Analysis of Lipophilicity and Pharmacokinetic Parameters of Dipyridothiazine Dimers with Anticancer Potency
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Chromatographic Procedure
2.3. Theoretical Lipophilicity, ADMET Parameters and Target Prediction
3. Results
4. Discussion
- Dimers 1a–4a RM0 = −89.395b − 0.4208 (r = 0.9952);
- Dimers 1b–4b RM0 = −106.55b − 1.1075 (r = 0.9957);
- Dimers 1c–4c RM0 = −93.8444b − 0.5607 (r = 0.9912);
- Dimers 1d–4d RM0 = −96.341b − 0.5183607 (r = 0.9933).
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ginex, T.; Vazquez, J.; Gilbert, E.; Herrero, E.; Luque, F.J. Lipophilicity in drug design: An overview of lipophilicity descriptors in 3D-QSAR studies. Future Med. Chem. 2019, 11, 1177–1193. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.W.; Gallego, R.A.; Edwards, M.P. Lipophilic efficiency as an important metric in drug design. J. Med. Chem. 2018, 61, 6401–6420. [Google Scholar] [CrossRef] [PubMed]
- Ditzinger, F.; Price, D.J.; Ilie, A.R.; Köhl, N.J.; Jankovic, S.; Tsakiridou, G.; Aleandri, S.; Kalantzi, L.; Holm, R.; Nair, A.; et al. Lipophilicity and hydrophobicity considerations in bioenabling oral formulations approaches—A PEARRL review. J. Pharm. Pharmacol. 2019, 71, 464–482. [Google Scholar] [CrossRef] [PubMed]
- Markovic, M.; Ben-Shabat, S.; Keinan, S.; Aponick, A.; Zimmermann, E.M.; Dahan, A. Lipidic prodrug approach for improved oral drug delivery and therapy. Med. Res. Rev. 2019, 39, 579–607. [Google Scholar] [CrossRef] [PubMed]
- Di, L.; Kerns, E.H. Profiling drug-like properties in discovery research. Curr. Opin. Chem. Biol. 2003, 7, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Manto Chagasa, C.; Mossa, S.; Alisaraie, L. Drug metabolites and their effects on the development of adverse reactions: Revisiting Lipinski’s Rule of Five. Int. J. Pharm. 2018, 549, 133–149. [Google Scholar] [CrossRef]
- Lobo, S. Is there enough focus on lipophilicity in drug discovery? Expert Opin. Drug Discov. 2020, 15, 261–263. [Google Scholar] [CrossRef]
- Alqahtani, M.S.; Kazi, M.; Alsenaidy, M.A.; Ahmad, M.Z. Advances in Oral Drug Delivery. Front. Pharmacol. 2021, 12, 618411. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Giaginis, G.; Tsopelas, F.; Tsantili-Kakoulidou, A. The Impact of Lipophilicity in Drug Discovery: Rapid Measurements by Means of Reversed-Phase HPLC. Methods Mol. Biol. 2018, 1824, 217–228. [Google Scholar] [CrossRef]
- Dulsat, J.; López-Nieto, B.; Estrada-Tejedor, R.; Borrell, J.I. Evaluation of free online ADMET tools for academic or small biotech environments. Molecules 2023, 28, 776. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Mei, L.; Quach, T.; Porter, C.; Trevaskis, N. Lipophilic Conjugates of Drugs: A Tool to Improve Drug Phar-macokinetic and Therapeutic Profiles. Pharm. Res. 2021, 38, 1497–1518. [Google Scholar] [CrossRef] [PubMed]
- Soares, J.X.; Santos, Á.; Fernandes, C.; Pinto, M.M.M. Liquid chromatography on the different methods for the determination of lipophilicity: An essential analytical tool in medicinal chemistry. Chemosensors 2022, 10, 340. [Google Scholar] [CrossRef]
- Roman, I.P.; Mastromichali, A.; Tyrovola, K.; Canals, A.; Psillakis, E. Rapid determination of octanol-water partition coefficient using vortex-assisted liquid-liquid microextraction. J. Chromatogr. Sci. 2014, 1330, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Toma, A.; Hapau, D.; Casoni, D.; Zaharia, V. Heterocycles 33: Lipophilicity of new class of thioethers estimated by reversed-phase thin-layer chromatography and different computational methods. J. Chromatogr. Sci. 2014, 52, 1302–1307. [Google Scholar] [CrossRef]
- Franke, U.; Munk, A.; Wiese, M. Ionization Constants and Distribution Coefficients of Phenothiazines as Calcium Channel Antagonists Determined by a pH-Metric Method and Correlation with Calculated Partition Coefficients. J. Pharm. Sci. 1999, 88, 89–95. [Google Scholar] [CrossRef]
- Hendrich, H.B.; Wesołowska, O.; Poła, A.; Motohashi, N.; Molnár, J.; Michalak, K. Neither lipophilicity nor membrane-perturbing potency of phenothiazinemaleates correlate with the ability to inhibit P-glycoprotein transport activity. Mol. Membr. Biol. 2003, 20, 53–60. [Google Scholar] [CrossRef]
- Morak-Młodawska, B.; Jeleń, M.; Pluta, K. Phenothiazines Modified with the Pyridine Ring as Promising Anticancer Agents. Life 2021, 11, 206. [Google Scholar] [CrossRef]
- Nycz-Empel, A.; Bober, K.; Wyszomirski, M.; Kisiel, E.; Zięba, A. The Application of CA and PCA to the evaluation of lipophilicity and physicochemical properties of tetracyclic diazaphenothiazine derivatives. J. Anal. Methods Chem. 2019, 2019, 8131235. [Google Scholar] [CrossRef]
- Morak-Młodawska, B.; Jeleń, M.; Martula, E.; Korlacki, R. Study of Lipophilicity and ADME Properties of 1,9-Diazaphenothiazines with Anticancer Action. Int. J. Mol. Sci. 2023, 24, 6970. [Google Scholar] [CrossRef]
- Martula, E.; Morak-Młodawska, B.; Jeleń, M.; Okechukwu, N.P.; Balachandran, A.; Tehirunavukarasu, P.; Anamalay, K.; Ulaganathan, V. Synthesis and structural characterization of novel dimers of dipyridothiazine as promising antiproliferative agents. Molecules 2023, 28, 7662. [Google Scholar] [CrossRef] [PubMed]
- Martula, E.; Morak-Młodawska, B.; Jeleń, M.; Strzyga-Łach, P.; Struga, M.; Żurawska, K.; Kasprzycka, A.; Bagrowska, W. Comparative Analysis of Structural Analogs of Dipyridothiazines with m-Xylene and a Lutidine Moiety—In Silico, In Vitro, and Docking Studies. Appl. Sci. 2024, 14, 7263. [Google Scholar] [CrossRef]
- Virtual Computational Chemistry Laboratory. Available online: https://vcclab.org/ (accessed on 11 July 2024).
- ChemDraw: ChemDraw Ultra, version 12. PerkinElmer Informatics. CambridgeSoft: Cambridge, MA, USA, 2007.
- SwissADME SwissDrugDesign. Available online: http://www.swissadme.ch/ (accessed on 11 July 2024).
- PreADMET. Home–Prediction of Metabolism|PreMetabo. Available online: https://premetabo.webservice.bmdrc.org/home-2/ (accessed on 11 July 2024).
- SwissTargetPrediction SwissDrugDesign. Available online: http://www.swisstargetprediction.ch/predict.php (accessed on 11 July 2024).
- Bodor, N.; Gabanyi, Z.; Wong, C.K. A new method for the estimation of partition coefficient. J. Am. Chem. Soc. 1989, 111, 3783–3786. [Google Scholar] [CrossRef]
- Mannhold, R.; Cruciani, G.; Dross, K.; Rekker, R. Multivariate analysis of experimental and computational descriptors of molecular lipophilicity. J. Comput.-Aided Mol. Des. 1998, 12, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, A.; Han, Y.; Hopfinger, A.J. Predicting Caco-2 Cell Permeation Coecients of Organic Molecules Using Membrane-Interaction QSAR Analysis. J. Chem. Inf. Comput. Sci. 2002, 42, 331–342. [Google Scholar] [CrossRef]
- Irvine, J.D.; Takahashi, L.; Lockhart, K.; Cheong, J.; Tolan, J.W.; Selick, H.E.; Grove, J.R. MDCK (Madin-Darby Canine Kidney) Cells: A Tool for Membrane Permeability Screening. J. Pharm. Sci. 1999, 88, 28–33. [Google Scholar] [CrossRef]
- Feher, M.; Schmidt, J.M. Property Distributions: Differences Between Drugs, Natural Products, and Molecules from Combinatorial Chemistry. J. Chem. Inf. Comput. Sci. 2003, 34, 218–227. [Google Scholar] [CrossRef]
- Dołowy, M. Comparative Study of the Lipophilicity of Selected Tauro-Conjugates Bile Acids Determined with the Use of RPTLC and RPHPTLC Methods. J. Liq. Chromatogr. Relat. Technol. 2009, 32, 2281–2292. [Google Scholar] [CrossRef]
- Kowalska, A.; Pluta, K. RP TLC Assay of The Lipophilicity of New Azathioprine Analogs. J. Liq. Chromatogr. Relat. Technol. 2012, 35, 1686–1696. [Google Scholar] [CrossRef]
- Nowak, M.; Pluta, K. Study of the Lipophilicity of Novel Diquinothiazinesa. J. Planar Chromatogr. 2006, 19, 157–160. [Google Scholar] [CrossRef]
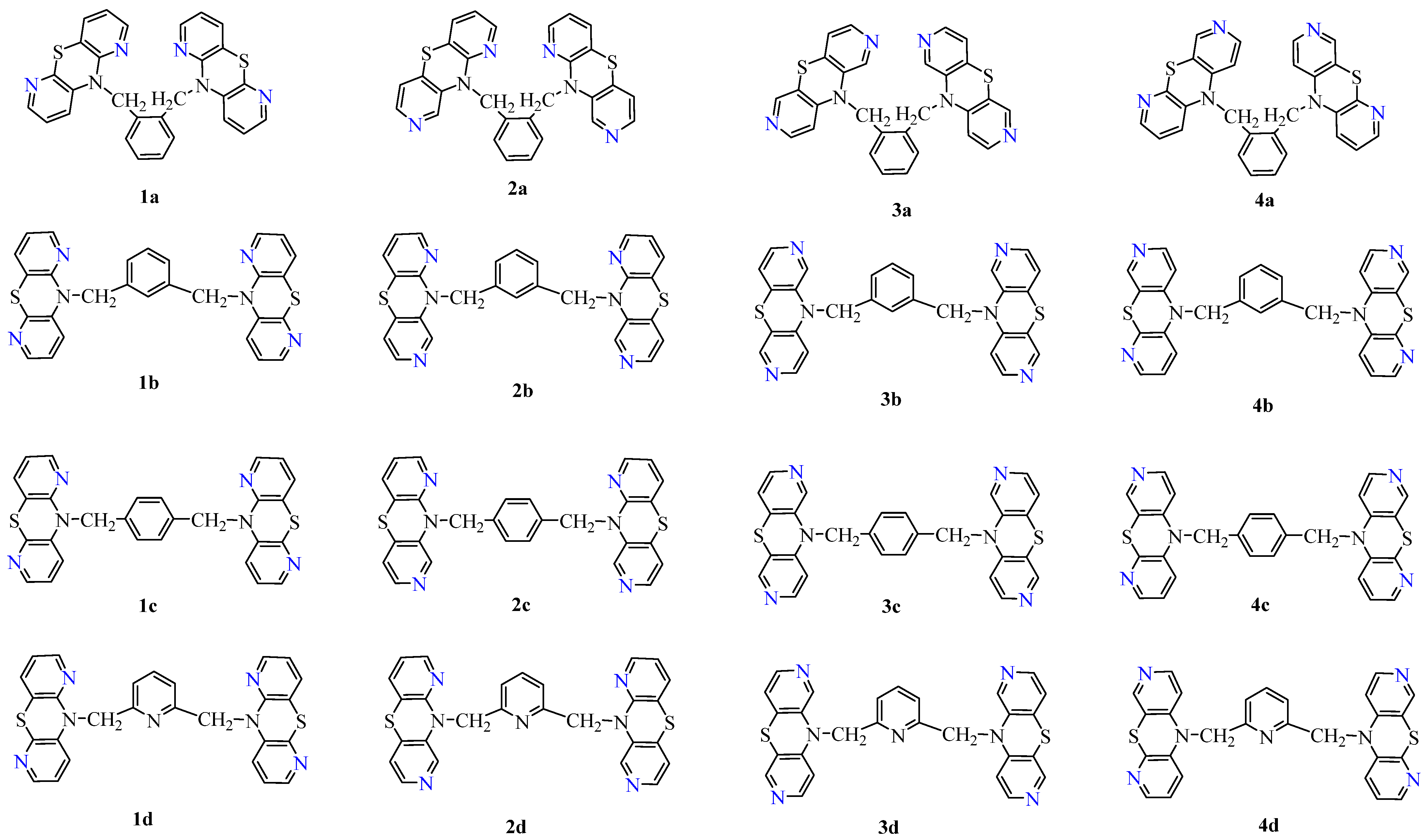
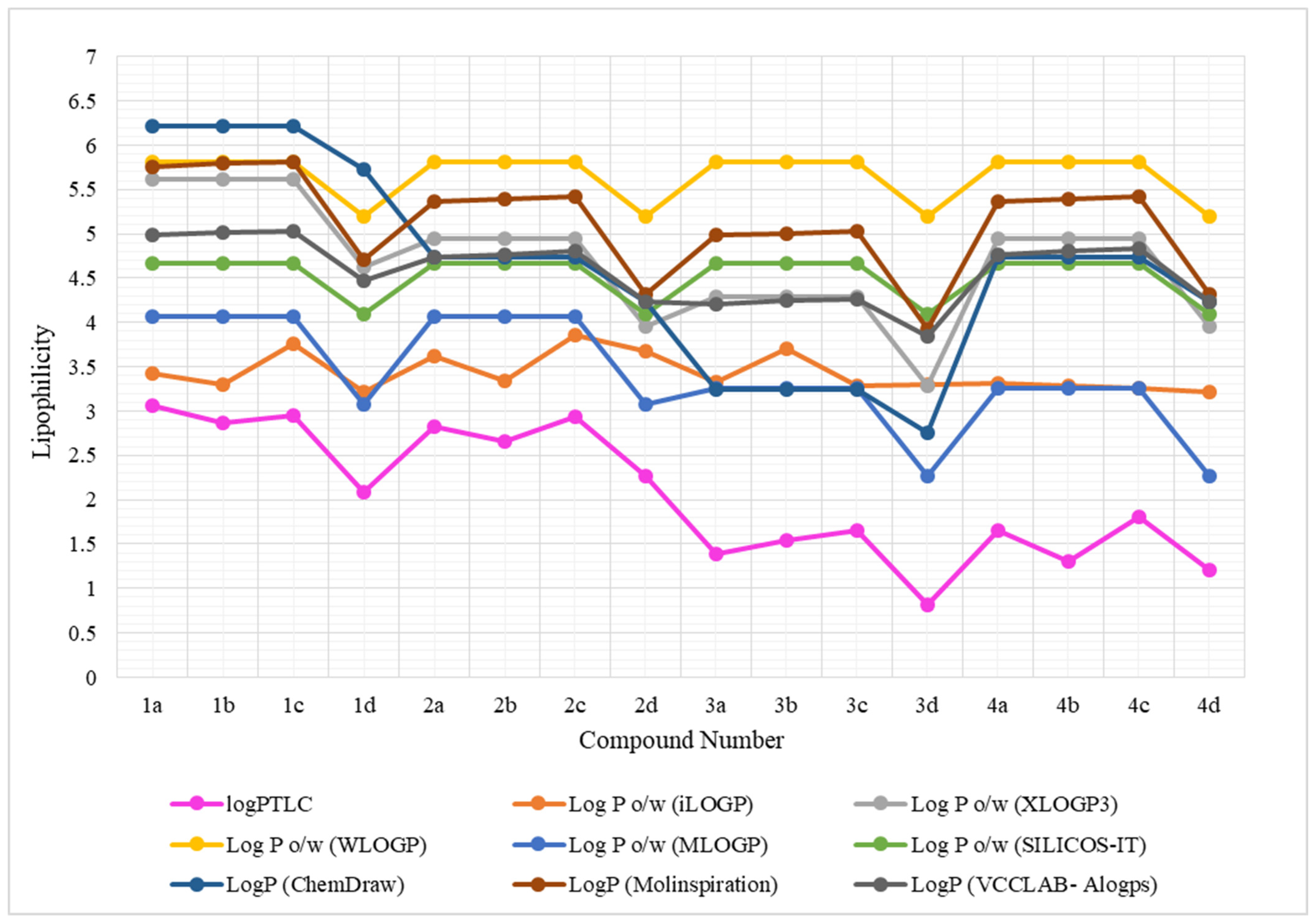
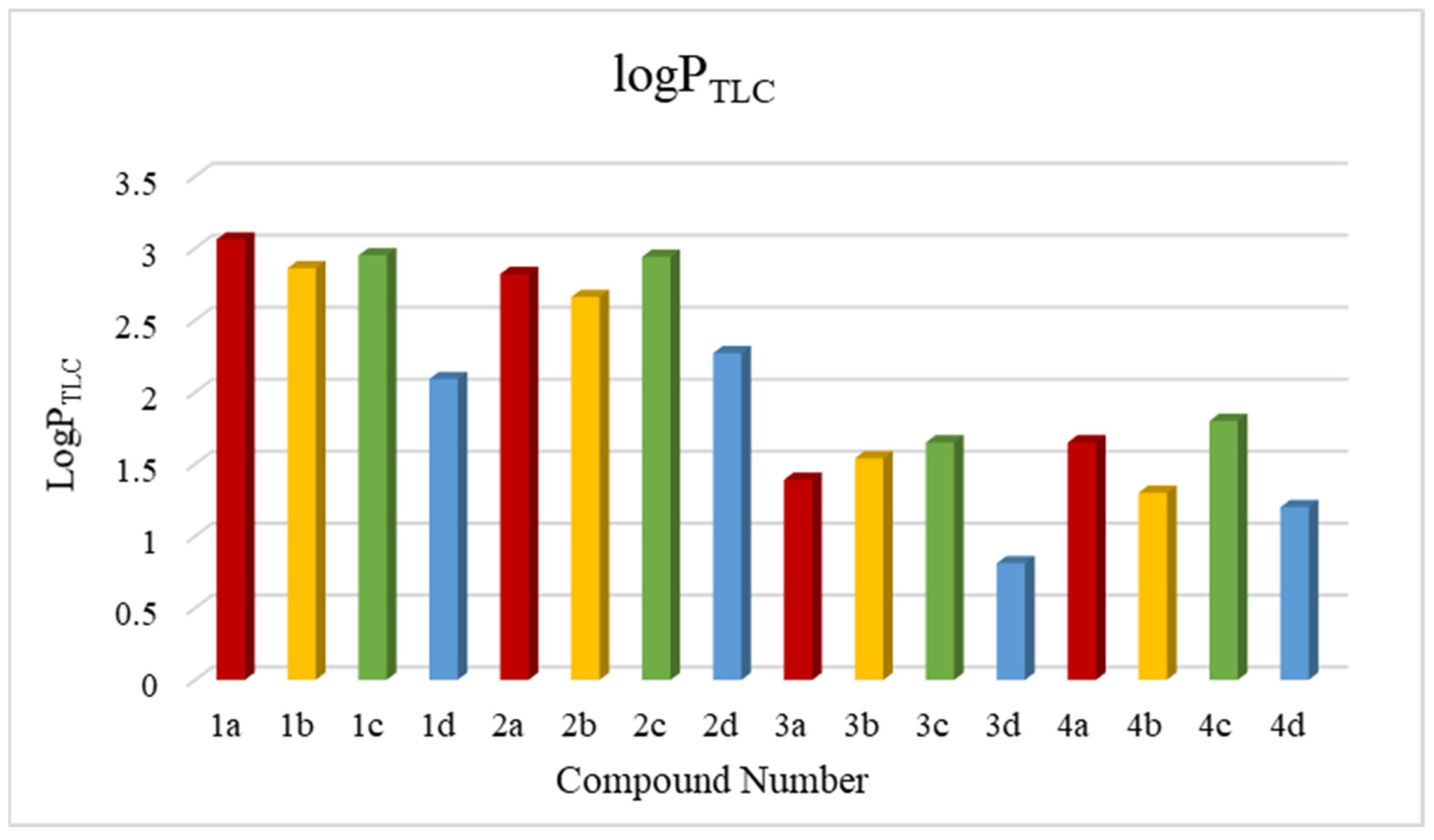
| No. | LogPcalcd. | |||||||
|---|---|---|---|---|---|---|---|---|
| iLOGP | XLOGP3 | WLOGP | MLOGP | SILICOS-IT | LogP (ChemDraw) | LogP (Mol Inspiration) | LogP (VCCLAB Alogps) | |
| 1a | 3.42 | 5.62 | 5.81 | 4.07 | 4.67 | 6.22 | 5.76 | 4.98 |
| 1b | 3.30 | 5.62 | 5.81 | 4.07 | 4.67 | 6.22 | 5.79 | 5.01 |
| 1c | 3.76 | 5.62 | 5.81 | 4.07 | 4.67 | 6.22 | 5.81 | 5.03 |
| 1d | 3.22 | 4.62 | 5.20 | 3.07 | 4.10 | 5.73 | 4.71 | 4.47 |
| 2a | 3.62 | 4.95 | 5.81 | 4.07 | 4.67 | 4.73 | 5.37 | 4.74 |
| 2b | 3.34 | 4.95 | 5.81 | 4.07 | 4.67 | 4.73 | 5.39 | 4.76 |
| 2c | 3.85 | 4.95 | 5.81 | 4.07 | 4.67 | 4.73 | 5.42 | 4.80 |
| 2d | 3.67 | 3.95 | 5.20 | 3.07 | 4.10 | 4.24 | 4.32 | 4.24 |
| 3a | 3.33 | 4.29 | 5.81 | 3.26 | 4.67 | 3.24 | 4.98 | 4.20 |
| 3b | 3.70 | 4.29 | 5.81 | 3.26 | 4.67 | 3.24 | 5.00 | 4.25 |
| 3c | 3.29 | 4.29 | 5.81 | 3.26 | 4.67 | 3.24 | 5.03 | 4.26 |
| 3d | 3.30 | 3.29 | 5.20 | 2.26 | 4.10 | 2.75 | 3.93 | 3.84 |
| 4a | 3.31 | 4.95 | 5.81 | 3.26 | 4.67 | 4.73 | 5.37 | 4.77 |
| 4b | 3.28 | 4.95 | 5.81 | 3.26 | 4.67 | 4.73 | 5.39 | 4.81 |
| 4c | 3.26 | 4.95 | 5.81 | 3.26 | 4.67 | 4.73 | 5.42 | 4.84 |
| 4d | 3.22 | 3.95 | 5.20 | 2.26 | 4.10 | 4.24 | 4.32 | 4.24 |
| No. | −b | RM0 | r |
|---|---|---|---|
| 1a | 0.045 | 3.5760 | 0.9973 |
| 1b | 0.0426 | 3.3468 | 0.9806 |
| 1c | 0.0432 | 3.4546 | 0.9979 |
| 1d | 0.0300 | 2.4253 | 0.9978 |
| 2a | 0.0409 | 3.3041 | 0.9983 |
| 2b | 0.0385 | 3.1117 | 0.9850 |
| 2c | 0.0418 | 3.4411 | 0.9972 |
| 2d | 0.0327 | 2.6375 | 0.9941 |
| 3a | 0.0217 | 1.6016 | 0.9856 |
| 3b | 0.0274 | 1.7742 | 0.9674 |
| 3c | 0.0252 | 1.9040 | 0.9850 |
| 3d | 0.0139 | 0.8993 | 0.9562 |
| 4a | 0.0274 | 1.9033 | 0.9933 |
| 4b | 0.0243 | 1.4866 | 0.9621 |
| 4c | 0.0296 | 2.0767 | 0.9983 |
| 4d | 0.0209 | 1.3580 | 0.9765 |
| Parameters | I | II | III | IV | V |
|---|---|---|---|---|---|
| LogPlit. | 0.64 [28] | 1.21 [29] | 1.58 [29] | 2.43 [29] | 4.45 [28] |
| RM0 | 0.5858 | 0.9275 | 1.5099 | 2.1803 | 2.6378 |
| −b | 0.0168 | 0.0181 | 0.0225 | 0.0288 | 0.0346 |
| r | 0.9954 | 0.9936 | 0.9920 | 0.9960 | 0.9930 |
| No. | 1a | 1b | 1c | 1d | 2a | 2b | 2c | 2d | 3a | 3b | 3c | 3d | 4a | 4b | 4c | 4d |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| logPTLC | 3.06 | 2.86 | 2.95 | 2.09 | 2.82 | 2.66 | 2.94 | 2.27 | 1.39 | 1.54 | 1.68 | 0.81 | 1.65 | 1.30 | 1.80 | 1.21 |
| No. | Molecular Mass (g/mol) | H-Bond Acceptors | H-Bond Donors | Rotatable Bonds | Molar Refractivity | TPSA [Å2] | P-gp Substrate | Lipinski’s Rules | Ghose’s Rules | Veber’s Rules | Muegge’s Rules |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1a | 504.63 | 4 | 0 | 4 | 149.60 | 108.64 | + | + | − | + | − |
| 1b | 504.63 | 4 | 0 | 4 | 149.60 | 108.64 | + | + | − | + | − |
| 1c | 504.63 | 4 | 0 | 4 | 149.60 | 108.64 | + | + | − | + | − |
| 1d | 505.62 | 5 | 0 | 4 | 147.40 | 121.53 | + | + | − | + | + |
| 2a | 504.63 | 4 | 0 | 4 | 149.60 | 108.64 | + | + | − | + | + |
| 2b | 504.63 | 4 | 0 | 4 | 149.60 | 108.64 | + | + | − | + | + |
| 2c | 504.63 | 4 | 0 | 4 | 149.60 | 108.64 | + | + | − | + | + |
| 2d | 505.62 | 5 | 0 | 4 | 147.40 | 121.53 | + | + | − | + | + |
| 3a | 504.63 | 4 | 0 | 4 | 149.60 | 108.64 | + | + | − | + | + |
| 3b | 504.63 | 4 | 0 | 4 | 149.60 | 108.64 | + | + | − | + | + |
| 3c | 504.63 | 4 | 0 | 4 | 149.60 | 108.64 | + | + | − | + | + |
| 3d | 505.62 | 5 | 0 | 4 | 147.40 | 121.53 | + | + | − | + | + |
| 4a | 504.63 | 4 | 0 | 4 | 149.60 | 108.64 | + | + | − | + | + |
| 4b | 504.63 | 4 | 0 | 4 | 149.60 | 108.64 | + | + | − | + | + |
| 4c | 504.63 | 4 | 0 | 4 | 149.60 | 108.64 | + | + | − | + | + |
| 4d | 505.62 | 5 | 0 | 4 | 147.40 | 121.53 | + | + | − | + | + |
| No. | Caco-2 Permeability (nm/s) | Skin Permeability (SP, log Kp) | BBB Permeability (C.brain/C.blood) | HIA (%) | MDCK (nm/s) | Plasma Protein Binding (PPB,%) |
|---|---|---|---|---|---|---|
| 1a | 35.7262 | −2.60631 | 1.783 | 97.816 | 0.275 | 100 |
| 1b | 30.5877 | −2.62873 | 0.208 | 97.816 | 0.073 | 100 |
| 1c | 32.9405 | −2.63079 | 0.202 | 97.816 | 3.944 | 100 |
| 1d | 28.8091 | −2.91305 | 0.415 | 98.019 | 0.205 | 98 |
| 2a | 29.7800 | −2.91532 | 0.905 | 97.816 | 0.1395 | 96 |
| 2b | 26.4756 | −2.94119 | 0.224 | 97.816 | 0.0607 | 97 |
| 2c | 27.9283 | −2.94361 | 0.311 | 97.816 | 1.399 | 95 |
| 2d | 25.5742 | −3.25587 | 0.205 | 98.019 | 0.114 | 91 |
| 3a | 29.3153 | −3.48028 | 0.391 | 97.816 | 0.151 | 91 |
| 3b | 25.9114 | −3.50753 | 0.400 | 97.816 | 0.064 | 90 |
| 3c | 27.4437 | −3.5101 | 0.201 | 97.816 | 1.636 | 90 |
| 3d | 25.1325 | −3.81394 | 0.183 | 98.019 | 0.121 | 87 |
| 4a | 32.0035 | −3.15846 | 0.258 | 97.816 | 4.062 | 97 |
| 4b | 29.4980 | −3.1559 | 0.938 | 97.816 | 0.072 | 99 |
| 4c | 32.0035 | −3.15846 | 0.258 | 97.816 | 4.062 | 97 |
| 4d | 28.0612 | −3.47613 | 0.403 | 98.019 | 0.203 | 91 |
| Doxorubicin | 17.7263 | −4.73786 | 0.036 | 56.841 | 1.204 | 31 |
| No. of Compounds | ADMET Activities | Equation | r |
|---|---|---|---|
| 1a,b,c,d–4a,b,c,d | Caco-2 | Caco-2 = 4.0763 RM03 − 26.748 RM02 + 54.697 RM0 − 6.1633 | 0.7288 |
| 1a,b,c,d–4a,b,c,d | SP | SP = 0.5685 RM03 − 3.7459 RM02 + 7.621 RM0 − 7.7475 | 0.3022 |
| 1a,b,c,d–4a,b,c,d | BBB | BBB = 0.4934 RM03 − 3.0831 RM02 + 5.8357 RM0 − 2.9445 | 0.6158 |
| 1a,b,c,d–4a,b,c,d | HIA | HIA = −0.118 RM03 + 0.7997 RM02 − 1.7015 RM0 + 99.01 | 0.6705 |
| 1a,b,c,d–4a,b,c,d | MDCK | MDCK = 0.7955 RM03 − 5.8698 RM02 + 13.621 RM0 − 8.7393 | 0.3143 |
| 1a,b,c,d–4a,b,c,d | PPB | PPB = −2.0899 RM03 − 14.54 RM02 + 34.436 RM0 + 66.883 | 0.6918 |
| No. of Compound | Target Prediction | ||
|---|---|---|---|
| 1a | Kinase | Family C G protein-coupled receptor | Phosphodiesterase |
| 1b | Ligand-gated ion channel | Cytochrome P450 | Phosphodiesterase |
| 1c | Kinase | Protease | Enzyme |
| 1d | Kinase | Family C G protein-coupled receptor | Phosphodiesterase |
| 2a | Kinase | Enzyme | Phosphodiesterase |
| 2b | Kinase | Histone deacetylase 1 | Phosphodiesterase |
| 2c | Kinase | Family C G protein-coupled receptor | Protease |
| 2d | Kinase | Family C G protein-coupled receptor | Phosphodiesterase |
| 3a | Kinase | Cytochrome P450 | Protease |
| 3b | Cytochrome P450 | Protease | Phosphodiesterase |
| 3c | Kinase | Enzyme | Protease |
| 3d | Reader | Family C G protein-coupled receptor | Protease |
| 4a | Phosphodiesterase | Family C G protein-coupled receptor | Protease |
| 4b | Kinase | Family A G protein-coupled receptor | Cytochrome P450 |
| 4c | Kinase | Family C G protein-coupled receptor | Voltage-gated ion channel |
| 4d | Phosphodiesterase | Bromodomain-containing protein 4,3,2 | Cytochrome P450 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martula, E.; Morak-Młodawska, B.; Jeleń, M.; Okechukwu, P.N. Analysis of Lipophilicity and Pharmacokinetic Parameters of Dipyridothiazine Dimers with Anticancer Potency. Pharmaceutics 2024, 16, 1235. https://doi.org/10.3390/pharmaceutics16091235
Martula E, Morak-Młodawska B, Jeleń M, Okechukwu PN. Analysis of Lipophilicity and Pharmacokinetic Parameters of Dipyridothiazine Dimers with Anticancer Potency. Pharmaceutics. 2024; 16(9):1235. https://doi.org/10.3390/pharmaceutics16091235
Chicago/Turabian StyleMartula, Emilia, Beata Morak-Młodawska, Małgorzata Jeleń, and Patrick Nwabueze Okechukwu. 2024. "Analysis of Lipophilicity and Pharmacokinetic Parameters of Dipyridothiazine Dimers with Anticancer Potency" Pharmaceutics 16, no. 9: 1235. https://doi.org/10.3390/pharmaceutics16091235
APA StyleMartula, E., Morak-Młodawska, B., Jeleń, M., & Okechukwu, P. N. (2024). Analysis of Lipophilicity and Pharmacokinetic Parameters of Dipyridothiazine Dimers with Anticancer Potency. Pharmaceutics, 16(9), 1235. https://doi.org/10.3390/pharmaceutics16091235






