Silver(I) Complexes Bearing S-Alkyl Thiosalicylic Acid Derivatives: DNA/BSA Binding and Antitumor Activity In Vitro and In Vivo
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemistry
Reagents and Instruments
2.2. Syntheses
2.2.1. General Procedure for the Synthesis of Bidentate Ligands—S-Alkyl Derivatives of Thiosalicylic Acid (L1–L5)
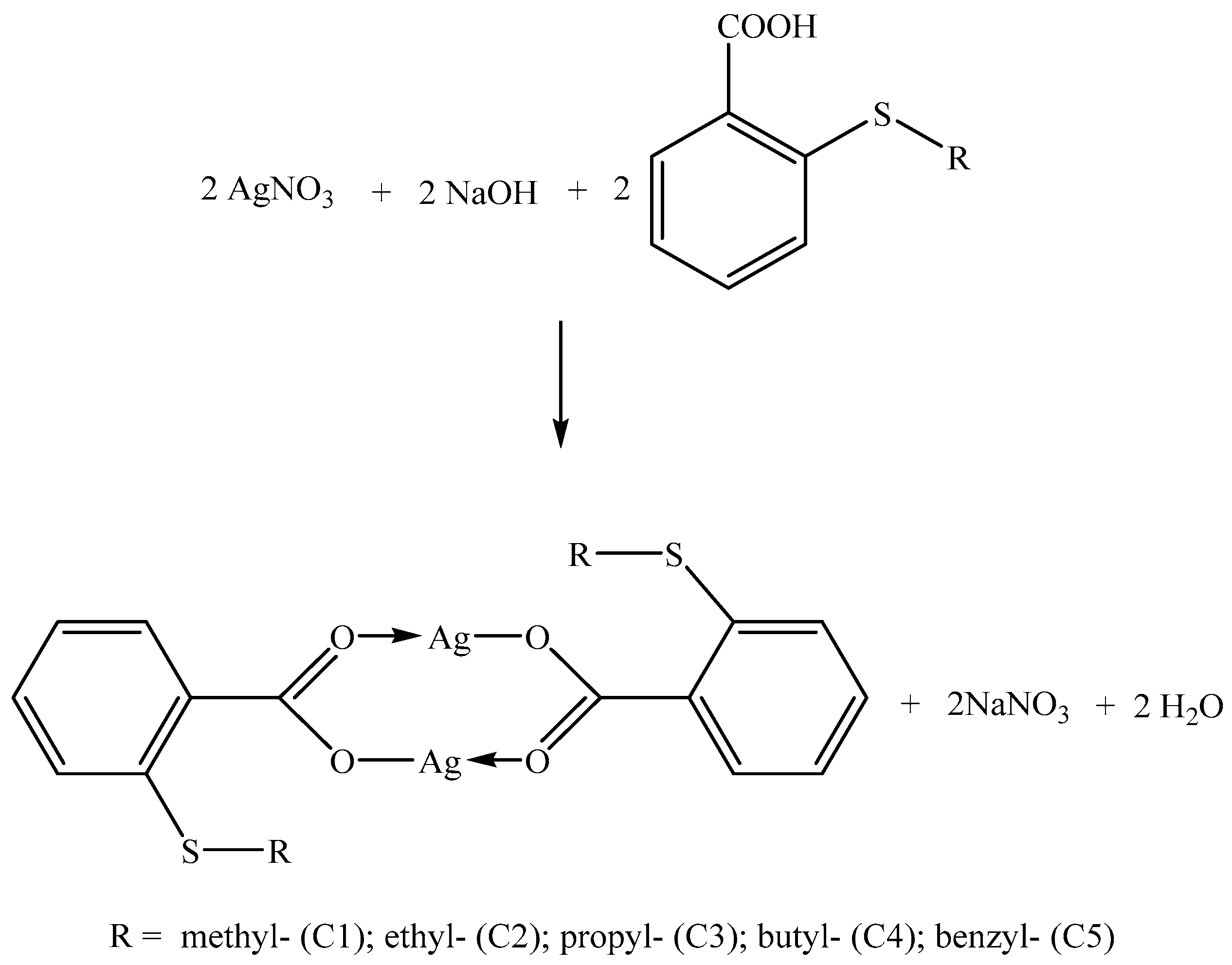
2.2.2. General Procedure for the Synthesis of Silver(I) Complexes with S-Alkyl Derivatives of Thiosalicylic Acid (C1–C5)
Preparation of the Complex [Ag2(S-Methyl-Thiosal)2] (C1)
Preparation of the Complex [Ag2(S-Ethyl-Thiosal)2] (C2)
Preparation of the Complex [Ag2(S-Propyl-Thiosal)2] (C3)
Preparation of the Complex [Ag2(S-Butyl-Thiosal)2] (C4)
Preparation of the Complex [Ag2(S-Benzyl-Thiosal)2], (C5)
2.3. Absorption Spectroscopic Studies
2.4. Fluorescence Quenching Measurements
2.5. Viscosity Measurements
2.6. Albumin-Binding Studies
2.7. In Silico Molecular Docking Studies
2.8. MTT Assay
2.9. Evaluation of Breast Cancer Development In Vivo
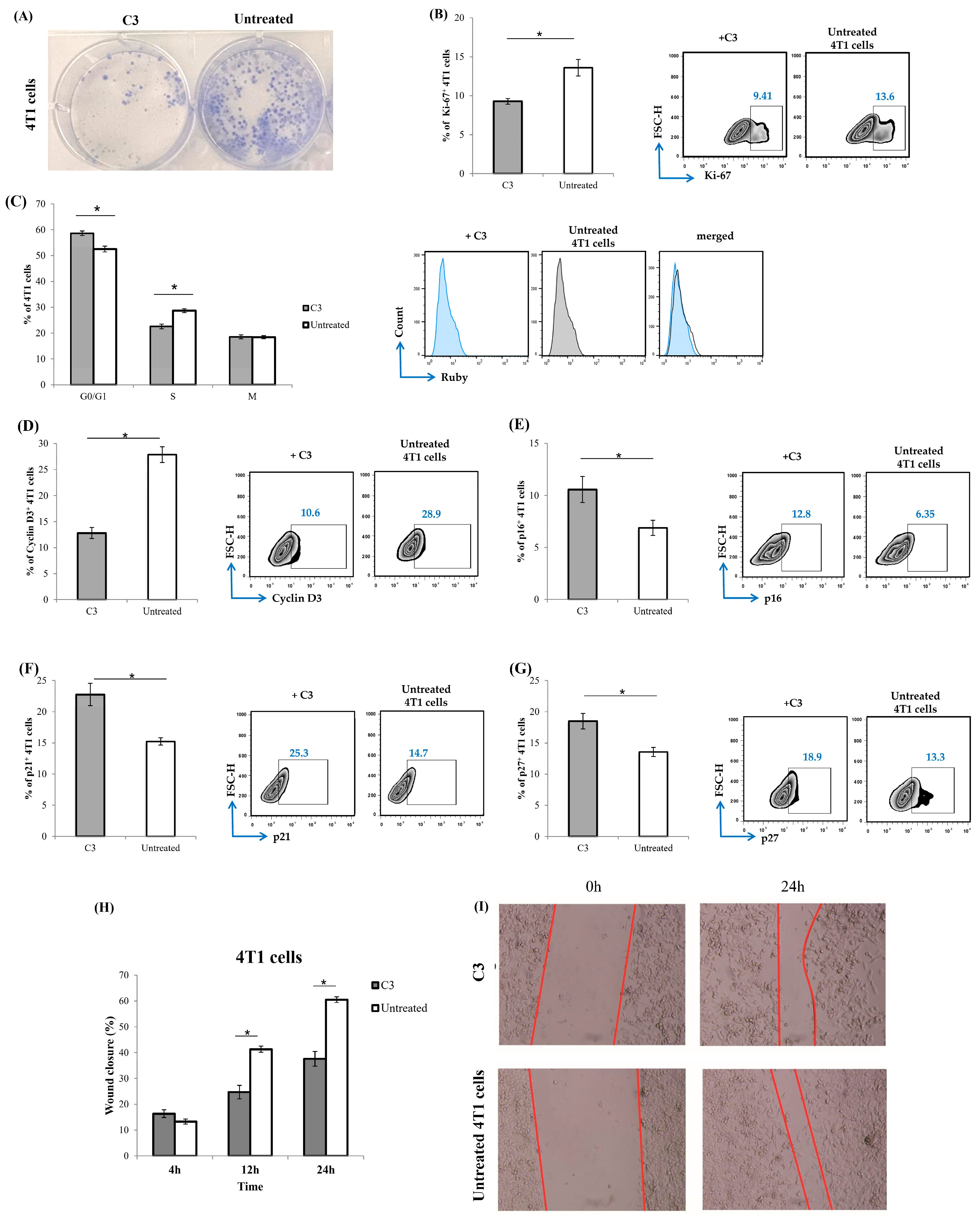
2.10. Colony Formation Assay
2.11. Analysis of Cell Death
2.12. Cell Cycle Distribution
2.13. Flow Cytometry Analyses
2.14. The Measurement of Mitochondrial Membrane Potential and ROS Levels
2.15. Scratch Assay
2.16. Statistical Analysis
3. Results and Discussion
3.1. Synthesis and Chemical Characterization
3.2. DNA-Binding Studies
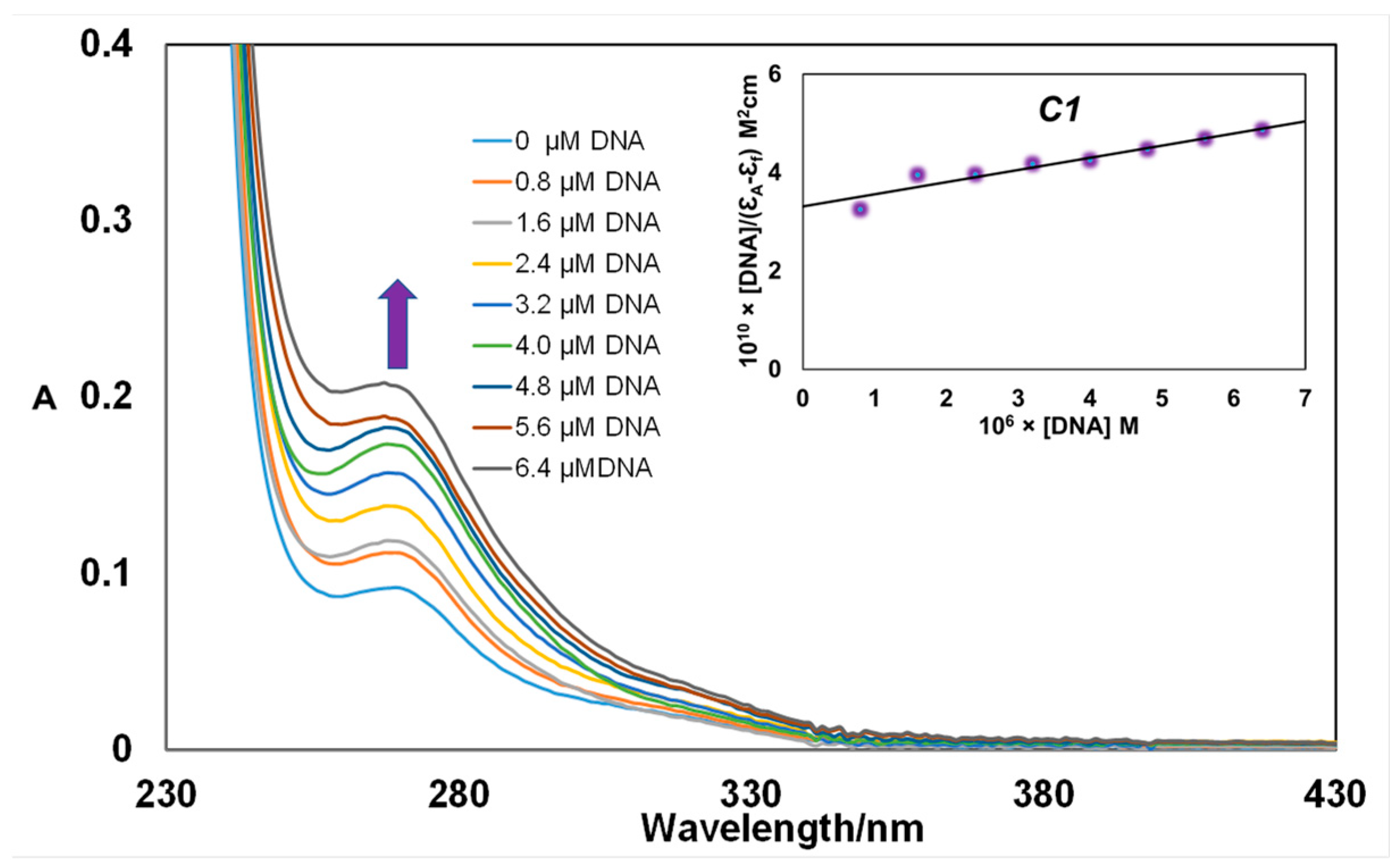
3.2.1. Absorption Spectroscopic Studies
3.2.2. Viscosity Measurements
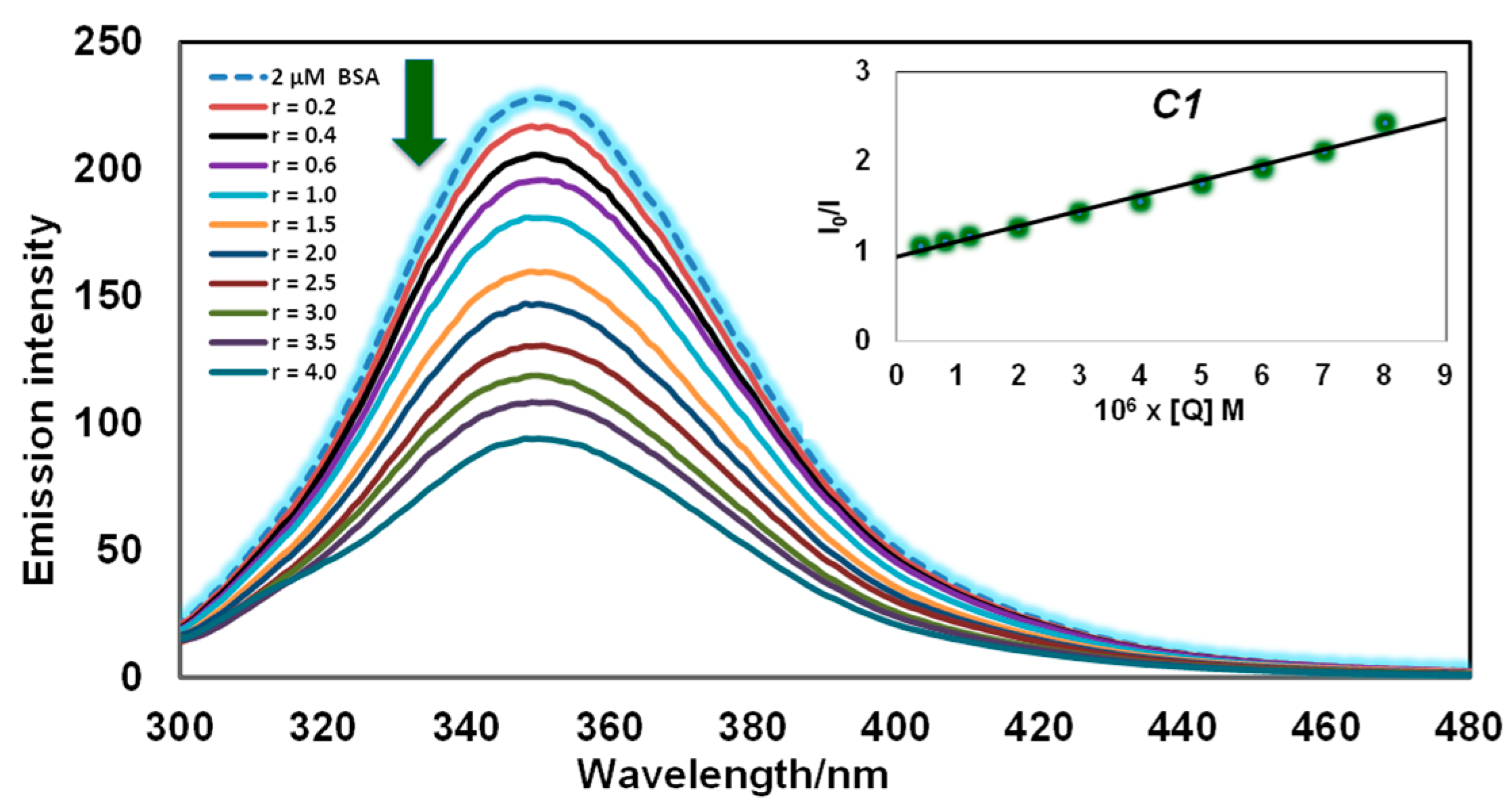
3.2.3. Fluorescence Quenching Measurements
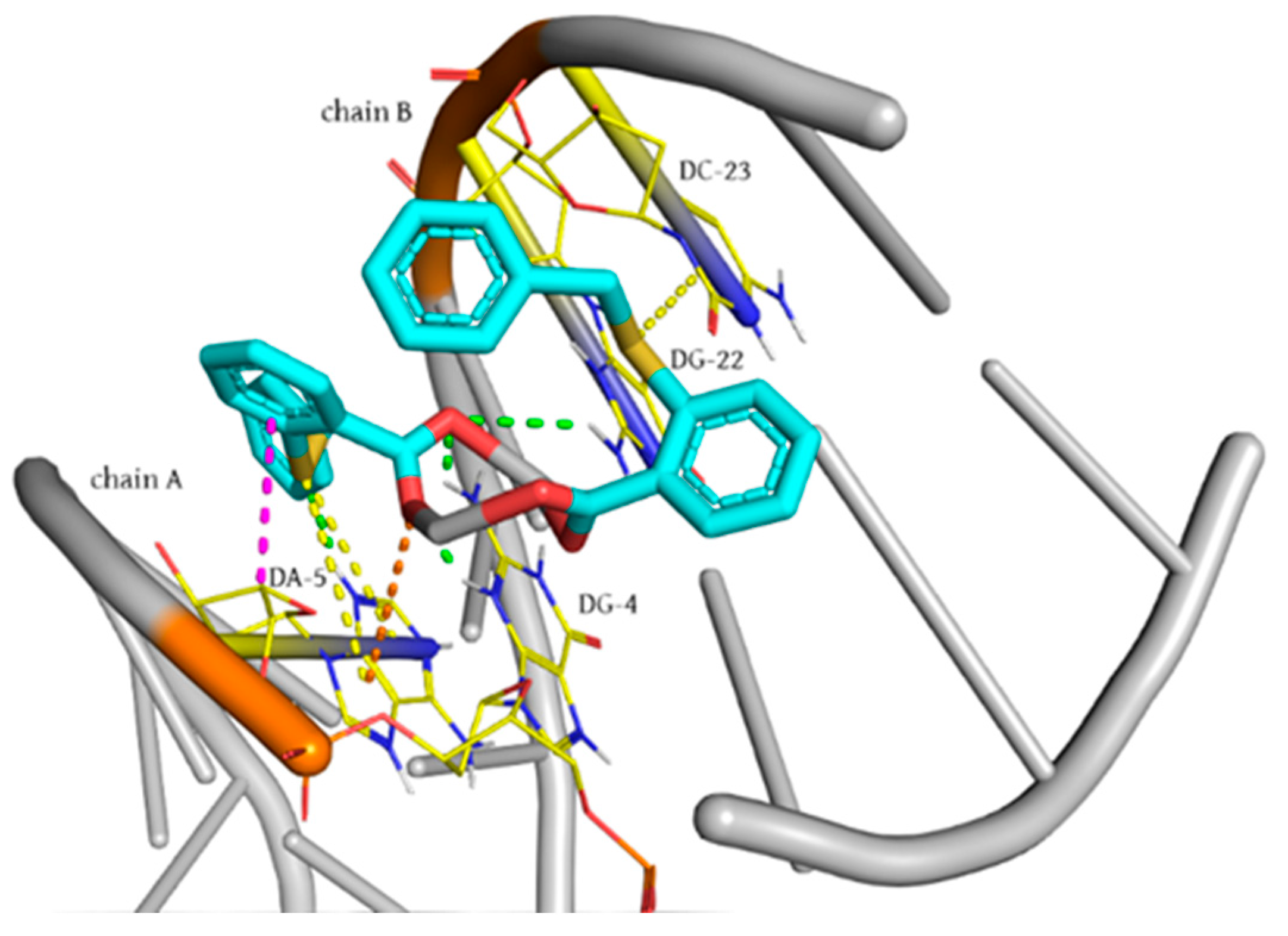
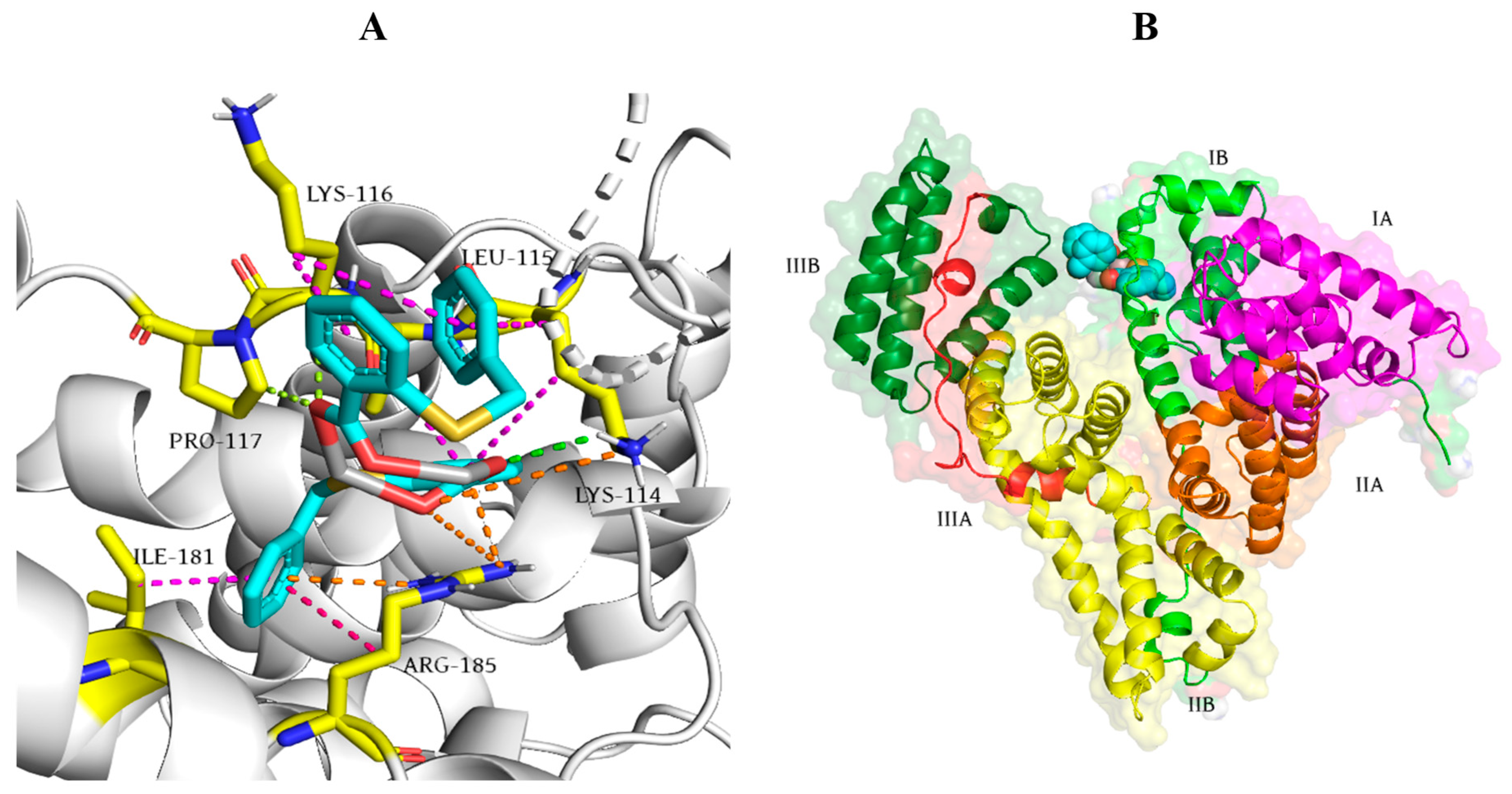
3.3. Molecular Docking Studies with DNA
3.4. Albumin-Binding Studies
3.5. Molecular Docking Studies with BSA
3.6. In Vitro Cytotoxicity of Silver(I) Complexes with S-Alkyl Derivatives of Thiosalicylic Acid (C1–C5)
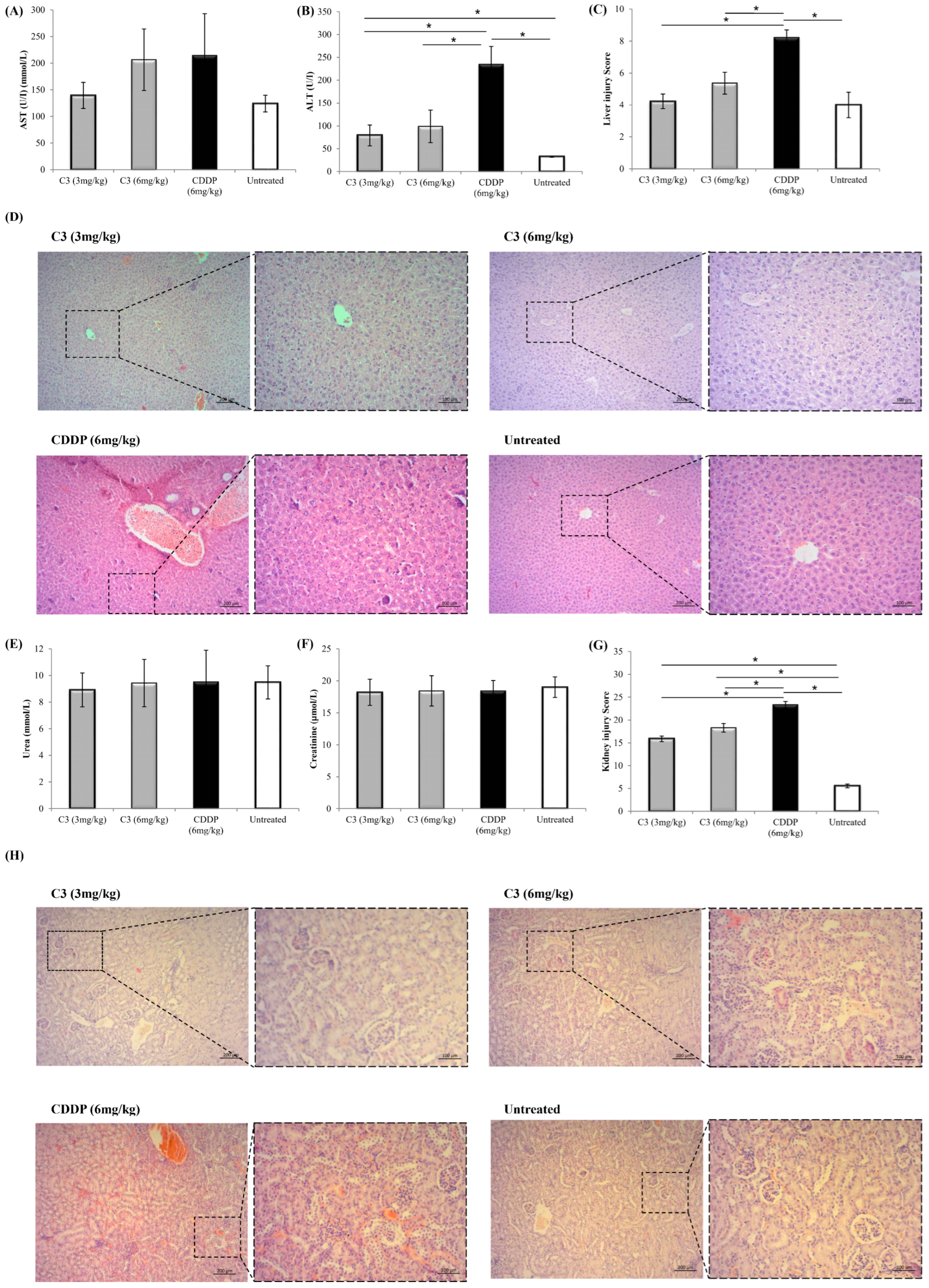
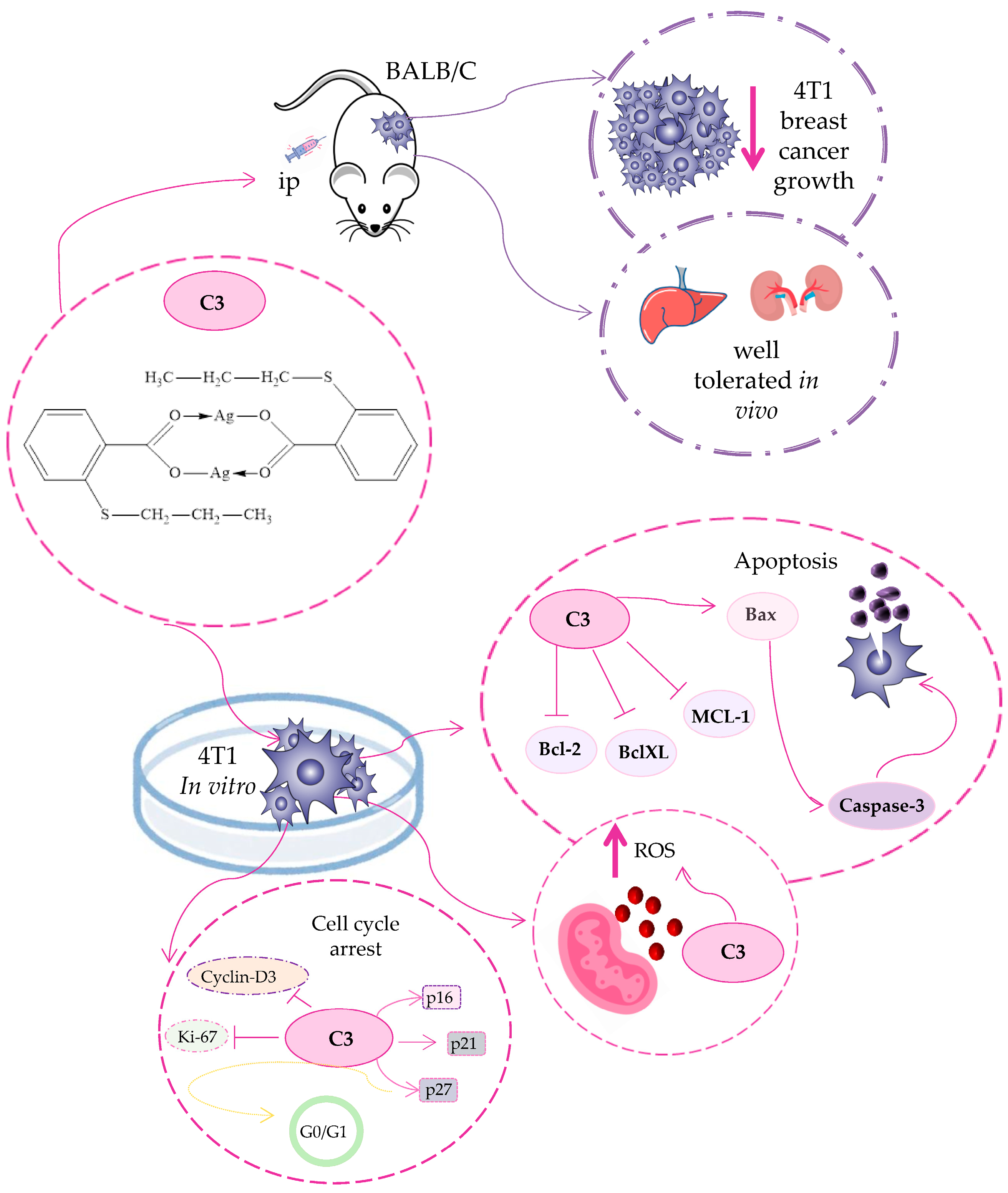
3.7. C3 Reduces Tumor Growth In Vivo
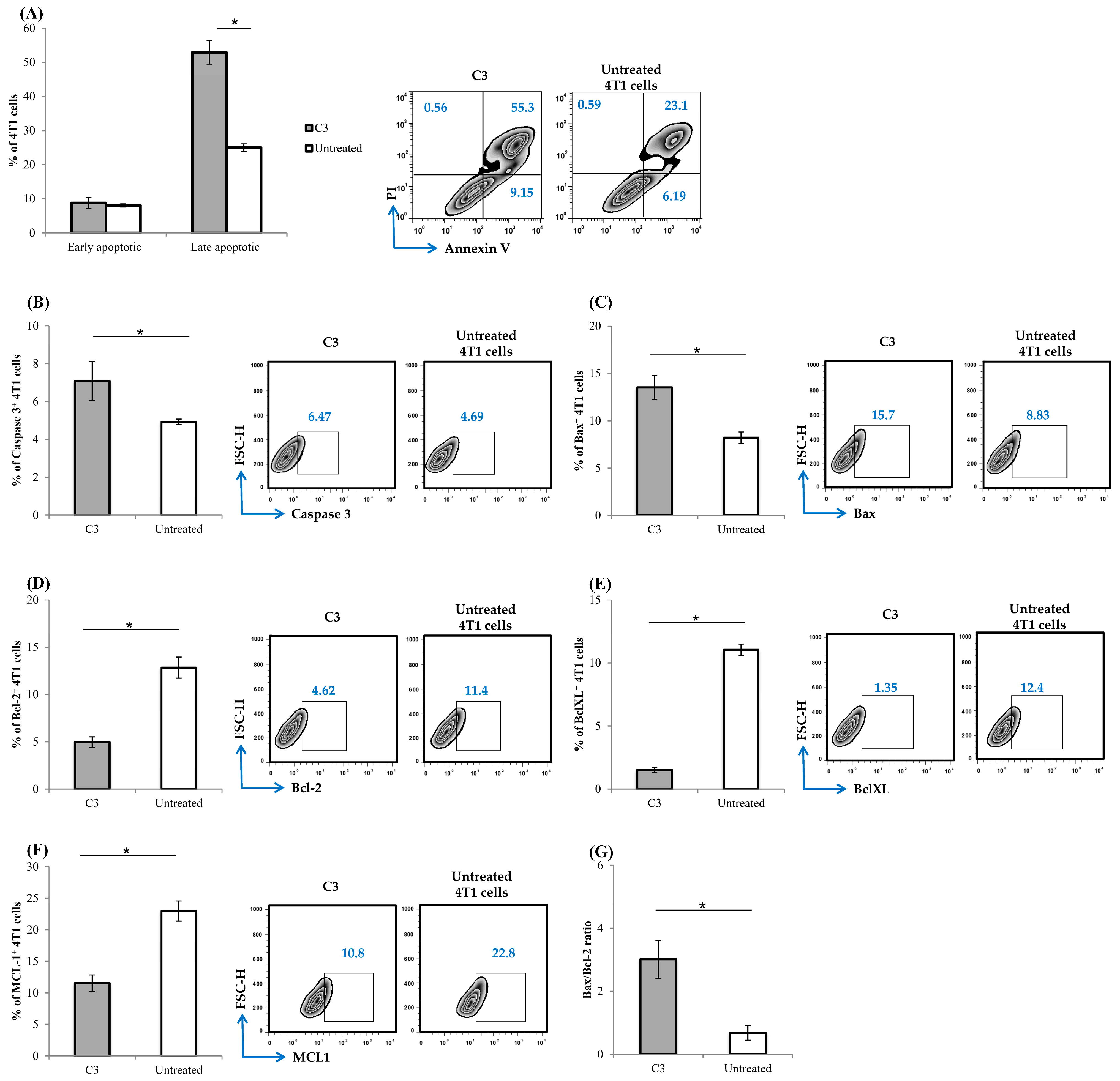
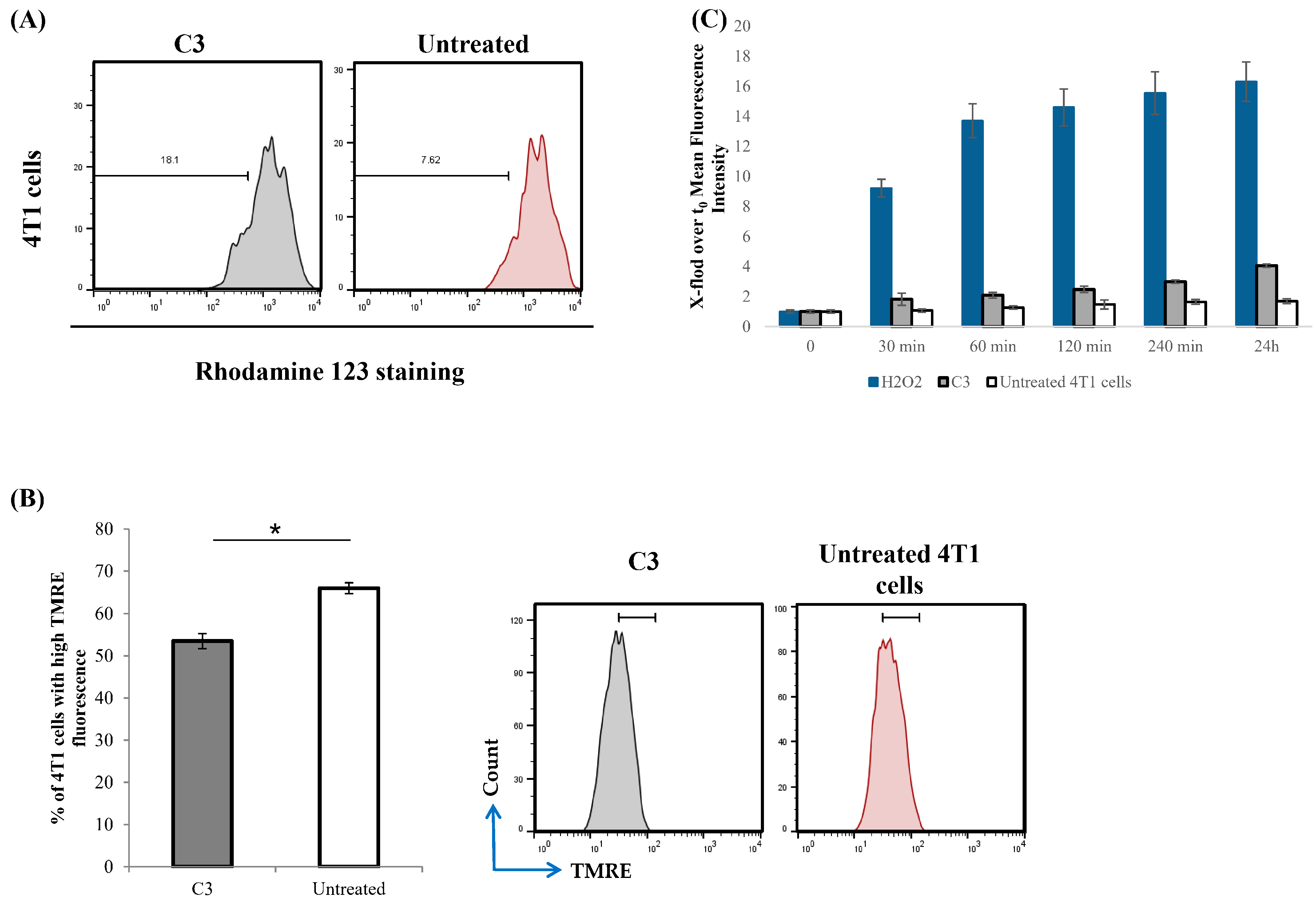
3.8. C3 Induces Apoptotic Cell Death and Interferes with Mitochondrial Function
3.9. C3 Suppresses Breast Cancer Cell Proliferation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Asif, M.; Iqbal, M.A.; Hussein, M.A.; Oon, C.E.; Haque, R.A.; Ahamed, M.B.K.; Majid, A.M.S.A. Human colon cancer targeted pro-apoptotic, anti-metastatic and cytostatic effects of binuclear Silver (I)–N-Heterocyclic carbene (NHC) complexes. Eur. J. Med. Chem. 2016, 108, 177–187. [Google Scholar] [CrossRef]
- Banti, C.N.; Hadjikakou, S.K. Anti-proliferative and anti-tumor activity of silver (I) compounds. Metallomics 2013, 5, 569–596. [Google Scholar] [CrossRef]
- Atiyeh, B.S.; Costagliola, M.; Hayek, S.N.; Dibo, S.A. Effect of silver on burn wound infection control and healing: Review of the literature. Burns 2007, 33, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Coyle, B.; McCann, M.; Kavanagh, K.; Devereux, M.; McKee, V.; Kayal, N.; Egan, D.; Deegan, C.; Finn, G.J. Synthesis, X-ray crystal structure, anti-fungal and anti-cancer activity of [Ag2(NH3)2(salH)2] (salH2 = salicylic acid). J. Inorg. Biochem. 2004, 98, 1361–1366. [Google Scholar] [CrossRef] [PubMed]
- Rowland, C.E.; Cantos, P.M.; Toby, B.H.; Frisch, M.; Deschamps, J.R.; Cahill, C.L. Controlling disulfide bond formation and crystal growth from 2-mercaptobenzoic acid. Cryst. Growth Des. 2011, 11, 1370–1374. [Google Scholar] [CrossRef]
- Gimple, R.C.; Wang, X. RAS: Striking at the Core of the Oncogenic Circuitry. Front. Oncol. 2019, 9, 965. [Google Scholar] [CrossRef]
- Smalley, K.S.; Eisen, T.G. Farnesyl thiosalicylic acid inhibits the growth of melanoma cells through a combination of cytostatic and pro-apoptotic effects. Int. J. Cancer 2002, 98, 514–522. [Google Scholar] [CrossRef]
- Bhattacharyya, M.; Girish, G.V.; Ghosh, R.; Chakraborty, S.; Sinha, A.K. Acetyl salicyclic acid (aspirin) improves synthesis of maspin and lowers incidence of metastasis in breast cancer patients. Cancer Sci. 2010, 101, 2105–2109. [Google Scholar] [CrossRef]
- Girish, G.V.; Sinha, N.; Chakraborty, K.; Bhattacharya, G.; Kahn, N.N.; Sinha, A.K. Restoration by aspirin of impaired plasma maspin level in human breast cancer. Acta Oncol. 2006, 45, 184–187. [Google Scholar] [CrossRef]
- Garcia-Albeniz, X.; Chan, A.T. Aspirin for the prevention of colorectal cancer. Best Pract. Res. Clin. Gastroenterol. 2011, 25, 461–472. [Google Scholar] [CrossRef]
- Sun, M.; Yu, J.; Wan, J.; Dou, X.; Chen, X.; Ye, F. Role of aspirin in cancer prevention. Cancer Treat. Res. Commun. 2025, 43, 100884. [Google Scholar] [CrossRef]
- Stark, L.A.; Din, F.V.N.; Zwacka, R.M.; Dunlop, M.G. Aspirin-induced activation of the NF-κB signaling pathway: A novel mechanism for aspirin-mediated apoptosis in colon cancer cells. FASEB J. 2001, 15, 1273–1275. [Google Scholar] [CrossRef]
- Benazic, S.; Silconi, Z.B.; Jevtovic, A.; Jurisevic, M.; Milovanovic, J.; Mijajlovic, M.; Nikolic, M.; Kanjevac, T.; Potočňák, I.; Samoľová, E.; et al. The Zn (S-pr-thiosal)2 complex attenuates murine breast cancer growth by inducing apoptosis and G1/S cell cycle arrest. Future Med. Chem. 2020, 12, 897–914. [Google Scholar] [CrossRef]
- Silconi, Z.B.; Rosic, V.; Benazic, S.; Radosavljevic, G.; Mijajlovic, M.; Pantic, J.; Ratkovic, Z.R.; Radic, G.; Arsenijevic, A.; Milovanovic, M.; et al. The Pt (S-pr-thiosal)2 and BCL1 Leukemia Lymphoma: Antitumor Activity In Vitro and In Vivo. Int. J. Mol. Sci. 2022, 23, 8161. [Google Scholar] [CrossRef]
- Liang, X.; Luan, S.; Yin, Z.; He, M.; He, C.; Yin, L.; Zou, Y.; Yuan, Z.; Li, L.; Song, X.; et al. Recent advances in the medical use of silver complex. Eur. J. Med. Chem. 2018, 157, 62–80. [Google Scholar] [CrossRef]
- Ou, Y.H.; Du, R.K.; Zhang, S.P.; Ling, Y.; Li, S.; Zhao, C.J.; Zhang, W.Z.; Zhang, L. Synthesis, crystal structure and in vitro antifungal activity of two-dimensional silver (I)-voriconazole coordination complexes. J. Mol. Struct. 2020, 1215, 128229. [Google Scholar] [CrossRef]
- Gil-Moles, M.; Olmos, M.E.; Monge, M.; Beltrán-Visiedo, M.; Marzo, I.; López-de-Luzuriaga, J.M.; Concepción Gimeno, M. Silver-Based Terpyridine Complexes as Antitumor Agents. Chem. Eur. J. 2023, 29, 202300116. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.E.; Becceneri, A.B.; Solcia, M.C.; Santiago, J.V.; Moreira, M.B.; Neto, J.A.G.; Pavan, F.R.; Cominetti, M.R.; Pereira, J.C.M.; Netto, A.V. Cytotoxic and apoptotic effects of ternary silver (I) complexes bearing 2-formylpyridine thiosemicarbazones and 1, 10-phenanthroline. Dalton Trans. 2020, 49, 5264–5275. [Google Scholar] [CrossRef]
- Monteiro, D.C.; Phillips, R.M.; Crossley, B.D.; Fielden, J.; Willans, C.E. Enhanced cytotoxicity of silver complexes bearing bidentate N-heterocyclic carbene ligands. Dalton Trans. 2012, 41, 3720–3725. [Google Scholar] [CrossRef]
- Allison, S.J.; Sadiq, M.; Baronou, E.; Cooper, P.A.; Dunnill, C.; Georgopoulos, N.T.; Latif, A.; Shepherd, D.; Shnyder, S.D.; Statford, I.J.; et al. Preclinical anti-cancer activity and multiple mechanisms of action of a cationic silver complex bearing N-heterocyclic carbene ligands. Cancer Lett. 2017, 403, 98–107. [Google Scholar] [CrossRef]
- Almeida, V.Y.; Rocha, J.S.; Felix, D.P.; Oliveira, G.P.; Lima, M.A.; Farias, R.L.; Zanetti, R.D.; Netto, A.V.; Zambom, C.R.; Garrido, S.S.; et al. Cytotoxicity and antibacterial activity of silver complexes bearing semicarbazones and triphenylphosphine. ChemistrySelect 2020, 5, 14559–14563. [Google Scholar] [CrossRef]
- Ota, A.; Tajima, M.; Mori, K.; Sugiyama, E.; Sato, V.H.; Sato, H. The selective cytotoxicity of silver thiosulfate, a silver complex, on MCF-7 breast cancer cells through ROS-induced cell death. Pharmacol. Rep. 2021, 73, 847–857. [Google Scholar] [CrossRef]
- Rocha, J.S.; Pereira, G.B.; Oliveira, G.P.; Lima, M.A.; Araujo-Neto, J.H.; Pinto, L.S.; Forim, M.R.; Zanetti, R.D.; Netto, A.V.; Castellano, E.E.; et al. Synthesis and characterization of silver (I) complexes bearing phenanthroline derivatives as ligands: Cytotoxicity and DNA interaction evaluation. Inorg. Chem. Commun. 2021, 131, 108757. [Google Scholar] [CrossRef]
- Caglar, S.; Altay, A.; Harurluoglu, B.; Yeniceri, E.K.; Caglar, B.; Şahin, O. Synthesis, structural characterization and evaluation of anticancer activity of polymeric silver (I) complexes based on niflumic acid/naproxen and picoline derivatives. J. Coord. Chem. 2022, 75, 178–196. [Google Scholar] [CrossRef]
- Altay, A.; Caglar, S.; Caglar, B. Silver (I) complexes containing diclofenac and niflumic acid induce apoptosis in human-derived cancer cell lines. Arch. Physiol. Biochem. 2022, 128, 69–79. [Google Scholar] [CrossRef]
- Harurluoglu, B.; Altay, A.; Caglar, S.; Yeniceri, E.K.K.; Caglar, B.; Şahin, Z.S. Binuclear silver (I) complexes with the non-steroidal anti-inflammatory drug tolfenamic acid: Synthesis, characterization, cytotoxic activity and evaluation of cellular mechanism of action. Polyhedron 2021, 202, 115189. [Google Scholar] [CrossRef]
- Dimiza, F.; Fountoulaki, S.; Papadopoulos, A.N.; Kontogiorgis, C.A.; Tangoulis, V.; Raptopoulou, C.P.; Psycharis, V.; Terzis, A.; Kessissoglou, D.P.; Psomas, G. Non-steroidal antiinflammatory drug–copper (II) complexes: Structure and biological perspectives. Dalton Trans. 2011, 40, 8555–8568. [Google Scholar] [CrossRef] [PubMed]
- Dimiza, F.; Perdih, F.; Tangoulis, V.; Turel, I.; Kessissoglou, D.P.; Psomas, G. Interaction of copper (II) with the non-steroidal anti-inflammatory drugs naproxen and diclofenac: Synthesis, structure, DNA-and albumin-binding. J. Inorg. Biochem. 2011, 105, 476–489. [Google Scholar] [CrossRef] [PubMed]
- Radić, G.P.; Glođović, V.V.; Radojević, I.D.; Stefanović, O.D.; Čomić, L.R.; Ratković, Z.R.; Valkonen, A.; Rissanen, K.; Trifunović, S.R. Synthesis, characterization and antimicrobial activity of palladium(II) complexes with some alkyl derivates of thiosalicylic acids: Crystal structure of the bis(S-benzyl-thiosalicylate)–palladium(II) complex, [Pd(S-bz-thiosal)2]. Polyhedron 2012, 31, 69–76. [Google Scholar] [CrossRef]
- Ganji, N.; Chityala, V.K.; Marri, P.K.; Aveli, R.; Narendrula, V.; Daravath, S. DNA incision evaluation, binding investigation and biocidal screening of Cu(II), Ni(II) and Co(II) complexes with isoxazole Schiff bases. J. Photochem. Photobiol. B 2017, 175, 132–140. [Google Scholar] [CrossRef]
- Pyle, A.M.; Rehmann, J.P.; Meshoyrer, R.; Kumar, C.V.; Turro, N.J.; Barton, J.K. Mixed-ligand complexes of ruthenium(II): Factors governing binding to DNA. J. Am. Chem. Soc. 1989, 111, 3051–3058. [Google Scholar] [CrossRef]
- Lakowicz, J.R.; Weber, G. Quenching of fluorescence by oxygen. Probe for structural fluctuations in macromolecules. Biochemistry 1973, 12, 4161–4170. [Google Scholar] [CrossRef]
- Wu, S.S.; Yuan, W.B.; Wang, H.Y.; Zhang, Q.; Liu, M.; Yu, K.B. Synthesis, crystal structure and interaction with DNA and HSA of (N, N′-dibenzylethane-1, 2-diamine) transition metal complexes. J. Inorg. Biochem. 2008, 102, 2026–2034. [Google Scholar] [CrossRef] [PubMed]
- Ćoćić, D.; Jovanović-Stević, S.; Jelić, R.; Matić, S.; Popović, S.; Djurdjević, P.; Baskić, B.; Petrović, B. Homo-and hetero-dinuclear Pt (II)/Pd (II) complexes: Studies of hydrolysis, nucleophilic substitution reactions, DNA/BSA interactions, DFT calculations, molecular docking and cytotoxic activity. Dalton Trans. 2020, 49, 14411–14431. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking With Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger, LLC.; DeLano, W. The PyMOLmolecular Graphics System, version 2.0; PyMOL; Schrödinger, LLC.: New York, NY, USA; Available online: https://www.pymol.org/pymol (accessed on 1 October 2025).
- Konovalov, B.; Franich, A.A.; Jovanović, M.; Jurisević, M.; Gajović, N.; Jovanović, M.; Arsenijević, N.; Marić, V.; Jovanović, I.; Živković, M.D.; et al. Synthesis, DNA-/bovine serum albumin-binding affinity, and cytotoxicity of dinuclear platinum (II) complexes with 1, 6-naphthyridine-bridging ligand. Appl. Organomet. Chem. 2021, 35, e6112. [Google Scholar] [CrossRef]
- Stojanović, M.N.D.; Franich, A.A.; Jurišević, M.M.; Gajović, N.M.; Arsenijević, N.N.; Jovanović, I.P.; Stojanović, B.S.; Mitrović, S.L.; Kljun, J.; Rajković, S.; et al. Platinum (II) complexes with malonic acids: Synthesis, characterization, in vitro and in vivo antitumor activity and interactions with biomolecules. J. Inorg. Biochem. 2022, 231, 111773. [Google Scholar] [CrossRef]
- Gajovic, N.; Jurisevic, M.; Pantic, J.; Radosavljevic, G.; Arsenijevic, N.; Lukic, M.L.; Jovanovic, I. Attenuation of NK cells facilitates mammary tumor growth in streptozotocin-induced diabetes in mice. Endocr. Relat. Cancer 2018, 25, 493–507. [Google Scholar] [CrossRef]
- Jurisevic, M.; Arsenijevic, A.; Pantic, J.; Gajovic, N.; Milovanovic, J.; Milovanovic, M.; Poljarevic, J.; Sabo, T.; Vojvodic, D.; Radosavljevic, G.D.; et al. The organic ester O, O’-diethyl-(S,S)-ethylenediamine-N,N’-di-2-(3-cyclohexyl) propanoate dihydrochloride attenuates murine breast cancer growth and metastasis. Oncotarget 2018, 9, 28195. [Google Scholar] [CrossRef]
- Ali, I.; Wani, W.A.; Saleem, K. Empirical formulae to molecular structures of metal complexes by molar conductance. Synth. React. Inorg. Met.-Org. Chem. 2013, 43, 1162–1170. [Google Scholar] [CrossRef]
- Grodzicki, A.; Łakomska, I.; Piszczek, P.; Szymańska, I.; Szłyk, E. Copper(I), silver(I) and gold(I) carboxylate complexes as precursors in chemical vapour deposition of thin metallic films. Coord. Chem. Rev. 2005, 249, 2232–2258. [Google Scholar] [CrossRef]
- Lewandowski, W.; Kalinowska, M.; Lewandowska, H. The influence of metals on the electronic system of biologically important ligands. Spectroscopic study of benzoates, salicylates, nicotinates and isoorotates. Review. J. Inorg. Biochem. 2005, 99, 1407–1423. [Google Scholar] [CrossRef]
- Azócar, M.I.; Muñoz, H.; Levin, P.; Dinamarca, N.; Gómez, G.; Ibañez, A.; Garland, M.T.; Paez, M.A. Synthesis and characterization of silver(I) complexes with ligands having anti-inflammatory properties. Inorg. Chem. Commun. 2013, 1, 19–21. [Google Scholar]
- Kellett, A.; Molphy, Z.; Slator, C.; McKee, V.; Farrell, N.P. Molecular methods for assessment of non-covalent metallodrug–DNA interactions. Chem. Soc. Rev. 2019, 48, 971–988. [Google Scholar] [CrossRef] [PubMed]
- Despaigne, A.A.R.; Da Silva, J.G.; Da Costa, P.R.; Dos Santos, R.G.; Beraldo, H. ROS-mediated cytotoxic effect of copper(II) hydrazone complexes against human glioma cells. Molecules 2014, 19, 17202–17220. [Google Scholar] [CrossRef] [PubMed]
- Hertzberg, R.P.; Dervan, P.B. Cleavage of double helical DNA by methidium-propyl-EDTA-iron (II). J. Am. Chem. Soc. 1982, 104, 313–315. [Google Scholar] [CrossRef]
- Franich, A.A.; Živković, M.D.; Milovanović, J.; Arsenijević, D.; Arsenijević, A.; Milovanović, M.; Djuranm, M.I.; Rajković, S. In vitro cytotoxic activities, DNA-and BSA-binding studies of dinuclear palladium(II) complexes with different pyridine-based bridging ligands. J. Inorg. Biochem. 2020, 210, 111158. [Google Scholar] [CrossRef]
- Timerbaev, A.R.; Hartinger, C.G.; Aleksenko, S.S.; Keppler, B.K. Interactions of antitumor metallodrugs with serum proteins: Advances in characterization using modern analytical methodology. Chem. Rev. 2006, 106, 2224–2248. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Zhang, H.M.; Zhang, G.C.; Tao, W.H.; Tang, S.H. Interaction of the flavonoid hesperidin with bovine serum albumin: A fluorescence quenching study. J. Lumin. 2007, 126, 211–218. [Google Scholar] [CrossRef]
- Chudzik, M.; Maciążek-Jurczyk, M.; Pawełczak, B.; Sułkowska, A. Spectroscopic studies on the molecular ageing of serum albumin. Molecules 2016, 22, 34. [Google Scholar] [CrossRef]
- Deepa, S.; Mishra, A.K. Fluorescence spectroscopic study of serum albumin–bromadiolone interaction: Fluorimetric determination of bromadiolone. J. Pharm. Biomed. Anal. 2005, 38, 556–563. [Google Scholar] [CrossRef]
- Jovanović, S.; Obrenčević, K.; Bugarčić, Ž.D.; Popović, I.; Žakula, J.; Petrović, B. New bimetallic palladium (II) and platinum (II) complexes: Studies of the nucleophilic substitution reactions, interactions with CT-DNA, bovine serum albumin and cytotoxic activity. Dalton Trans. 2016, 45, 12444–12457. [Google Scholar] [CrossRef]
- Ćoćić, D.; Jovanović, S.; Radisavljević, S.; Korzekwa, J.; Scheurer, A.; Puchta, R.; Baskić, D.; Todorović, D.; Popović, S.; Matić, S.; et al. New monofunctional platinum(II) and palladium(II) complexes: Studies of the nucleophilic substitution reactions, DNA/BSA interaction, and cytotoxic activity. J. Inorg. Biochem. 2018, 189, 91–102. [Google Scholar] [CrossRef]
- Nikolić, M.V.; Mijajlović, M.Ž.; Jevtić, V.V.; Ratković, Z.R.; Novaković, S.B.; Bogdanović, G.A.; Milovanović, J.; Arsenijević, A.; Stojanović, B.; Trifunović, S.R.; et al. Cytotoxicity of copper(II)-complexes with some S-alkyl derivatives of thiosalicylic acid. Crystal structure of the binuclear copper (II)-complex with S-ethyl derivative of thiosalicylic acid. J. Mol. Struct. 2016, 1116, 264–271. [Google Scholar] [CrossRef]
- Mijajlović, M.Ž.; Nikolić, M.V.; Jevtić, V.V.; Ratković, Z.R.; Marković, B.S.; Volarević, V.; Arsenijević, N.N.; Novaković, S.B.; Bogdanović, G.A.; Trifunović, S.R.; et al. Cytotoxicity of palladium(II) complexes with some alkyl derivates of thiosalicylic acid. Crystal structure of the bis (S-butyl-thiosalicylate) palladium(II) complex,[Pd(S-bu-thiosal)2]. Polyhedron 2015, 90, 34–40. [Google Scholar] [CrossRef]
- Besser Silconi, Z.; Benazic, S.; Milovanovic, J.; Jurisevic, M.; Djordjevic, D.; Nikolic, M.; Mijajlovic, M.; Ratkovic, Z.; Radić, G.; Radisavljevic, S.; et al. DNA binding and antitumor activities of platinum(IV) and zinc(II) complexes with some S-alkyl derivatives of thiosalicylic acid. Transit. Met. Chem. 2018, 43, 719–729. [Google Scholar] [CrossRef]
- Popović, A.; Nikolić, M.; Mijajlović, M.; Ratković, Z.; Jevtić, V.; Trifunović, S.R.; Radić, G.; Zarić, M.; Canović, P.; Milovanović, M.; et al. DNA binding and antitumor activities of zinc (II) complexes with some S-alkenyl derivatives of thiosalicylic acid. Transit. Met. Chem. 2019, 44, 219–228. [Google Scholar] [CrossRef]
- Varna, D.; Geromichalou, E.; Papachristou, E.; Papi, R.; Hatzidimitriou, A.G.; Panteris, E.; Psomas, G.; Geromichalos, G.D.; Aslanidis, P.; Choli-Papadopoulou, T.; et al. Biocompatible silver(I) complexes with heterocyclic thioamide ligands for selective killing of cancer cells and high antimicrobial activity—A combined in vitro and in silico study. J. Inorg. Biochem. 2022, 228, 111695. [Google Scholar] [CrossRef]
- Jaros, S.W.; Komarnicka, U.K.; Kyzioł, A.; Pucelik, B.; Nesterov, D.S.; Kirillov, A.M.; Smolenski, P. Therapeutic potential of a water-soluble silver-diclofenac coordination polymer on 3D pancreatic cancer spheroids. J. Med. Chem. 2022, 65, 11100–11110. [Google Scholar] [CrossRef]
- Şahin-Bölükbaşı, S.; Cantürk-Kılıçkaya, P.; Kılıçkaya, O. Silver (I)-N-heterocyclic carbene complexes challenge cancer; evaluation of their anticancer properties and in silico studies. Drug Dev. Res. 2021, 82, 907–926. [Google Scholar] [CrossRef]
- Ceramella, J.; Catalano, A.; Mariconda, A.; D’Amato, A.; Aquila, S.; Saturnino, C.; Rosano, C.; Sinicropi, M.S.; Longo, P. Silver N-heterocyclic carbene (NHC) complexes as antimicrobial and/or anticancer agents. Pharmaceuticals 2024, 18, 9. [Google Scholar] [CrossRef]
- Roberts, K.E.; Engelbrecht, Z.; Potgieter, K.; Meijboom, R.; Cronjé, M.J. Silver (I) bromide phosphines induce mitochondrial-mediated apoptosis in malignant human colorectal cells. Biomedicines 2023, 11, 2794. [Google Scholar] [CrossRef]
- Gandin, V.; Fernandes, A.P. Metal-and semimetal-containing inhibitors of thioredoxin reductase as anticancer agents. Molecules 2015, 20, 12732–12756. [Google Scholar] [CrossRef] [PubMed]
- Stefanache, A.; Miftode, A.M.; Constantin, M.; Bogdan Goroftei, R.E.; Olaru, I.; Gutu, C.; Vornicu, A.; Lungu, I.I. Noble Metal Complexes in Cancer Therapy: Unlocking Redox Potential for Next-Gen Treatments. Inorganics 2025, 13, 64. [Google Scholar] [CrossRef]
- Félix-Piña, P.; Franco Molina, M.A.; García Coronado, P.L.; Prado-Garcia, H.; Zarate-Triviño, D.G.; Castro-Valenzuela, B.E.; Moreno-Amador, K.A.; Uscanga Palomeque, A.C.; Rodríguez Padilla, C. β-D-glucose-reduced silver nanoparticles remodel the tumor microenvironment in a murine model of triple-negative breast cancer. Int. J. Mol. Sci. 2024, 25, 8432. [Google Scholar] [CrossRef]
- Azizi, M.; Ghourchian, H.; Yazdian, F.; Bagherifam, S.; Bekhradnia, S.; Nyström, B. Anti-cancerous effect of albumin coated silver nanoparticles on MDA-MB 231 human breast cancer cell line. Sci. Rep. 2017, 7, 5178. [Google Scholar] [CrossRef]
- Ferdous, Z.; Nemmar, A. Health impact of silver nanoparticles: A review of the biodistribution and toxicity following various routes of exposure. Int. J. Mol. Sci. 2020, 21, 2375. [Google Scholar] [CrossRef]
- Wiemann, M.; Vennemann, A.; Blaske, F.; Sperling, M.; Karst, U. Silver nanoparticles in the lung: Toxic effects and focal accumulation of silver in remote organs. Nanomaterials 2017, 7, 441. [Google Scholar] [CrossRef]
- Raju, S.K.; Karunakaran, A.; Kumar, S.; Sekar, P.; Murugesan, M.; Karthikeyan, M. Silver complexes as anticancer agents: A perspective review. Ger. J. Pharm. Biomater. 2022, 1, 6–28. [Google Scholar] [CrossRef]
- Mustafa, M.; Ahmad, R.; Tantry, I.Q.; Ahmad, W.; Siddiqui, S.; Alam, M.; Abbas, K.; Moinuddin; Hassan, M.I.; Habib, S.; et al. Apoptosis: A comprehensive overview of signaling pathways, morphological changes, and physiological significance and therapeutic implications. Cells 2024, 13, 1838. [Google Scholar] [CrossRef]
- Meijboom, R.; Iroegbu, A.O.C.; Ray, S.S. Advancing cancer therapy: The role of silver(I) phosphine complexes in overcoming resistance and toxicity. Discov. Oncol. 2025, 16, 792. [Google Scholar] [CrossRef] [PubMed]
- Elsayed, S.A.; Saleh, E.E.; Aboelnga, M.M.; Toson, E.A. Experimental and computational studies of silver(I) dibenzoylmethane-based complexes, interaction with DNA/RNA/BSA biomolecules, and in vitro cytotoxic activity. J. Inorg. Biochem. 2023, 241, 112132. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.J.; Huang, X.; Li, X.Y.; Tang, S.L.; Li, D.J.; Cheng, Y.Z.; Azam, M.; Zhang, L.P.; Sun, D. Synthesis, structure and biological activity of silver(I) complexes containing triphenylphosphine and non-steroidal anti-inflammatory drug ligands. J. Inorg. Biochem. 2024, 250, 112404. [Google Scholar] [CrossRef] [PubMed]
- Niu, Z.L.; Wu, T.T.; Wu, Y.Y.; Zhou, S.H.; Li, Z.; Guo, J.C.; Zhou, S.M.; Deng, S.H.; Xu, J.Y.; Xie, M.J. Two novel silver(I) phenanthroline derivative complexes induce G2/M phase cycle arrest and apoptosis of MDA-MB-231 cancer cells by multiple mechanisms. Appl. Organomet. Chem. 2023, 37, e7123. [Google Scholar] [CrossRef]
- Gottlieb, E.; Armour, S.M.; Harris, M.H.; Thompson, C.B. Mitochondrial membrane potential regulates matrix configuration and cytochrome c release during apoptosis. Cell Death Differ. 2003, 10, 709–717. [Google Scholar] [CrossRef]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014, 94, 909–950. [Google Scholar] [CrossRef]
- Dammak, K.; Porchia, M.; De Franco, M.; Zancato, M.; Naïli, H.; Gandin, V.; Marzano, C. Antiproliferative homoleptic and heteroleptic phosphino silver (I) complexes: Effect of ligand combination on their biological mechanism of action. Molecules 2020, 25, 5484. [Google Scholar] [CrossRef]
- Vahabirad, M.; Daei, S.; Abbasalipourkabir, R.; Ziamajidi, N. Anticancer action of silver nanoparticles in SKBR3 breast cancer cells through promotion of oxidative stress and apoptosis. BioMed Res. Int. 2024, 2024, 7145339. [Google Scholar] [CrossRef]
- Chaudhry, G.E.S.; Md Akim, A.; Sung, Y.Y.; Sifzizul, T.M.T. Cancer and apoptosis: The apoptotic activity of plant and marine natural products and their potential as targeted cancer therapeutics. Front. Pharmacol. 2022, 13, 842376. [Google Scholar] [CrossRef]
- Suski, J.M.; Braun, M.; Strmiska, V.; Sicinski, P. Targeting cell-cycle machinery in cancer. Cancer Cell 2021, 39, 759–778. [Google Scholar] [CrossRef]
- Heinke, L. Mitochondrial ROS drive cell cycle progression. Nat. Rev. Mol. Cell Biol. 2022, 23, 581. [Google Scholar] [CrossRef]
- Sun, X.; Kaufman, P.D. Ki-67: More than a proliferation marker. Chromosoma 2018, 127, 175–186. [Google Scholar] [CrossRef]
- Saleban, M.; Harris, E.L.; Poulter, J.A. D-type cyclins in development and disease. Genes 2023, 14, 1445. [Google Scholar] [CrossRef]
- Kuzderová, G.; Rendošová, M.; Gyepes, R.; Sovová, S.; Sabolová, D.; Vilková, M.; Olejníková, P.; Bačová, I.; Stokič, S.; Kello, M.; et al. Antimicrobial and anticancer application of silver(I) dipeptide complexes. Molecules 2021, 26, 6335. [Google Scholar] [CrossRef]
- Novikov, N.M.; Zolotaryova, S.Y.; Gautreau, A.M.; Denisov, E.V. Mutational drivers of cancer cell migration and invasion. Br. J. Cancer 2021, 124, 102–114. [Google Scholar] [CrossRef]


| Compound | Carboxylate Group | ||
|---|---|---|---|
| νasym COO− | νsym COO− | ∆ν | |
| C1 | 1537 cm−1 | 1369 cm−1 | 168 |
| C2 | 1537 cm−1 | 1370 cm−1 | 167 |
| C3 | 1547 cm−1 | 1366 cm−1 | 181 |
| C4 | 1548 cm−1 | 1368 cm−1 | 180 |
| C5 | 1550 cm−1 | 1377 cm−1 | 173 |
| Compound | Kb (M−1) | ΔG (KJ mol−1) | Ksv EB (M−1) |
|---|---|---|---|
| C1 | (7.5 ± 0.1) × 104 | −27.8 | (6.0 ± 0.1) × 103 |
| C2 | (9.1 ± 0.1) × 104 | −28.3 | (1.1 ± 0.1) × 104 |
| C3 | (2.2 ± 0.1) × 105 | −30.5 | (1.4 ± 0.1) × 104 |
| C4 | (3.5 ± 0.1) × 105 | −31.6 | (4.0 ± 0.1) × 104 |
| C5 | (6.6 ± 0.2) × 105 | −33.2 | (6.8 ± 0.1) × 104 |
| PDB ID | Complex | ΔG (kcal/mol) | ΔGvdw + hbond + desolv (kcal/mol) | ΔGelec (kcal/mol) | ΔGIntermol. Energy (kcal/mol) | ΔGtotal (kcal/mol) | ΔGtor (kcal/mol) | ΔGunb (kcal/mol) |
|---|---|---|---|---|---|---|---|---|
| 1Z3F | C1 | −6.79 | −7.72 | −0.17 | −7.89 | −0.57 | 1.10 | −0.57 |
| C2 | −6.50 | −8.09 | −0.05 | −8.15 | −0.82 | 1.65 | −0.82 | |
| C3 | −6.23 | −8.39 | −0.03 | −8.42 | −0.98 | 2.20 | −0.98 | |
| C4 | −5.95 | −8.55 | −0.14 | −8.69 | −1.51 | 2.74 | −1.51 | |
| C5 | −7.62 | −9.89 | 0.07 | −9.81 | −2.23 | 2.20 | −2.23 | |
| 1BNA | C1 | −8.18 | −10.88 | −0.04 | −10.92 | −1.50 | 2.74 | −1.50 |
| C2 | −8.30 | −10.38 | −0.11 | −10.49 | −2.09 | 2.20 | −2.09 | |
| C3 | −8.50 | −10.56 | −0.13 | −10.69 | −0.88 | 2.20 | −0.88 | |
| C4 | −9.07 | −10.14 | −0.04 | −10.17 | −0.56 | 1.10 | −0.56 | |
| C5 | −9.28 | −10.88 | −0.04 | −10.92 | −0.69 | 1.65 | −0.69 |
| Compound | Ksv (M−1) | kq (M−1 s−1) | K (M−1) | n |
|---|---|---|---|---|
| C5 | (5.8 ± 0.1) × 104 | (5.8 ± 0.1) × 1012 | (8.3 ± 0.1) × 104 | 0.8 |
| C4 | (6.3 ± 0.1) × 104 | (6.3 ± 0.1) × 1012 | (8.9 ± 0.1) × 104 | 0.8 |
| C3 | (9.6 ± 0.1) × 104 | (9.6 ± 0.1) × 1012 | (1.1 ± 0.1) × 105 | 1.1 |
| C2 | (1.4 ± 0.1) × 105 | (1.4 ± 0.1) × 1013 | (1.3 ± 0.1) × 105 | 1.1 |
| C1 | (1.7 ± 0.1) × 105 | (1.7 ± 0.1) × 1013 | (1.7 ± 0.1) × 105 | 0.8 |
| PDB ID | Complex | ΔG (kcal/mol) | ΔGvdw + hbond + desolv (kcal/mol) | ΔGelec (kcal/mol) | ΔGIntermol. Energy (kcal/mol) | ΔGtotal (kcal/mol) | ΔGtor (kcal/mol) | ΔGunb (kcal/mol) |
|---|---|---|---|---|---|---|---|---|
| C1 | −8.55 | −10.73 | −0.01 | −10.74 | −1.13 | 2.20 | −1.13 | |
| C2 | −8.07 | −9.18 | 0.01 | −9.17 | −0.73 | 1.10 | −0.73 | |
| 4F5S | C3 | −8.02 | −9.71 | 0.04 | −9.67 | −1.06 | 1.65 | −1.06 |
| C4 | −7.94 | −10.72 | 0.03 | −10.69 | −0.97 | 2.74 | −0.97 | |
| C5 | −8.87 | −11.12 | 0.05 | −11.07 | −2.50 | 2.20 | −2.50 |
| Cell Line | Time | C1 | C2 | C3 | C4 | C5 | L1 | L2 | L3 | L4 | L5 | CDDP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4T1 | 24 h | 0.33 ± 0.21 | 0.57 ± 0.15 | 0.25 ± 0.19 | 0.64 ± 0.28 | 18.65 ± 1.15 | 1420.66 ± 68.58 | 628.32 ± 40.51 | 954.56 ± 63.34 | 406.83 ± 35.1 | 290.96 ± 20.47 | 11.98 ± 1.98 |
| 48 h | 4.00 ± 0.95 | 4.84 ± 0.92 | 2.13 ± 1.17 | 4.55 ± 0.88 | 4.20 ± 1.48 | 442.84 ± 30.52 | 499.79 ± 7.60 | 499.69 ± 30.48 | 416.85 ± 61.42 | 375.12 ± 40.37 | 5.17 ± 0.98 | |
| MDA-MB-468 | 24 h | 4.19 ± 0.98 | 8.67 ± 1.13 | 15.47 ± 1.21 | 15.06 ± 1.56 | 18.77 ± 0.69 | 885.89 ± 35.22 | 721.63 ± 55.32 | 612.78 ± 49.57 | 899.17 ± 29.55 | 457,12 ± 45.99 | 4.06 ± 0.97 |
| 48 h | 4.37 ± 0.41 | 5.43 ± 0.34 | 8.76 ± 1.24 | 9.88 ± 0.962 | 8.60 ± 1.65 | 511.35 ± 74.55 | 767.78 ± 28.84 | 528.51 ± 51.96 | 407.51 ± 12.74 | 337.18 ± 18.23 | 3.47 ± 0.47 | |
| CT26 | 24 h | 2.03 ± 0.59 | 1.08 ± 0.03 | 1.58 ± 0.82 | 7.71 ± 0.21 | 29.94 ± 2.12 | 664.54 ± 28.84 | 708.81 ± 83.91 | 533.77 ± 27.72 | 647.14 ± 11.14 | 389.16 ± 9.59 | 16.94 ± 1.42 |
| 48 h | 1.24 ± 0.47 | 3.21 ± 0.39 | 3.29 ± 1.28 | 4.45 ± 0.37 | 7.19 ± 0.38 | 701.74 ± 61.08 | 464.56 ± 39.52 | 683.57 ± 61.21 | 367.83 ± 58.31 | 369.88 ± 18.69 | 1.29 ± 0.11 | |
| HCT 116 | 24 h | 3.24 ± 0.67 | 3.01 ± 1.31 | 1.19 ± 0.21 | 16.73 ± 2.79 | 44.45 ± 3.68 | 736.60 ± 60.02 | 713.96 ± 30.01 | 649.43 ± 46.58 | 669.11 ± 43.11 | 246.99 ± 19.02 | 63.31 ± 3.21 |
| 48 h | 1.24 ± 0.065 | 1.04 ± 0.06 | 1.46 ± 0.83 | 11.28 ± 2.13 | 3.96 ± 0.97 | 1341.94 ± 136.14 | 1161.57 ± 98.7 | 1335.21 ± 54.16 | 951.03 ± 32.64 | 208.38 ± 28.84 | 15.78 ± 1.28 | |
| LLC1 | 24 h | 39.75 ± 1.07 | 18.25 ± 0.93 | 41.58 ± 1.27 | 57.77 ± 1.64 | 29.09 ± 0.90 | 590.55 ± 47.85 | 566.5 ± 17.67 | 805.5 ± 31.66 | 1489.10 ± 71.82 | 741.99 ± 37.81 | 21.82 ± 1.11 |
| 48 h | 32.02 ± 1.47 | 20.21 ± 0.79 | 20.31 ± 1.18 | 46.53 ± 1.32 | 24.23 ± 0.78 | 947.42 ± 41.37 | 960.29 ± 63.64 | 681.48 ± 32.23 | 670.09 ± 48.13 | 860.98 ± 60.96 | 3.51 ± 0.92 | |
| A549 | 24 h | 58.27 ± 1.91 | 37.75 ± 1.01 | 55.77 ± 1.83 | 48.61 ± 1.92 | 24.35 ± 1.57 | 1059.61 ± 75.52 | 723.31 ± 37.99 | 1414.53 ± 75.31 | 494.87 ± 3.58 | 394.57 ± 39.06 | 23.22 ± 1.77 |
| 48 h | 48.62 ± 1.057 | 50.23 ± 1.074 | 66.27 ± 1.18 | 99.03 ± 3.85 | 59.92 ± 2.02 | 1529.22 ± 82.21 | 540.11 ± 34.32 | 499.94 ± 47.29 | 489.72 ± 22.01 | 643.56 ± 26.17 | 14.00 ± 1.44 | |
| mMSC | 24 h | 2.84 ± 0.21 | 4.29 ± 0.92 | 2.81 ± 0.5 | 9.41 ± 1.13 | 28.65 ± 1.02 | 815.29 ± 36.59 | 1203.02 ± 67.98 | 843.33 ± 31.09 | 540.67 ± 9.85 | 434.82 ± 10.09 | 17.14 ± 1.15 |
| 48 h | 2.54 ± 0.26 | 4.52 ± 0.48 | 4.22 ± 0.33 | 1.67 ± 0.27 | 2.16 ± 0.62 | 697.5 ± 7.78 | 843.33 ± 31.09 | 1139.57 ± 41.61 | 1884.32 ± 96.77 | 636.60 ± 41.88 | 3.11 ± 0.93 | |
| MRC-5 | 24 h | 46.3 ± 1.52 | 38.04 ± 1.91 | 38.1 ± 1.13 | 26.49 ± 0.91 | 46.04 ± 1.37 | 658.33 ± 53.23 | 538.45 ± 20.63 | 637.81 ± 50.98 | 947.93 ± 50.03 | 331.54 ± 29.02 | 40.5 ± 0.57 |
| 48 h | 53.25 ± 1.27 | 43.17 ± 1.23 | 43.34 ± 1.1 | 15.34 ± 0.73 | 46.65 ± 1.87 | 487.71 ± 41.56 | 384.10 ± 20.53 | 628.26 ± 60.49 | 718.50 ± 39.71 | 447.87 ± 36.04 | 27.35 ± 0.72 |
| Selectivity Index (IC50 mMSC or MRC-5/ IC50 Cancer Cell) | |||||||
|---|---|---|---|---|---|---|---|
| Cell Line | Time | C1 | C2 | C3 | C4 | C5 | CDDP |
| 4T1 | 24 h | 8.60 | 7.53 | 11.24 | 14.70 | 1.549 | 1.43 |
| 48 h | 0.64 | 0.93 | 1.98 | 0.37 | 0.51 | 0.60 | |
| CT26 | 24 h | 1.40 | 3.97 | 1.78 | 1.22 | 0.96 | 1.01 |
| 48 h | 2.04 | 1.41 | 1.28 | 0.38 | 0.30 | 2.41 | |
| LLC1 | 24 h | 0.07 | 0.24 | 0.07 | 0.16 | 0.98 | 0.79 |
| 48 h | 0.08 | 0.22 | 0.21 | 0.04 | 0.09 | 0.89 | |
| MDA-MB-468 | 24 h | 11.05 | 4.39 | 2.46 | 1.76 | 2.45 | 9.98 |
| 48 h | 12.18 | 7.95 | 4.95 | 1.55 | 5.42 | 7.88 | |
| HCT-116 | 24 h | 14.29 | 12.64 | 32.02 | 1.58 | 1.04 | 0.64 |
| 48 h | 42.94 | 41.51 | 29.68 | 1.36 | 11.78 | 1.73 | |
| A549 | 24 h | 0.79 | 1.01 | 0.68 | 0.55 | 1.89 | 1.74 |
| 48 h | 1.10 | 0.86 | 0.65 | 0.15 | 0.78 | 1.95 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marinković, J.; Jurišević, M.; Jovanović, M.; Milosavljević, M.; Jovanović, I.; Stević, S.J.; Vesović, M.; Nikolić, M.; Nedeljković, N.; Živanović, A.; et al. Silver(I) Complexes Bearing S-Alkyl Thiosalicylic Acid Derivatives: DNA/BSA Binding and Antitumor Activity In Vitro and In Vivo. Pharmaceutics 2025, 17, 1340. https://doi.org/10.3390/pharmaceutics17101340
Marinković J, Jurišević M, Jovanović M, Milosavljević M, Jovanović I, Stević SJ, Vesović M, Nikolić M, Nedeljković N, Živanović A, et al. Silver(I) Complexes Bearing S-Alkyl Thiosalicylic Acid Derivatives: DNA/BSA Binding and Antitumor Activity In Vitro and In Vivo. Pharmaceutics. 2025; 17(10):1340. https://doi.org/10.3390/pharmaceutics17101340
Chicago/Turabian StyleMarinković, Jovana, Milena Jurišević, Marina Jovanović, Miloš Milosavljević, Ivan Jovanović, Snežana Jovanović Stević, Marina Vesović, Miloš Nikolić, Nikola Nedeljković, Ana Živanović, and et al. 2025. "Silver(I) Complexes Bearing S-Alkyl Thiosalicylic Acid Derivatives: DNA/BSA Binding and Antitumor Activity In Vitro and In Vivo" Pharmaceutics 17, no. 10: 1340. https://doi.org/10.3390/pharmaceutics17101340
APA StyleMarinković, J., Jurišević, M., Jovanović, M., Milosavljević, M., Jovanović, I., Stević, S. J., Vesović, M., Nikolić, M., Nedeljković, N., Živanović, A., Tomović, D., Bukonjić, A., Radić, G., & Gajović, N. (2025). Silver(I) Complexes Bearing S-Alkyl Thiosalicylic Acid Derivatives: DNA/BSA Binding and Antitumor Activity In Vitro and In Vivo. Pharmaceutics, 17(10), 1340. https://doi.org/10.3390/pharmaceutics17101340









