, J.; Zolotareva, O.; List, M. Cancer driver drug interaction explorer. Nucleic Acids Res. 2022, 50, W138–W144. [Google Scholar] [CrossRef]



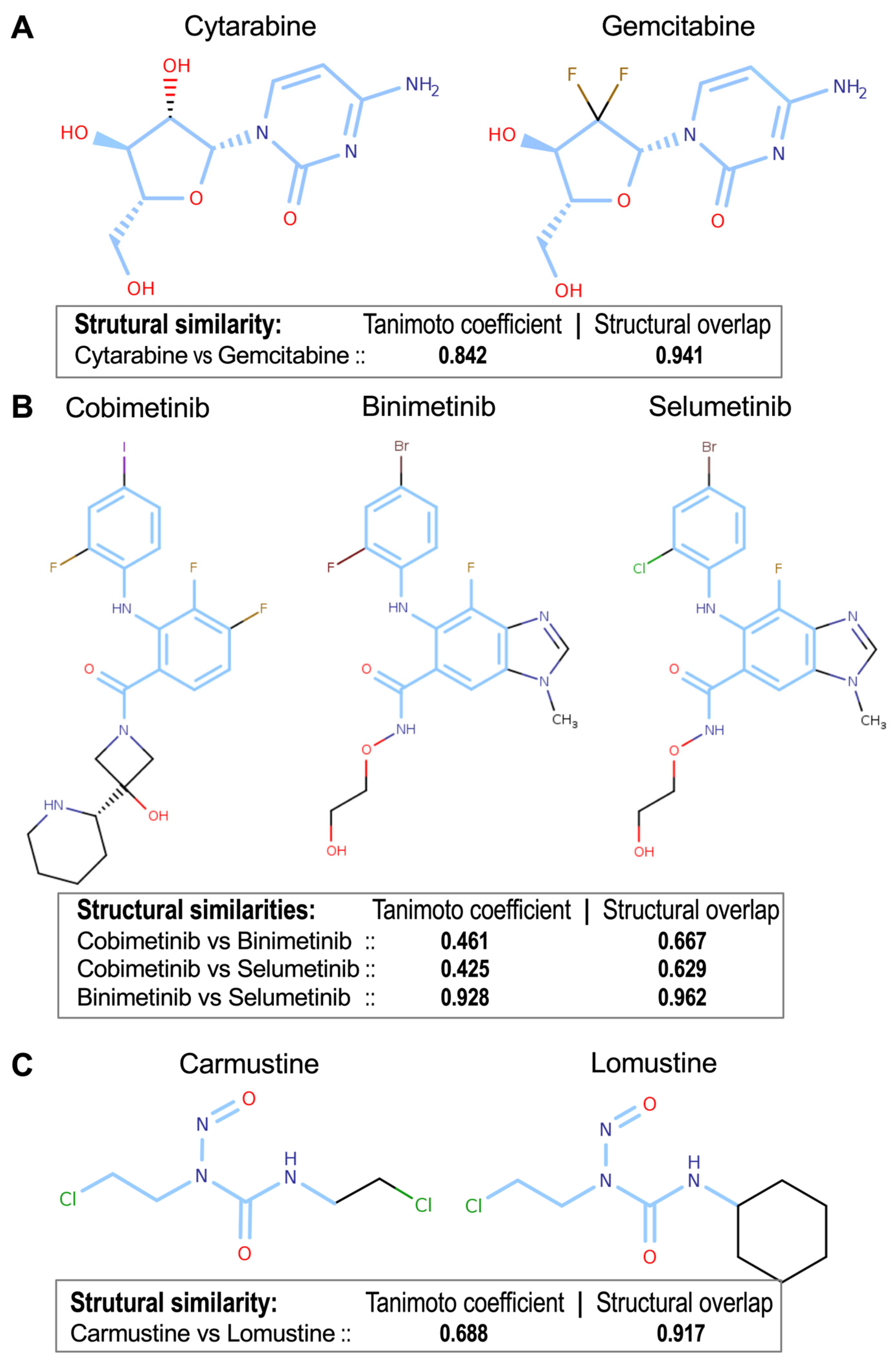

| Drug Pair | B-Index | Jaccard Index | Tanimoto Coefficient | Structural Overlap (MCS Score) | Shared Targets | Drug Class |
|---|---|---|---|---|---|---|
| Cytarabine—Gemcitabine | 0.542 | 0.300 | 0.842 | 0.941 | 3 | Nucleoside analogs |
| Cobimetinib—Binimetinib | 0.857 | 0.733 | 0.461 | 0.667 | 22 | MEK inhibitors |
| Cobimetinib—Selumetinib | 0.858 | 0.750 | 0.425 | 0.629 | 23 | MEK inhibitors |
| Binimetinib—Selumetinib | 0.805 | 0.667 | 0.928 | 0.962 | 19 | MEK inhibitors |
| Carmustine—Lomustine | 0.715 | 0.556 | 0.688 | 0.917 | 20 | Nitrosoureas |
| Afatinib—Dacomitinib | 0.555 | 0.385 | 0.811 | 0.909 | 5 | TKIs 1 |
| Afatinib—Neratinib | 0.694 | 0.500 | 0.644 | 0.853 | 5 | TKIs 1 |
| Dacomitinib—Neratinib | 0.417 | 0.250 | 0.622 | 0.848 | 3 | TKIs 1 |
| Neratinib—Tucatinib | 0.000 | 0.000 | 0.357 | 0.555 | 0 | TKIs 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berral-González, A.; Arroyo, M.M.; Alonso-López, D.; Rivas-López, M.J.; Sánchez-Santos, J.M.; De Las Rivas, J. Pharmacogenomic Drug–Target Network Analysis Reveals Similarity Profiles Among FDA–Approved Cancer Drugs. Pharmaceutics 2025, 17, 1421. https://doi.org/10.3390/pharmaceutics17111421
Berral-González A, Arroyo MM, Alonso-López D, Rivas-López MJ, Sánchez-Santos JM, De Las Rivas J. Pharmacogenomic Drug–Target Network Analysis Reveals Similarity Profiles Among FDA–Approved Cancer Drugs. Pharmaceutics. 2025; 17(11):1421. https://doi.org/10.3390/pharmaceutics17111421
Chicago/Turabian StyleBerral-González, Alberto, Monica M. Arroyo, Diego Alonso-López, María Jesús Rivas-López, José Manuel Sánchez-Santos, and Javier De Las Rivas. 2025. "Pharmacogenomic Drug–Target Network Analysis Reveals Similarity Profiles Among FDA–Approved Cancer Drugs" Pharmaceutics 17, no. 11: 1421. https://doi.org/10.3390/pharmaceutics17111421
APA StyleBerral-González, A., Arroyo, M. M., Alonso-López, D., Rivas-López, M. J., Sánchez-Santos, J. M., & De Las Rivas, J. (2025). Pharmacogenomic Drug–Target Network Analysis Reveals Similarity Profiles Among FDA–Approved Cancer Drugs. Pharmaceutics, 17(11), 1421. https://doi.org/10.3390/pharmaceutics17111421







