Advancing Precision Medicine: A Review of Innovative In Silico Approaches for Drug Development, Clinical Pharmacology and Personalized Healthcare
Abstract
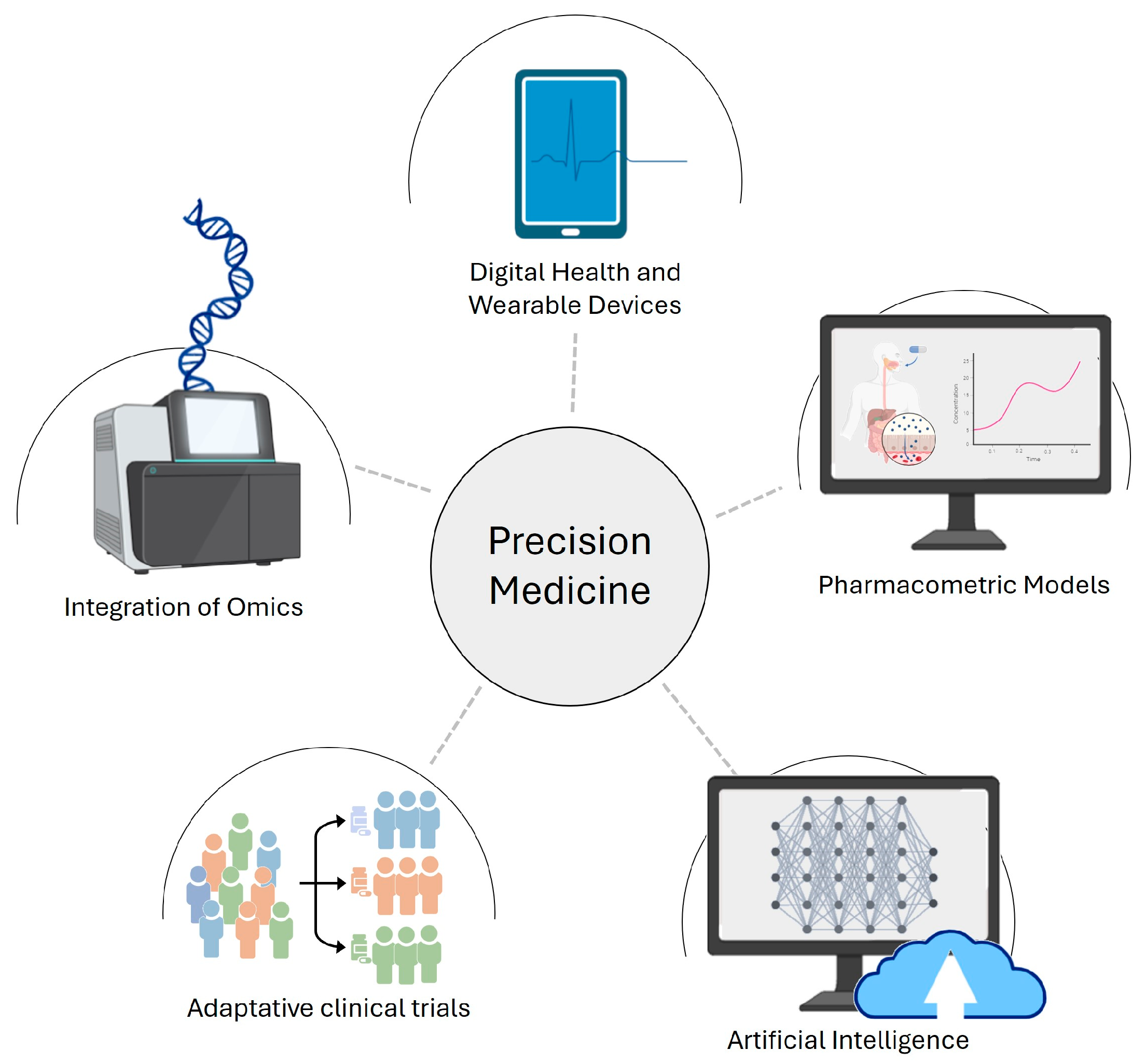
1. Introduction
2. OMICS in Advancing Clinical Decision-Making
2.1. Pharmacogenomics: Tailoring Treatment to Genetic Profiles
2.2. Challenges and Considerations in Integrating OMICS: Navigating the Road to Precision Medicine
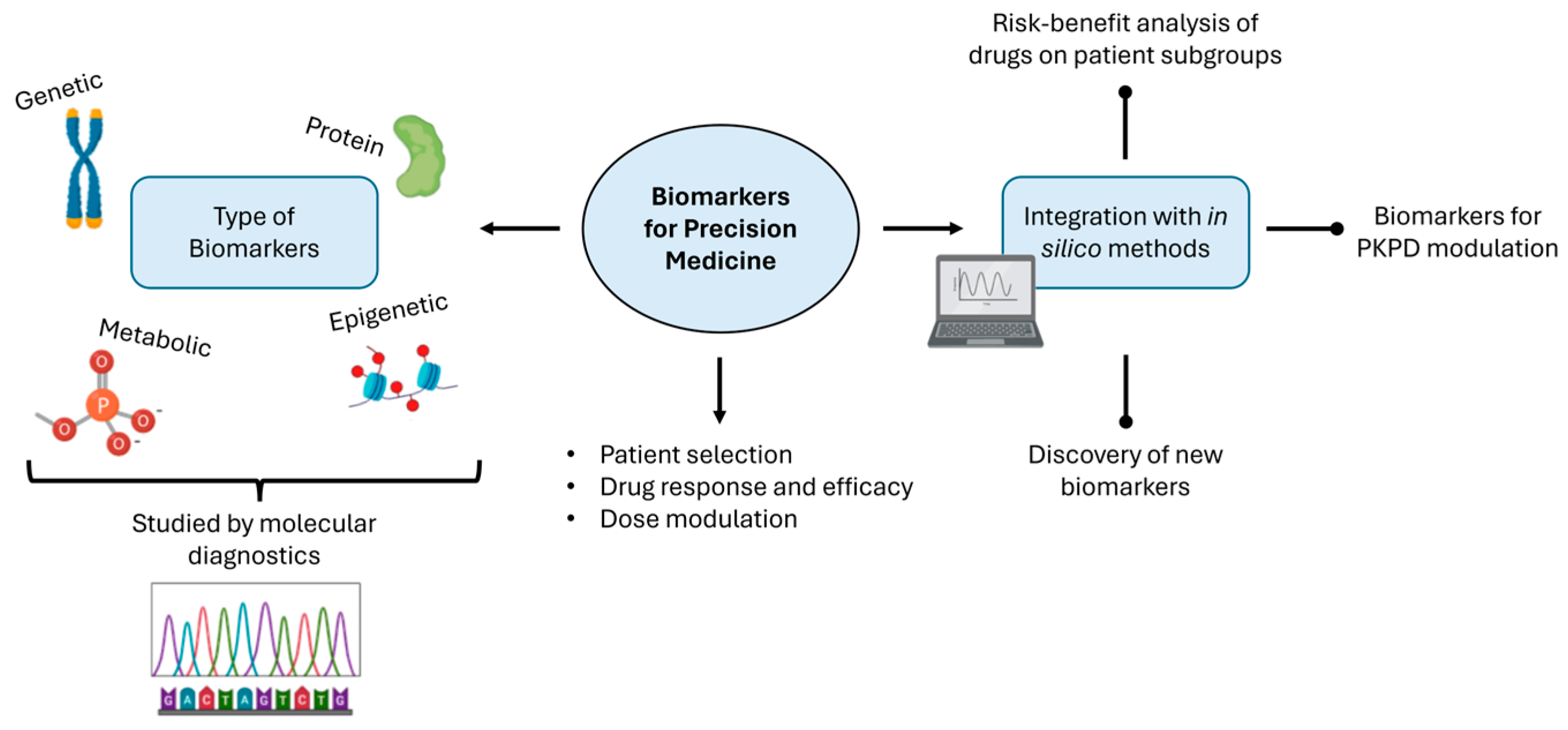
3. Biomarkers and Molecular Diagnostics
3.1. Harnessing Biomarkers for Precision Drug Development and Treatment Optimization
3.2. Challenges in Implementing Biomarkers and Molecular Diagnostics in Precision Medicine
4. Pharmacometrics Tools: Significance and Challenges in Precision Medicine
4.1. A Triad of Precision: PKPD, PBPK, and Population PK Models in Pharmacological Insights
4.2. Challenges of Quantitative Drug Modeling
5. Data Integration and Analytics: Data-Driven Approaches in Pharmacokinetic Modeling
5.1. Unraveling Complexity: Data-Driven Pharmacokinetic Modeling in Combination Therapy
5.2. Challenges and Regulatory Considerations in Data-Driven Pharmacokinetic Modeling
6. Artificial Intelligence: Integration of Machine Learning in Pharmacometrics
6.1. Examples of ML Approaches That Can Address Unique Challenges and Opportunities within Pharmacometrics
6.2. Challenges and Future Directions
- Expanded applications of PBPK models, informing clinical study design and predicting drug interactions.
- Pediatric dosing regimen prediction to ensure safer and more effective treatments for pediatric patients.
- Utilization of PBPK models for predicting drug exposure in patients with organ impairment.
- Estimation of maternal–fetal drug disposition during pregnancy.
- Prediction of pH-mediated drug interactions using PBPK models.
- Improved predictive performance of popPK models by focusing on data adequacy.
- Integration of generic PBPK models for extrapolations and continuous updates.
7. Digital Health and Wearable Technologies
8. Clinical Trials and Study Design
9. Future Perspectives: Integration of In Silico Tools in Hospital Settings
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Visvikis-Siest, S.; Theodoridou, D.; Kontoe, M.S.; Kumar, S.; Marschler, M. Milestones in Personalized Medicine: From the Ancient Time to Nowadays—The Provocation of COVID-19. Front. Genet. 2020, 11, 569175. [Google Scholar] [CrossRef]
- The Changing Landscape of Precision Medicine. Available online: https://www.astrazeneca.com/what-science-can-do/topics/technologies/precision-medicine-history.html (accessed on 10 October 2023).
- Akhoon, N. Precision Medicine: A New Paradigm in Therapeutics. Int. J. Prev. Med. 2021, 12, 12. [Google Scholar]
- Gameiro, G.R.; Sinkunas, V.; Liguori, G.R.; Auler-Júnior, J.O.C. Precision Medicine: Changing the Way We Think about Healthcare. Clinics 2018, 73, e723. [Google Scholar] [CrossRef]
- Denny, J.C.; Collins, F.S. Precision Medicine in 2030—Seven Ways to Transform Healthcare. Cell 2021, 184, 1415–1419. [Google Scholar] [CrossRef]
- Grissinger, M. The Five Rights: A Destination Without a Map. Pharm. Ther. 2010, 35, 542. [Google Scholar]
- National Research Council. Toward Precision Medicine; National Academies Press: Cambridge, MA, USA, 2011; ISBN 0309222222. [Google Scholar]
- Delpierre, C.; Lefèvre, T. Precision and Personalized Medicine: What Their Current Definition Says and Silences about the Model of Health They Promote. Implication for the Development of Personalized Health. Front. Sociol. 2023, 8, 1112159. [Google Scholar] [CrossRef]
- Baiardini, I.; Heffler, E. The Patient-Centered Decision System as per the 4Ps of Precision Medicine; Elsevier Inc.: Amsterdam, The Netherlands, 2018; ISBN 9780128134719. [Google Scholar]
- Kim, H.J.; Kim, H.J.; Park, Y.; Lee, W.S.; Lim, Y.; Kim, J.H. Clinical Genome Data Model (CGDM) Provides Interactive Clinical Decision Support for Precision Medicine. Sci. Rep. 2020, 10, 1414. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.P. The Wholeness in Suffix -Omics, -Omes, and the Word Om. J. Biomol. Tech. 2007, 18, 277. [Google Scholar]
- Hasanzad, M.; Sarhangi, N.; Chimeh, S.E.; Ayati, N.; Afzali, M.; Khatami, F.; Nikfar, S.; Meybodi, H. Precision Medicine Journey through Omics Approach. J. Diabetes Metab. Disord. 2022, 21, 881–888. [Google Scholar] [CrossRef] [PubMed]
- De Maria Marchiano, R.; Di Sante, G.; Piro, G.; Carbone, C.; Tortora, G.; Boldrini, L.; Pietragalla, A.; Daniele, G.; Tredicine, M.; Cesario, A.; et al. Translational Research in the Era of Precision Medicine: Where We Are and Where We Will Go. J. Pers. Med. 2021, 11, 216. [Google Scholar] [CrossRef] [PubMed]
- Tebani, A.; Afonso, C.; Marret, S.; Bekri, S. Omics-Based Strategies in Precision Medicine: Toward a Paradigm Shift in Inborn Errors of Metabolism Investigations. Int. J. Mol. Sci. 2016, 17, 1555. [Google Scholar] [CrossRef]
- Ahmed, Z. Precision Medicine with Multi-Omics Strategies, Deep Phenotyping, and Predictive Analysis. Prog. Mol. Biol. Transl. Sci. 2022, 190, 101–125. [Google Scholar] [CrossRef]
- Kwon, Y.W.; Jo, H.S.; Bae, S.; Seo, Y.; Song, P.; Song, M.; Yoon, J.H. Application of Proteomics in Cancer: Recent Trends and Approaches for Biomarkers Discovery. Front. Med. 2021, 8, 747333. [Google Scholar] [CrossRef]
- Giannitsis, E.; Katus, H.A. Biomarkers for Clinical Decision-Making in the Management of Pulmonary Embolism. Clin. Chem. 2017, 63, 91–100. [Google Scholar] [CrossRef]
- Wafi, A.; Mirnezami, R. Translational –Omics: Future Potential and Current Challenges in Precision Medicine. Methods 2018, 151, 3–11. [Google Scholar] [CrossRef]
- Hu, C.; Jia, W. Multi-Omics Profiling: The Way toward Precision Medicine in Metabolic. J. Mol. Cell Biol. 2021, 13, 576. [Google Scholar] [CrossRef]
- Pirmohamed, M. Pharmacogenomics: Current Status and Future Perspectives. Nat. Rev. Genet. 2023, 24, 350–362. [Google Scholar] [CrossRef] [PubMed]
- Badary, O.A. Pharmacogenomics and COVID-19: Clinical Implications of Human Genome Interactions with Repurposed Drugs. Pharmacogenom. J. 2021, 21, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Miteva-Marcheva, N.N.; Ivanov, H.Y.; Dimitrov, D.K.; Stoyanova, V.K. Application of Pharmacogenetics in Oncology. Biomark. Res. 2020, 8, 32. [Google Scholar] [CrossRef] [PubMed]
- Licinio, J.; Wong, M.-L. Pharmacogenomics of Antidepressant Treatment Effects. Dialogues Clin. Neurosci. 2011, 13, 63–71. [Google Scholar] [CrossRef] [PubMed]
- McDonough, C.W. Pharmacogenomics in Cardiovascular Diseases. Curr. Protoc. 2021, 1, e189. [Google Scholar] [CrossRef]
- Mallal, S.; Phillips, E.; Carosi, G.; Molina, J.-M.; Workman, C.; Tomažič, J.; Jägel-Guedes, E.; Rugina, S.; Kozyrev, O.; Cid, J.F.; et al. HLA-B*5701 Screening for Hypersensitivity to Abacavir. N. Engl. J. Med. 2008, 358, 568–579. [Google Scholar] [CrossRef] [PubMed]
- Lecomte, T.; Ferraz, J.-M.; Zinzindohoué, F.; Loriot, M.-A.; Tregouet, D.-A.; Landi, B.; Berger, A.; Cugnenc, P.-H.; Jian, R.; Beaune, P.; et al. Thymidylate Synthase Gene Polymorphism Predicts Toxicity in Colorectal Cancer Patients Receiving 5-Fluorouracil-Based Chemotherapy. Clin. Cancer Res. 2004, 10, 5880–5888. [Google Scholar] [CrossRef]
- Flockhart, D.A.; O’Kane, D.; Williams, M.S.; Watson, M.S.; Flockhart, D.A.; Gage, B.; Gandolfi, R.; King, R.; Lyon, E.; Nussbaum, R.; et al. Pharmacogenetic Testing of CYP2C9 and VKORC1 Alleles for Warfarin. Genet. Med. 2008, 10, 139–150. [Google Scholar] [CrossRef]
- Ferrell, P.B.; McLeod, H.L. Carbamazepine, HLA-B*1502 and Risk of Stevens–Johnson Syndrome and Toxic Epidermal Necrolysis: US FDA Recommendations. Pharmacogenomics 2008, 9, 1543–1546. [Google Scholar] [CrossRef] [PubMed]
- de Souza, J.A.; Olopade, O.I. CYP2D6 Genotyping and Tamoxifen: An Unfinished Story in the Quest for Personalized Medicine. Semin. Oncol. 2011, 38, 263–273. [Google Scholar] [CrossRef] [PubMed]
- D’Adamo, G.L.; Widdop, J.T.; Giles, E.M. The Future Is Now? Clinical and Translational Aspects of “Omics” Technologies. Immunol. Cell Biol. 2021, 99, 168–176. [Google Scholar] [CrossRef]
- Vogeser, M.; Bendt, A.K. From Research Cohorts to the Patient—A Role for “Omics” in Diagnostics and Laboratory Medicine? Clin. Chem. Lab. Med. 2023, 61, 974–980. [Google Scholar] [CrossRef]
- Castaneda, C.; Nalley, K.; Mannion, C.; Bhattacharyya, P.; Blake, P.; Pecora, A.; Goy, A.; Suh, K.S. Clinical Decision Support Systems for Improving Diagnostic Accuracy and Achieving Precision Medicine. J. Clin. Bioinform. 2015, 5, 4. [Google Scholar] [CrossRef]
- Sperber, N.R.; Dong, O.M.; Roberts, M.C.; Dexter, P.; Elsey, A.R.; Ginsburg, G.S.; Horowitz, C.R.; Johnson, J.A.; Levy, K.D.; Ong, H.; et al. Strategies to Integrate Genomic Medicine into Clinical Care: Evidence from the IGNITE Network. J. Pers. Med. 2021, 11, 647. [Google Scholar] [CrossRef]
- FDA Label Search. Available online: https://labels.fda.gov/ (accessed on 16 October 2023).
- The Personalized Medicine Coalition. Available online: https://www.personalizedmedicinecoalition.org/ (accessed on 16 October 2023).
- Precision Medicine|FDA. Available online: https://www.fda.gov/medical-devices/in-vitro-diagnostics/precision-medicine (accessed on 10 October 2023).
- Nimmesgern, E.; Benediktsson, I.; Norstedt, I. Personalized Medicine in Europe. Clin. Transl. Sci. 2017, 10, 61–63. [Google Scholar] [CrossRef]
- Aronson, J.K.; Ferner, R.E. Biomarkers—A General Review. Curr. Protoc. Pharmacol. 2017, 2017, 9.23.1–9.23.17. [Google Scholar] [CrossRef] [PubMed]
- Ho, D.; Quake, S.R.; McCabe, E.R.B.; Chng, W.J.; Chow, E.K.; Ding, X.; Gelb, B.D.; Ginsburg, G.S.; Hassenstab, J.; Ho, C.M.; et al. Enabling Technologies for Personalized and Precision Medicine. Trends Biotechnol. 2020, 38, 497–518. [Google Scholar] [CrossRef]
- Mokondjimobe, E.; Longo-Mbenza, B.; Akiana, J.; Ndalla, U.O.; Dossou-Yovo, R.; Mboussa, J.; Parra, H.J. Biomarkers of Oxidative Stress and Personalized Treatment of Pulmonary Tuberculosis: Emerging Role of Gamma-Glutamyltransferase. Adv. Pharmacol. Sci. 2012, 2012, 465634. [Google Scholar] [CrossRef]
- Kirkwood, S.C.; Hockett, R.D. Pharmacogenomic Biomarkers. Dis. Markers 2002, 18, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Mendrick, D.L. Genomic and Genetic Biomarkers of Toxicity. Toxicology 2008, 245, 175–181. [Google Scholar] [CrossRef]
- Karaulov, A.V.; Garib, V.; Garib, F.; Valenta, R. Protein Biomarkers in Asthma. Int. Arch. Allergy Immunol. 2018, 175, 189–208. [Google Scholar] [CrossRef]
- Sigdel, T.K.; Gao, X.; Sarwal, M.M. Protein and Peptide Biomarkers in Organ Transplantation. Biomark. Med. 2012, 6, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Garulacan, L.A.; Storm, S.M.; Hefta, S.A.; Opiteck, G.J.; Lin, J.H.; Moulin, F.; Dambach, D.M. Identification of in Vitro Protein Biomarkers of Idiosyncratic Liver Toxicity. Toxicol. Vitr. 2004, 18, 533–541. [Google Scholar] [CrossRef]
- Grondman, I.; Pirvu, A.; Riza, A.; Ioana, M.; Netea, M.G. Biomarkers of Inflammation and the Etiology of Sepsis. Biochem. Soc. Trans. 2020, 48, 1–14. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond Biomarkers and towards Mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Costa-pinheiro, P.; Montezuma, D. Diagnostic and Prognostic Epigenetic Biomarkers in Cancer. Epigenomics 2015, 7, 1003–1015. [Google Scholar] [CrossRef] [PubMed]
- Hoque, M.O.; Begum, S.; Topaloglu, O.; Jeronimo, C.; Mambo, E.; Westra, W.H.; Califano, J.A.; Sidransky, D. Quantitative Detection of Promoter Hypermethylation of Multiple Genes in the Tumor, Urine, and Serum DNA of Patients with Renal Cancer. Cancer Res. 2004, 64, 5511–5517. [Google Scholar] [CrossRef]
- Javitt, G.H.; Vollebregt, E.R. Regulation of Molecular Diagnostics. Annu. Rev. Genom. Hum. Genet. 2022, 23, 653–673. [Google Scholar] [CrossRef]
- Sun, L.; Pfeifer, J.D. Pitfalls in Molecular Diagnostics. Semin. Diagn. Pathol. 2019, 36, 342–354. [Google Scholar] [CrossRef] [PubMed]
- Chien, J.Y.; Friedrich, S.; Heathman, M.A.; de Alwis, D.P.; Sinha, V. Pharmacokinetics/Pharmacodynamics and the Stages of Drug Development: Role of Modeling and Simulation. AAPS J. 2005, 7, E544–E559. [Google Scholar] [CrossRef]
- McComb, M.; Ramanathan, M. Generalized Pharmacometric Modeling, a Novel Paradigm for Integrating Machine Learning Algorithms: A Case Study of Metabolomic Biomarkers. Clin. Pharmacol. Ther. 2020, 107, 1343–1351. [Google Scholar] [CrossRef]
- Goetz, L.H.; Schork, N.J. Personalized Medicine: Motivation, Challenges, and Progress. Fertil. Steril. 2018, 109, 952–963. [Google Scholar] [CrossRef]
- Rigatti, S.J. Random Forest. J. Insur. Med. 2017, 47, 31–39. [Google Scholar] [CrossRef]
- Al-kaabawi, Z.; Wei, Y.; Moyeed, R. Bayesian Hierarchical Models for Linear Networks. J. Appl. Stat. 2022, 49, 1421–1448. [Google Scholar] [CrossRef]
- Leil, T.A.; Kasichayanula, S.; Boulton, D.W.; LaCreta, F. Evaluation of 4β-Hydroxycholesterol as a Clinical Biomarker of CYP3A4 Drug Interactions Using a Bayesian Mechanism-Based Pharmacometric Model. CPT Pharmacomet. Syst. Pharmacol. 2014, 3, 1–10. [Google Scholar] [CrossRef]
- Diczfalusy, U.; Nylén, H.; Elander, P.; Bertilsson, L. 4β-Hydroxycholesterol, an Endogenous Marker of CYP3A4/5 Activity in Humans. Br. J. Clin. Pharmacol. 2011, 71, 183–189. [Google Scholar] [CrossRef]
- Kathman, S.J.; Williams, D.H.; Hodge, J.P.; Dar, M. A Bayesian Population PK-PD Model of Ispinesib-Induced Myelosuppression. Clin. Pharmacol. Ther. 2007, 81, 88–94. [Google Scholar] [CrossRef]
- Bauer, R.J.; Guzy, S.; Ng, C. A Survey of Population Analysis Methods and Software for Complex Pharmacokinetic and Pharmacodynamic Models with Examples. AAPS J. 2007, 9, E60–E83. [Google Scholar] [CrossRef]
- Terranova, N.; Venkatakrishnan, K.; Benincosa, L.J. Application of Machine Learning in Translational Medicine: Current Status and Future Opportunities. AAPS J. 2021, 23, 1–10. [Google Scholar] [CrossRef]
- Wang, R.; Shao, X.; Zheng, J.; Saci, A.; Qian, X.; Pak, I.; Roy, A.; Bello, A.; Rizzo, J.I.; Hosein, F.; et al. A Machine-Learning Approach to Identify a Prognostic Cytokine Signature That Is Associated With Nivolumab Clearance in Patients With Advanced Melanoma. Clin. Pharmacol. Ther. 2019, 107, 978–987. [Google Scholar] [CrossRef]
- Feng, Y.; Wang, X.; Bajaj, G.; Agrawal, S.; Bello, A.; Lestini, B.; Finckenstein, F.G.; Park, J.; Roy, A. Nivolumab Exposure—Response Analyses of Ef Fi Cacy and Safety in Previously Treated Squamous or Nonsquamous Non—Small Cell Lung Cancer. Clin. Cancer Res. 2017, 23, 5394–5406. [Google Scholar] [CrossRef] [PubMed]
- Data, T.A.; Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More Than. Radiology 2016, 278, 563–577. [Google Scholar]
- Terranova, N.; Girard, P.; Ioannou, K.; Klinkhardt, U.; Munafo, A. Assessing Similarity among Individual Tumor Size Lesion Dynamics: The CICIL Methodology. CPT Pharmacomet. Syst. Pharmacol. 2018, 7, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Terranova, N.; Girard, P.; Klinkhardt, U.; Munafo, A. Resistance Development: A Major Piece in the Jigsaw Puzzle of Tumor Size Modeling. CPT Pharmacomet. Syst. Pharmacol. 2015, 4, 320–323. [Google Scholar] [CrossRef] [PubMed]
- Sands, B.E.; Chen, J.; Feagan, B.G.; Penney, M.; Rees, W.A.; Ph, D.; Danese, S.; Higgins, P.D.R. Efficacy and Safety of MEDI2070, an Antibody Against Interleukin 23, Patients with Moderate to Severe Crohn’s Disease: A Phase 2a Study; Elsevier Inc.: Amsterdam, The Netherlands, 2017; ISBN 6465378647. [Google Scholar]
- Zhang, N.; Liang, M.; Jing, C.; Philip, L.; Bo, Z.B.; Vainshtein, I.; Roskos, L.K.; Faggioni, R.; Savic, R.M. Combining Pharmacometric Models with Predictive and Prognostic Biomarkers for Precision Therapy in Crohn’s Disease: A Case Study of Brazikumab. CPT Pharmacomet. Syst. Pharmacol. 2023, 12, 1945–1959. [Google Scholar] [CrossRef] [PubMed]
- Best, W.R.; Becktel, J.M.; Signleton, J.W.; Kern, F. Development of a Crohn’s Disease Activity Index. National Cooperative Crohn’s Disease Study. Gastroenterology 1976, 70, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, T.; Watanabe, K.; Kojima, K.; Koshiba, R.; Fujimoto, K.; Sato, T.; Kawai, M.; Kamikozuru, K.; Yokoyama, Y.; Hida, N.; et al. Efficacies and Related Issues of Ustekinumab in Japanese Patients with Crohn’s Disease: A Preliminary Study. Digestion 2019, 101, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Hendrikse, N.M.; Llinares Garcia, J.; Vetter, T.; Humphreys, A.J.; Ehmann, F. Biomarkers in Medicines Development—From Discovery to Regulatory Qualification and Beyond. Front. Med. 2022, 9, 878942. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Ma, Y.; Uzzi, B.; Loscalzo, J. Importance of Scientific Collaboration in Contemporary Drug Discovery and Development: A Detailed Network Analysis. BMC Biol. 2020, 18, 138. [Google Scholar] [CrossRef]
- Initiative, I.M. IMI Mission and Objectives. Available online: https://www.imi.europa.eu/about-imi/mission-objectives (accessed on 22 September 2023).
- Zheng, Q.S.; Li, L.J. Pharmacometrics: A Quantitative Tool of Pharmacological Research. Acta Pharmacol. Sin. 2012, 33, 1337–1338. [Google Scholar] [CrossRef]
- Bandeira, L.C.; Pinto, L.; Carneiro, C.M. Pharmacometrics: The Already-Present Future of Precision Pharmacology. Ther. Innov. Regul. Sci. 2022, 57, 57–69. [Google Scholar] [CrossRef]
- Himstedt, A.; Bäckman, P.; Borghardt, J.M. Physiologically-Based Pharmacokinetic Modeling after Drug Inhalation. In Inhaled Medicines: Optimizing Development through Integration of In Silico, In Vitro and In Vivo Approaches; Academic Press: Cambridge, MA, USA, 2021; pp. 319–358. [Google Scholar] [CrossRef]
- Usman, M.; Rasheed, H.; Pharmacokinetics, P.B.; Creation, D. Pharmacometrics and Its Application in Clinical Practice. Encycl. Pharm. Pract. Clin. Pharm. 2019, 3B, 227–238. [Google Scholar]
- Division of Pharmacometrics|FDA. Available online: https://www.fda.gov/about-fda/center-drug-evaluation-and-research-cder/division-pharmacometrics (accessed on 10 October 2023).
- Sheiner, L.B.; Rosenberg, B.; Marathe, V.V. Estimation of Population Characteristics of Pharmacokinetic Parameters from Routine Clinical Data. J. Pharmacokinet. Biopharm. 1977, 5, 445–479. [Google Scholar] [CrossRef]
- Dollery, C.T. Clinical Pharmacology—The First 75 Years and a View of the Future. Br. J. Clin. Pharmacol. 2006, 61, 650–665. [Google Scholar] [CrossRef]
- Usman, M.; Khadka, S.; Saleem, M.; Rasheed, H.; Kunwar, B.; Ali, M. Pharmacometrics: A New Era of Pharmacotherapy and Drug Development in Low- and Middle-Income Countries. Adv. Pharmacol. Pharm. Sci. 2023, 2023, 3081422. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B. Sheiner Lecturer Award. Available online: https://go-isop.org/awards/lewis-b-sheiner-award/ (accessed on 10 October 2023).
- Dagenais, S.; Russo, L.; Madsen, A.; Webster, J.; Becnel, L. Use of Real-World Evidence to Drive Drug Development Strategy and Inform Clinical Trial Design. Clin. Pharmacol. Ther. 2022, 111, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Ette, E.I.; Williams, P.J. Population Pharmacokinetics I: Background, Concepts, and Models. Ann. Pharmacother. 2004, 38, 1702–1706. [Google Scholar] [CrossRef] [PubMed]
- Abouir, K.; Samer, C.F.; Gloor, Y.; Desmeules, J.A.; Daali, Y. Reviewing Data Integrated for PBPK Model Development to Predict Metabolic Drug-Drug Interactions: Shifting Perspectives and Emerging Trends. Front. Pharmacol. 2021, 12, 708299. [Google Scholar] [CrossRef]
- Siebinga, H.; de Wit-Van der Veen, B.J.; Stokkel, M.D.M.; Huitema, A.D.R.; Hendrikx, J.J.M.A. Current Use and Future Potential of (Physiologically Based) Pharmacokinetic Modelling of Radiopharmaceuticals: A Review. Theranostics 2022, 12, 7804–7820. [Google Scholar] [CrossRef]
- Pfister, M.; D’Argenio, D.Z. The Emerging Scientific Discipline of Pharmacometrics. J. Clin. Pharmacol. 2010, 50, 6S. [Google Scholar] [CrossRef] [PubMed]
- Stone, J.A.; Banfield, C.; Pfister, M.; Tannenbaum, S.; Allerheiligen, S.; Wetherington, J.D.; Krishna, R.; Grasela, D.M. Model-Based Drug Development Survey Finds Pharmacometrics Impacting Decision Making in the Pharmaceutical Industry. J. Clin. Pharmacol. 2010, 50, 20S–30S. [Google Scholar] [CrossRef]
- Zou, H.; Banerjee, P.; Leung, S.S.Y.; Yan, X. Application of Pharmacokinetic-Pharmacodynamic Modeling in Drug Delivery: Development and Challenges. Front. Pharmacol. 2020, 11, 997. [Google Scholar] [CrossRef]
- Meibohm, B.; Derendorf, H. Basic Concepts of Pharmacokinetic/Pharmacodynamic (PK/PD) Modelling. Int. J. Clin. Pharmacol. Ther. 1997, 35, 401–413. [Google Scholar]
- Upton, R.N.; Mould, D.R. Basic Concepts in Population Modeling, Simulation, and Model-Based Drug Development: Part 3-Introduction to Pharmacodynamic Modeling Methods. CPT Pharmacomet. Syst. Pharmacol. 2014, 3, 1–16. [Google Scholar] [CrossRef]
- Salahudeen, M.S.; Nishtala, P.S. An Overview of Pharmacodynamic Modelling, Ligand-Binding Approach and Its Application in Clinical Practice. Saudi Pharm. J. 2017, 25, 165–175. [Google Scholar] [CrossRef]
- Felmlee, M.A.; Morris, M.E.; Mager, D.E. Mechanism-Based Pharmacodynamic Modeling. Comput. Toxicol. 2012, I, 583–600. [Google Scholar] [CrossRef]
- Lin, L.H.; Ghasemi, M.; Burke, S.M.; Mavis, C.K.; Nichols, J.R.; Torka, P.; Mager, D.E.; Hernandez-Ilizaliturri, F.J.; Goey, A.K.L. Population Pharmacokinetics and Pharmacodynamics of Carfilzomib in Combination with Rituximab, Ifosfamide, Carboplatin, and Etoposide in Adult Patients with Relapsed/Refractory Diffuse Large B Cell Lymphoma. Target Oncol. 2023, 18, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Palmer, M.E.; Andrews, L.J.; Abbey, T.C.; Dahlquist, A.E.; Wenzler, E. The Importance of Pharmacokinetics and Pharmacodynamics in Antimicrobial Drug Development and Their Influence on the Success of Agents Developed to Combat Resistant Gram Negative Pathogens: A Review. Front. Pharmacol. 2022, 13, 888079. [Google Scholar] [CrossRef] [PubMed]
- Derendorf, H.; Möllmann, H.; Hochhaus, G.; Meibohm, B.; Barth, J. Clinical PK/PD Modelling as a Tool in Drug Development of Corticosteroids. Int. J. Clin. Pharmacol. Ther. 1997, 35, 481–488. [Google Scholar] [PubMed]
- Tuntland, T.; Ethell, B.; Kosaka, T.; Blasco, F.; Zang, R.; Jain, M.; Gould, T.; Hoffmaster, K. Implementation of Pharmacokinetic and Pharmacodynamic Strategies in Early Research Phases of Drug Discovery and Development at Novartis Institute of Biomedical Research. Front. Pharmacol. 2014, 5, 174. [Google Scholar] [CrossRef] [PubMed]
- Qusai, U.; Hameed, A.; Rasheed, K.H. Compartmental and Non-Compartmental Pharmacokinetic Analysis of Extended Release Diclofenac Sodium Tablet. Coll. Eng. J. 2016, 19, 161–165. [Google Scholar]
- Gabrielsson, J.; Weiner, D. Non-Compartmental Analysis. Comput. Toxicol. 2012, 929, 377–389. [Google Scholar] [CrossRef]
- Foster, D.M. Noncompartmental versus Compartmental Approaches to Pharmacokinetic Analysis, 2nd ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2006; ISBN 9780123694171. [Google Scholar]
- Noncompartmental vs. Compartmental PK Analysis. Available online: https://www.allucent.com/resources/blog/what-noncompartmental-pharmacokinetic-analysis (accessed on 10 October 2023).
- Osipova, N.; Budko, A.; Maksimenko, O.; Shipulo, E.; Vanchugova, L.; Chen, W.; Gelperina, S.; Wacker, M.G. Comparison of Compartmental and Non-Compartmental Analysis to Detect Biopharmaceutical Similarity of Intravenous Nanomaterial-Based Rifabutin Formulations. Pharmaceutics 2023, 15, 1258. [Google Scholar] [CrossRef]
- Hosseini, I.; Gajjala, A.; Bumbaca Yadav, D.; Sukumaran, S.; Ramanujan, S.; Paxson, R.; Gadkar, K. GPKPDSim: A SimBiology®-Based GUI Application for PKPD Modeling in Drug Development. J. Pharmacokinet. Pharmacodyn. 2018, 45, 259–275. [Google Scholar] [CrossRef]
- World Health Organization. Characterization and Application of Physiologically Based Pharmacokinetic Models. Int. Programme Chem. Saf. 2010, 9, 16–37. [Google Scholar]
- Tan, Y.M.; Worley, R.R.; Leonard, J.A.; Fisher, J.W. Challenges Associated with Applying Physiologically Based Pharmacokinetic Modeling for Public Health Decision-Making. Toxicol. Sci. 2018, 162, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Teorell, T. Kinetics of Distribution of Substances Administered to the Body, I: The Extravascular Modes of Administration. Arch. Int. Pharmacodyn. Ther. 1937, 57, 205–225. [Google Scholar]
- Zhuang, X.; Lu, C. PBPK Modeling and Simulation in Drug Research and Development. Acta Pharm. Sin. B 2016, 6, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Jones, H.M.; Rowland-Yeo, K. Basic Concepts in Physiologically Based Pharmacokinetic Modeling in Drug Discovery and Development. CPT Pharmacomet. Syst. Pharmacol. 2013, 2, 1–12. [Google Scholar] [CrossRef]
- Umehara, K.; Huth, F.; Jin, Y.; Schiller, H.; Aslanis, V.; Heimbach, T.; He, H. Drug-Drug Interaction (DDI) Assessments of Ruxolitinib, a Dual Substrate of CYP3A4 and CYP2C9, Using a Verified Physiologically Based Pharmacokinetic (PBPK) Model to Support Regulatory Submissions. Drug Metab. Pers. Ther. 2019, 34, 20180042. [Google Scholar] [CrossRef] [PubMed]
- Marques, L.; Vale, N. Prediction of CYP-Mediated Drug Interaction Using Physiologically Based Pharmacokinetic Modeling: A Case Study of Salbutamol and Fluvoxamine. Pharmaceutics 2023, 15, 1586. [Google Scholar] [CrossRef]
- Zamir, A.; Rasool, M.F.; Imran, I.; Saeed, H.; Khalid, S.; Majeed, A.; Rehman, A.U.; Ahmad, T.; Alasmari, F.; Alqahtani, F. Physiologically Based Pharmacokinetic Model To Predict Metoprolol Disposition in Healthy and Disease Populations. ACS Omega 2023, 8, 29302–29313. [Google Scholar] [CrossRef]
- Amaeze, O.U.; Isoherranen, N. Application of a Physiologically Based Pharmacokinetic Model to Predict Isoniazid Disposition during Pregnancy. Clin. Transl. Sci. 2023, 16, 2163–2176. [Google Scholar] [CrossRef]
- Mould, D.R.; Upton, R.N. Basic Concepts in Population Modeling, Simulation, and Model-Based Drug Development—Part 2: Introduction to Pharmacokinetic Modeling Methods. CPT Pharmacomet. Syst. Pharmacol. 2013, 2, 1–14. [Google Scholar] [CrossRef]
- Li, A.; Mak, W.Y.; Ruan, T.; Dong, F.; Zheng, N.; Gu, M.; Guo, W.; Zhang, J.; Cheng, H.; Ruan, C.; et al. Population Pharmacokinetics of Amisulpride in Chinese Patients with Schizophrenia with External Validation: The Impact of Renal Function. Front. Pharmacol. 2023, 14, 1215065. [Google Scholar] [CrossRef]
- He, S.; Zhao, J.; Bian, J.; Zhao, Y.; Li, Y.; Guo, N.; Hu, L.; Liu, B.; Shao, Q.; He, H.; et al. Population Pharmacokinetics and Pharmacogenetics Analyses of Dasatinib in Chinese Patients with Chronic Myeloid Leukemia. Pharm. Res. 2023, 40, 2413–2422. [Google Scholar] [CrossRef]
- Verma, M.; Gall, L.; Biasetti, J.; Di Veroli, G.Y.; Pichardo-Almarza, C.; Gibbs, M.A.; Kimko, H. Quantitative Systems Modeling Approaches towards Model-Informed Drug Development: Perspective through Case Studies. Front. Syst. Biol. 2023, 2, 1063308. [Google Scholar] [CrossRef]
- Chen, Y.; Li, J.; Li, D.; Hu, C. Pharmacokinetic Modeling and Predictive Performance: Practical Considerations for Therapeutic Monoclonal Antibodies. Eur. J. Drug Metab. Pharmacokinet. 2021, 46, 595–600. [Google Scholar] [CrossRef]
- Krivelevich, I.; Lin, S. Visualization of Sparse PK Concentration Sampling Data, Step by Step (Improvement by Improvement) STEP 1: STARTING BOXPLOT First, Let’s Draw a Simple Boxplot as a Starting Point. Appl. Below Simple SAS Code PROC 2021, 1, 1–14. [Google Scholar]
- Choi, L.; Crainiceanu, C.M.; Caffo, B.S. Practical Recommendations for Population PK Studies with Sampling Time Errors. Eur. J. Clin. Pharmacol. 2013, 69, 2055. [Google Scholar] [CrossRef][Green Version]
- Alizadeh, E.A.; Rast, G.; Cantow, C.; Schiwon, J.; Krause, F.; De Meyer, G.R.Y.; Guns, P.J.; Guth, B.D.; Markert, M. Optimization of Bioanalysis of Dried Blood Samples. J. Pharmacol. Toxicol. Methods 2023, 123, 107296. [Google Scholar] [CrossRef] [PubMed]
- Sheiner, L.B.; Beal, S.L. Evaluation of Methods for Estimating Population Pharmacokinetic Parameters. III. Monoexponential Model: Routine Clinical Pharmacokinetic Data. J. Pharmacokinet. Biopharm. 1983, 11, 303–319. [Google Scholar] [CrossRef] [PubMed]
- Sheiner, L.B.; Beal, S.L. Evaluation of Methods for Estimating Population Pharmacokinetic Parameters II. Biexponential Model and Experimental Pharmacokinetic Data. J. Pharmacokinet. Biopharm. 1981, 9, 635–651. [Google Scholar] [CrossRef] [PubMed]
- Brocks, D.; Hamdy, D. Bayesian Estimation of Pharmacokinetic Parameters: An Important Component to Include in the Teaching of Clinical Pharmacokinetics and Therapeutic Drug Monitoring. Res. Pharm. Sci. 2020, 15, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Gennemark, P.; Danis, A.; Nyberg, J.; Hooker, A.C.; Tucker, W. Optimal Design in Population Kinetic Experiments by Set-Valued Methods. AAPS J. 2011, 13, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Sherwin, C.M.T.; Kiang, T.K.L.; Spigarelli, M.G.; Ensom, M.H.H. Fundamentals of Population Pharmacokinetic Modelling. Clin. Pharmacokinet. 2012, 51, 573–590. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Kang, J.J. Challenges and Strategies in PKPD Programming PKNCA Data Other Deliverables CHALLENGES IN PKPD PROGRAMMING Challenges Due to Source Data Multiple Data Sources; Merck & Co., Inc.: Rahway, NJ, USA, 2018; pp. 1–6. [Google Scholar]
- Schmidt, H.; Radivojevic, A. Enhancing Population Pharmacokinetic Modeling Efficiency and Quality Using an Integrated Workflow. J. Pharmacokinet. Pharmacodyn. 2014, 41, 319–334. [Google Scholar] [CrossRef]
- Lin, W.; Chen, Y.; Unadkat, J.D.; Zhang, X.; Wu, D.; Heimbach, T. Applications, Challenges, and Outlook for PBPK Modeling and Simulation: A Regulatory, Industrial and Academic Perspective. Pharm. Res. 2022, 39, 1701–1731. [Google Scholar] [CrossRef]
- Peters, S.A.; Dolgos, H. Requirements to Establishing Confidence in Physiologically Based Pharmacokinetic (PBPK) Models and Overcoming Some of the Challenges to Meeting Them. Clin. Pharmacokinet. 2019, 58, 1355–1371. [Google Scholar] [CrossRef]
- Binuya, M.A.E.; Engelhardt, E.G.; Schats, W.; Schmidt, M.K.; Steyerberg, E.W. Methodological Guidance for the Evaluation and Updating of Clinical Prediction Models: A Systematic Review. BMC Med. Res. Methodol. 2022, 22, 316. [Google Scholar] [CrossRef] [PubMed]
- Cook, S.F.; Bies, R.R. Disease Progression Modeling: Key Concepts and Recent Developments. Curr. Pharmacol. Rep. 2016, 2, 221–230. [Google Scholar] [CrossRef]
- Tyson, R.J.; Park, C.C.; Powell, J.R.; Patterson, J.H.; Weiner, D.; Watkins, P.B.; Gonzalez, D. Precision Dosing Priority Criteria: Drug, Disease, and Patient Population Variables. Front. Pharmacol. 2020, 11, 420. [Google Scholar] [CrossRef]
- Arida-Moody, L.; Moody, J.B.; Renaud, J.M.; Poitrasson-Rivière, A.; Hagio, T.; Smith, A.M.; Ficaro, E.P.; Murthy, V.L. Effects of Two Patient-Specific Dosing Protocols on Measurement of Myocardial Blood Flow with 3D 82Rb Cardiac PET. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 3835–3846. [Google Scholar] [CrossRef]
- Reyner, E.; Lum, B.; Jing, J.; Kagedal, M.; Ware, J.A.; Dickmann, L.J. Intrinsic and Extrinsic Pharmacokinetic Variability of Small Molecule Targeted Cancer Therapy. Clin. Transl. Sci. 2020, 13, 410–418. [Google Scholar] [CrossRef]
- Fabbiani, M.; Di Giambenedetto, S.; Bracciale, L.; Bacarelli, A.; Ragazzoni, E.; Cauda, R.; Navarra, P.; De Luca, A. Pharmacokinetic Variability of Antiretroviral Drugs and Correlation with Virological Outcome: 2 Years of Experience in Routine Clinical Practice. J. Antimicrob. Chemother. 2009, 64, 109–117. [Google Scholar] [CrossRef]
- Rao, P.S.; Modi, N.; Nguyen, N.T.T.; Vu, D.H.; Xie, Y.L.; Gandhi, M.; Gerona, R.; Metcalfe, J.; Heysell, S.K.; Alffenaar, J.W.C. Alternative Methods for Therapeutic Drug Monitoring and Dose Adjustment of Tuberculosis Treatment in Clinical Settings: A Systematic Review. Clin. Pharmacokinet. 2023, 62, 375–398. [Google Scholar] [CrossRef]
- Kriegova, E.; Kudelka, M.; Radvansky, M.; Gallo, J. A Theoretical Model of Health Management Using Data-Driven Decision-Making: The Future of Precision Medicine and Health. J. Transl. Med. 2021, 19, 68. [Google Scholar] [CrossRef] [PubMed]
- Martínez-García, M.; Hernández-Lemus, E. Data Integration Challenges for Machine Learning in Precision Medicine. Front. Med. 2022, 8, 784455. [Google Scholar] [CrossRef] [PubMed]
- Naithani, N.; Sinha, S.; Misra, P.; Vasudevan, B.; Sahu, R. Precision Medicine: Concept and Tools. Med. J. Armed Forces India 2021, 77, 249–257. [Google Scholar] [CrossRef]
- Giordano, C.; Brennan, M.; Mohamed, B.; Rashidi, P.; Modave, F.; Tighe, P. Accessing Artificial Intelligence for Clinical Decision-Making. Front. Digit. Health 2021, 3, 645232. [Google Scholar] [CrossRef] [PubMed]
- Ghaffar Nia, N.; Kaplanoglu, E.; Nasab, A. Evaluation of Artificial Intelligence Techniques in Disease Diagnosis and Prediction. Discov. Artif. Intell. 2023, 3, 5. [Google Scholar] [CrossRef]
- Xie, Y.; Meng, W.Y.; Li, R.Z.; Wang, Y.W.; Qian, X.; Chan, C.; Yu, Z.F.; Fan, X.X.; Pan, H.D.; Xie, C. Early Lung Cancer Diagnostic Biomarker Discovery by Machine Learning Methods. Transl. Oncol. 2021, 14, 100907. [Google Scholar] [CrossRef] [PubMed]
- Goenka, N.; Tiwari, S. Deep Learning for Alzheimer Prediction Using Brain Biomarkers; Springer: Dordrecht, The Netherlands, 2021; Volume 54, ISBN 1046202110016. [Google Scholar]
- Jarada, T.N.; Rokne, J.G.; Alhajj, R. A Review of Computational Drug Repositioning: Strategies, Approaches, Opportunities, Challenges, and Directions. J. Cheminform. 2020, 12, 46. [Google Scholar] [CrossRef]
- Lauschke, V.M.; Zhou, Y.; Ingelman-Sundberg, M. Novel Genetic and Epigenetic Factors of Importance for Inter-Individual Differences in Drug Disposition, Response and Toxicity. Pharmacol. Ther. 2019, 197, 122–152. [Google Scholar] [CrossRef]
- Dagliati, A.; Tibollo, V.; Sacchi, L.; Malovini, A.; Limongelli, I.; Gabetta, M.; Napolitano, C.; Mazzanti, A.; De Cata, P.; Chiovato, L.; et al. Big Data as a Driver for Clinical Decision Support Systems: A Learning Health Systems Perspective. Front. Digit. Humanit. 2018, 5, 8. [Google Scholar] [CrossRef]
- Sarker, I.H. Machine Learning: Algorithms, Real-World Applications and Research Directions. SN Comput. Sci. 2021, 2, 160. [Google Scholar] [CrossRef]
- Vermeulen, E.; van den Anker, J.N.; Della Pasqua, O.; Hoppu, K.; van der Lee, J.H. How to Optimise Drug Study Design: Pharmacokinetics and Pharmacodynamics Studies Introduced to Paediatricians. J. Pharm. Pharmacol. 2017, 69, 439–447. [Google Scholar] [CrossRef]
- Wedagedera, J.R.; Afuape, A.; Chirumamilla, S.K.; Momiji, H.; Leary, R.; Dunlavey, M.; Matthews, R.; Abduljalil, K.; Jamei, M.; Bois, F.Y. Population PBPK Modeling Using Parametric and Nonparametric Methods of the Simcyp Simulator, and Bayesian Samplers. CPT Pharmacomet. Syst. Pharmacol. 2022, 11, 755–765. [Google Scholar] [CrossRef]
- Ménard, T.; Barmaz, Y.; Koneswarakantha, B.; Bowling, R.; Popko, L. Enabling Data-Driven Clinical Quality Assurance: Predicting Adverse Event Reporting in Clinical Trials Using Machine Learning. Drug Saf. 2019, 42, 1045–1053. [Google Scholar] [CrossRef] [PubMed]
- Phillips, R.; Sauzet, O.; Cornelius, V. Statistical Methods for the Analysis of Adverse Event Data in Randomised Controlled Trials: A Scoping Review and Taxonomy. BMC Med. Res. Methodol. 2020, 20, 288. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, F.; Chauvin, J.; Deville, J.L.; Ciccolini, J. Adaptive Dosing of Sunitinib in a Metastatic Renal Cell Carcinoma Patient: When in Silico Modeling Helps to Go Quicker to the Point. Cancer Chemother. Pharmacol. 2022, 89, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, F.; Chauvin, J.; De Victor, B.; Lacarelle, B.; Deville, J.L.; Ciccolini, J. Clinical-Based vs. Model-Based Adaptive Dosing Strategy: Retrospective Comparison in Real-World MRCC Patients Treated with Sunitinib. Pharmaceuticals 2021, 14, 494. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Gao, W.; Hu, H.; Zhou, S. Why 90% of Clinical Drug Development Fails and How to Improve It? Acta Pharm. Sin. B 2022, 12, 3049–3062. [Google Scholar] [CrossRef] [PubMed]
- Polasek, T.M.; Kirkpatrick, C.M.J.; Rostami-Hodjegan, A. Precision Dosing to Avoid Adverse Drug Reactions. Ther. Adv. Drug Saf. 2019, 10, 2042098619894147. [Google Scholar] [CrossRef] [PubMed]
- Miller, N.A.; Reddy, M.B.; Heikkinen, A.T.; Lukacova, V.; Parrott, N. Physiologically Based Pharmacokinetic Modelling for First-In-Human Predictions: An Updated Model Building Strategy Illustrated with Challenging Industry Case Studies. Clin. Pharmacokinet. 2019, 58, 727–746. [Google Scholar] [CrossRef]
- Mao, J.; Chen, Y.; Xu, L.; Chen, W.; Chen, B.; Fang, Z.; Qin, W.; Zhong, M. Applying Machine Learning to the Pharmacokinetic Modeling of Cyclosporine in Adult Renal Transplant Recipients: A Multi-Method Comparison. Front. Pharmacol. 2022, 13, 1016399. [Google Scholar] [CrossRef] [PubMed]
- Phe, K.; Heil, E.L.; Tam, V.H. Optimizing Pharmacokinetics-Pharmacodynamics of Antimicrobial Management in Patients with Sepsis: A Review. J. Infect. Dis. 2021, 222, S132–S141. [Google Scholar] [CrossRef] [PubMed]
- Pallmann, P.; Bedding, A.W.; Choodari-Oskooei, B.; Dimairo, M.; Flight, L.; Hampson, L.V.; Holmes, J.; Mander, A.P.; Odondi, L.; Sydes, M.R.; et al. Adaptive Designs in Clinical Trials: Why Use Them, and How to Run and Report Them. BMC Med. 2018, 16, 29. [Google Scholar] [CrossRef] [PubMed]
- Shortliffe, E.H.; Buchanan, B.G. A Model of Inexact Reasoning in Medicine. Math. Biosci. 1975, 23, 351–379. [Google Scholar] [CrossRef]
- Miller, R.A.; Pople, H.E.; Myers, J.D. Internist, an Experimental Computer-Based Diagnostic Consultant for General Internal Medicine. N. Engl. J. Med. 1982, 307, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Poweleit, E.A.; Vinks, A.A.; Mizuno, T. Artificial Intelligence and Machine Learning Approaches to Facilitate Therapeutic Drug Management and Model-Informed Precision Dosing. Ther. Drug Monit. 2023, 45, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Keutzer, L.; You, H.; Farnoud, A.; Nyberg, J.; Wicha, S.G.; Maher-Edwards, G.; Vlasakakis, G.; Moghaddam, G.K.; Svensson, E.M.; Menden, M.P.; et al. Machine Learning and Pharmacometrics for Prediction of Pharmacokinetic Data: Differences, Similarities and Challenges Illustrated with Rifampicin. Pharmaceutics 2022, 14, 1530. [Google Scholar] [CrossRef]
- Mould, D.R.; Upton, R.N. Basic Concepts in Population Modeling, Simulation, and Model-Based Drug Development. CPT Pharmacomet. Syst. Pharmacol. 2012, 1, e6. [Google Scholar] [CrossRef]
- Gobburu, J.V.S.; Chen, E.P. Artificial Neural Networks as a Novel Approach to Integrated Pharmacokinetic-Pharmacodynamic Analysis. J. Pharm. Sci. 1996, 85, 505–510. [Google Scholar] [CrossRef]
- Veng-Pedersen, P.; Modi, N.B. Neural Networks in Pharmacodynamic Modeling. Is Current Modeling Practice of Complex Kinetic Systems at a Dead End? J. Pharmacokinet. Biopharm. 1992, 20, 397–412. [Google Scholar] [CrossRef] [PubMed]
- Cucurull-Sanchez, L.; Chappell, M.J.; Chelliah, V.; Amy Cheung, S.Y.; Derks, G.; Penney, M.; Phipps, A.; Malik-Sheriff, R.S.; Timmis, J.; Tindall, M.J.; et al. Best Practices to Maximize the Use and Reuse of Quantitative and Systems Pharmacology Models: Recommendations From the United Kingdom Quantitative and Systems Pharmacology Network. CPT Pharmacomet. Syst. Pharmacol. 2019, 8, 259–272. [Google Scholar] [CrossRef] [PubMed]
- McComb, M.; Bies, R.; Ramanathan, M. Machine Learning in Pharmacometrics: Opportunities and Challenges. Br. J. Clin. Pharmacol. 2022, 88, 1482–1499. [Google Scholar] [CrossRef] [PubMed]
- Collin, C.B.; Gebhardt, T.; Golebiewski, M.; Karaderi, T.; Hillemanns, M.; Khan, F.M.; Salehzadeh-Yazdi, A.; Kirschner, M.; Krobitsch, S.; Kuepfer, L. Computational Models for Clinical Applications in Personalized Medicine—Guidelines and Recommendations for Data Integration and Model Validation. J. Pers. Med. 2022, 12, 166. [Google Scholar] [CrossRef] [PubMed]
- Niazi, S.K. The Coming of Age of AI/ML in Drug Discovery, Development, Clinical Testing, and Manufacturing: The FDA Perspectives. Drug Des. Dev. Ther. 2023, 17, 2691–2725. [Google Scholar] [CrossRef]
- El-Alti, L.; Sandman, L.; Munthe, C. Person Centered Care and Personalized Medicine: Irreconcilable Opposites or Potential Companions? Health Care Anal. 2019, 27, 45–59. [Google Scholar] [CrossRef]
- Vicente, A.M.; Ballensiefen, W.; Jönsson, J.I. How Personalised Medicine Will Transform Healthcare by 2030: The ICPerMed Vision. J. Transl. Med. 2020, 18, 180. [Google Scholar] [CrossRef]
- Brnabic, A.; Hess, L.M. Systematic Literature Review of Machine Learning Methods Used in the Analysis of Real-World Data for Patient-Provider Decision Making. BMC Med. Inform. Decis. Mak. 2021, 21, 54. [Google Scholar] [CrossRef] [PubMed]
- Freriksen, J.J.M.; van der Heijden, J.E.M.; de Hoop-Sommen, M.A.; Greupink, R.; de Wildt, S.N. Physiologically Based Pharmacokinetic (PBPK) Model-Informed Dosing Guidelines for Pediatric Clinical Care: A Pragmatic Approach for a Special Population. Paediatr. Drugs 2023, 25, 5–11. [Google Scholar] [CrossRef]
- Weissler, E.H.; Naumann, T.; Andersson, T.; Ranganath, R.; Elemento, O.; Luo, Y.; Freitag, D.F.; Benoit, J.; Hughes, M.C.; Khan, F.; et al. The Role of Machine Learning in Clinical Research: Transforming the Future of Evidence Generation. Trials 2021, 22, 537. [Google Scholar] [CrossRef]
- Gallo, J.M. Pharmacokinetic/ Pharmacodynamic-Driven Drug Development. Mount Sinai J. Med. 2010, 77, 381–388. [Google Scholar] [CrossRef]
- Gao, H.; Wang, W.; Dong, J.; Ye, Z.; Ouyang, D. An Integrated Computational Methodology with Data-Driven Machine Learning, Molecular Modeling and PBPK Modeling to Accelerate Solid Dispersion Formulation Design. Eur. J. Pharm. Biopharm. 2021, 158, 336–346. [Google Scholar] [CrossRef]
- Joerger, M. Covariate Pharmacokinetic Model Building in Oncology and Its Potential Clinical Relevance. AAPS J. 2012, 14, 119–132. [Google Scholar] [CrossRef]
- Zhu, X.; Zhang, M.; Wen, Y.; Shang, D. Machine Learning Advances the Integration of Covariates in Population Pharmacokinetic Models: Valproic Acid as an Example. Front. Pharmacol. 2022, 13, 994665. [Google Scholar] [CrossRef]
- Fendt, R.; Hofmann, U.; Schneider, A.R.P.; Schaeffeler, E.; Burghaus, R.; Yilmaz, A.; Blank, L.M.; Kerb, R.; Lippert, J.; Schlender, J.F.; et al. Data-Driven Personalization of a Physiologically Based Pharmacokinetic Model for Caffeine: A Systematic Assessment. CPT Pharmacomet. Syst. Pharmacol. 2021, 10, 782–793. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, J.; Lehne, M.; Schepers, J.; Prasser, F.; Thun, S. The Use of Machine Learning in Rare Diseases: A Scoping Review. Orphanet J. Rare Dis. 2020, 15, 145. [Google Scholar] [CrossRef]
- Weaver, R.J.; Valentin, J.P. Today’s Challenges to De-Risk and Predict Drug Safety in Human “Mind-The-Gap”. Toxicol. Sci. 2019, 167, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Trifirò, G.; Crisafulli, S. A New Era of Pharmacovigilance: Future Challenges and Opportunities. Front. Drug Saf. Regul. 2022, 2, 2020–2023. [Google Scholar] [CrossRef]
- Kolluri, S.; Lin, J.; Liu, R.; Zhang, Y.; Zhang, W. Machine Learning and Artificial Intelligence in Pharmaceutical Research and Development: A Review. AAPS J. 2022, 24, 19. [Google Scholar] [CrossRef] [PubMed]
- Seyhan, A.A. Lost in Translation: The Valley of Death across Preclinical and Clinical Divide—Identification of Problems and Overcoming Obstacles. Transl. Med. Commun. 2019, 4, 18. [Google Scholar] [CrossRef]
- Cole, S.; Hay, J.L.; Luzon, E.; Nordmark, A.; Rusten, I.S. European Regulatory Perspective on Pediatric Physiologically Based Pharmacokinetic Models. Int. J. Pharmacokinet. 2017, 2, 113–124. [Google Scholar] [CrossRef]
- Wu, F.; Shah, H.; Li, M.; Duan, P.; Zhao, P.; Suarez, S.; Raines, K.; Zhao, Y.; Wang, M.; Lin, H.P.; et al. Biopharmaceutics Applications of Physiologically Based Pharmacokinetic Absorption Modeling and Simulation in Regulatory Submissions to the U.S. Food and Drug Administration for New Drugs. AAPS J. 2021, 23, 31. [Google Scholar] [CrossRef] [PubMed]
- Woillard, J.B.; Labriffe, M.; Prémaud, A.; Marquet, P. Estimation of Drug Exposure by Machine Learning Based on Simulations from Published Pharmacokinetic Models: The Example of Tacrolimus. Pharmacol. Res. 2021, 167, 105578. [Google Scholar] [CrossRef] [PubMed]
- Woillard, J.B.; Labriffe, M.; Debord, J.; Marquet, P. Tacrolimus Exposure Prediction Using Machine Learning. Clin. Pharmacol. Ther. 2021, 110, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Woillard, J.B.; Labriffe, M.; Debord, J.; Marquet, P. Mycophenolic Acid Exposure Prediction Using Machine Learning. Clin. Pharmacol. Ther. 2021, 110, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Uster, D.W.; Stocker, S.L.; Carland, J.E.; Brett, J.; Marriott, D.J.E.; Day, R.O.; Wicha, S.G. A Model Averaging/Selection Approach Improves the Predictive Performance of Model-Informed Precision Dosing: Vancomycin as a Case Study. Clin. Pharmacol. Ther. 2021, 109, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Bououda, M.; Uster, D.W.; Sidorov, E.; Labriffe, M.; Marquet, P.; Wicha, S.G.; Woillard, J.B. A Machine Learning Approach to Predict Interdose Vancomycin Exposure. Pharm. Res. 2022, 39, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Huang, W.; Lu, H.; Wang, Z.; Ni, X.; Hu, J.; Deng, S.; Tan, Y.; Li, L.; Zhang, M.; et al. A Machine Learning Approach to Personalized Dose Adjustment of Lamotrigine Using Noninvasive Clinical Parameters. Sci. Rep. 2021, 11, 5568. [Google Scholar] [CrossRef]
- Roche-Lima, A.; Roman-Santiago, A.; Feliu-Maldonado, R.; Rodriguez-Maldonado, J.; Nieves-Rodriguez, B.G.; Carrasquillo-Carrion, K.; Ramos, C.M.; Da Luz Sant’Ana, I.; Massey, S.E.; Duconge, J. Machine Learning Algorithm for Predicting Warfarin Dose in Caribbean Hispanics Using Pharmacogenetic Data. Front. Pharmacol. 2020, 10, 1550. [Google Scholar] [CrossRef]
- Gill, J.; Moullet, M.; Martinsson, A.; Miljković, F.; Williamson, B.; Arends, R.H.; Pilla Reddy, V. Evaluating the Performance of Machine-Learning Regression Models for Pharmacokinetic Drug-Drug Interactions. CPT Pharmacomet. Syst. Pharmacol. 2023, 12, 122–134. [Google Scholar] [CrossRef]
- Harun, R.; Yang, E.; Kassir, N.; Zhang, W.; Lu, J. Machine Learning for Exposure-Response Analysis: Methodological Considerations and Confirmation of Their Importance via Computational Experimentations. Pharmaceutics 2023, 15, 1381. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Chen, Y.; Min, Q.; Sun, Q.; Ye, K.; Zhou, C.; Yuan, S.; Sun, Z.; Liao, J. Similarity-Based Machine Learning Support Vector Machine Predictor of Drug-Drug Interactions with Improved Accuracies. J. Clin. Pharm. Ther. 2019, 44, 268–275. [Google Scholar] [CrossRef]
- Liu, C.; Xu, Y.; Liu, Q.; Zhu, H.; Wang, Y. Application of Machine Learning Based Methods in Exposure–Response Analysis. J. Pharmacokinet. Pharmacodyn. 2022, 49, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Bonate, P.L.; Barrett, J.S.; Ait-Oudhia, S.; Brundage, R.; Corrigan, B.; Duffull, S.; Gastonguay, M.; Karlsson, M.O.; Kijima, S.; Krause, A.; et al. Training the next Generation of Pharmacometric Modelers: A Multisector Perspective. J. Pharmacokinet. Pharmacodyn. 2023, 51, 5–31. [Google Scholar] [CrossRef]
- Karatza, E.; Yakovleva, T.; Adams, K.; Rao, G.G.; Ait-Oudhia, S. Knowledge Dissemination and Central Indexing of Resources in Pharmacometrics: An ISOP Education Working Group Initiative. J. Pharmacokinet. Pharmacodyn. 2022, 49, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Ismail, M.; Sale, M.; Yu, Y.; Pillai, N.; Liu, S.; Pflug, B.; Bies, R. Development of a Genetic Algorithm and NONMEM Workbench for Automating and Improving Population Pharmacokinetic/Pharmacodynamic Model Selection. J. Pharmacokinet. Pharmacodyn. 2022, 49, 243–256. [Google Scholar] [CrossRef]
- Sibieude, E.; Khandelwal, A.; Girard, P.; Hesthaven, J.S.; Terranova, N. Population Pharmacokinetic Model Selection Assisted by Machine Learning. J. Pharmacokinet. Pharmacodyn. 2022, 49, 257–270. [Google Scholar] [CrossRef]
- Liu, Q.; Huang, R.; Hsieh, J.; Zhu, H.; Tiwari, M.; Liu, G.; Jean, D.; ElZarrad, M.K.; Fakhouri, T.; Berman, S.; et al. Landscape Analysis of the Application of Artificial Intelligence and Machine Learning in Regulatory Submissions for Drug Development From 2016 to 2021. Clin. Pharmacol. Ther. 2023, 113, 771–774. [Google Scholar] [CrossRef]
- Mallon, A.M.; Häring, D.A.; Dahlke, F.; Aarden, P.; Afyouni, S.; Delbarre, D.; El Emam, K.; Ganjgahi, H.; Gardiner, S.; Kwok, C.H.; et al. Advancing Data Science in Drug Development through an Innovative Computational Framework for Data Sharing and Statistical Analysis. BMC Med. Res. Methodol. 2021, 21, 250. [Google Scholar] [CrossRef]
- Danese, M.D.; Halperin, M.; Duryea, J.; Duryea, R. The Generalized Data Model for Clinical Research. BMC Med. Inform. Decis. Mak. 2019, 19, 117. [Google Scholar] [CrossRef]
- Danilov, G.; Kotik, K.; Shifrin, M.; Strunina, Y.; Pronkina, T.; Tsukanova, T.; Nepomnyashiy, V.; Konovalov, N.; Danilov, V.; Potapov, A. Data Quality Estimation Via Model Performance: Machine Learning as a Validation Tool. Stud. Health Technol. Inform. 2023, 305, 369–372. [Google Scholar] [CrossRef]
- Castro-Alamancos, M.A. A System to Easily Manage Metadata in Biomedical Research Labs Based on Open-Source Software. Bio Protoc. 2022, 12, e4404. [Google Scholar] [CrossRef]
- Xiang, D.; Cai, W. Privacy Protection and Secondary Use of Health Data: Strategies and Methods. Biomed. Res. Int. 2021, 2021, 6967166. [Google Scholar] [CrossRef]
- Schmidt, B.M.; Colvin, C.J.; Hohlfeld, A.; Leon, N. Definitions, Components and Processes of Data Harmonisation in Healthcare: A Scoping Review. BMC Med. Inform. Decis. Mak. 2020, 20, 222. [Google Scholar] [CrossRef]
- Aldoseri, A.; Al-Khalifa, K.N.; Magid Hamouda, A. Re-Thinking Data Strategy and Integration for Artificial Intelligence: Concepts, Opportunities, and Challenges. Appl. Sci. 2023, 13, 7082. [Google Scholar] [CrossRef]
- Chiruvella, V.; Guddati, A.K. Ethical Issues in Patient Data Ownership. Interact. J. Med. Res. 2021, 10, e22269. [Google Scholar] [CrossRef]
- Siala, H.; Wang, Y. SHIFTing Artificial Intelligence to Be Responsible in Healthcare: A Systematic Review. Soc. Sci. Med. 2022, 296, 114782. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Jeong, J.; Jung, S.; Moon, J.; Rho, S. Verification of De-Identification Techniques for Personal Information Using Tree-Based Methods with Shapley Values. J. Pers. Med. 2022, 12, 190. [Google Scholar] [CrossRef] [PubMed]
- Hassija, V.; Chamola, V.; Mahapatra, A.; Singal, A.; Goel, D.; Huang, K.; Scardapane, S.; Spinelli, I.; Mahmud, M.; Hussain, A. Interpreting Black-Box Models: A Review on Explainable Artificial Intelligence. Cognit Comput. 2023, 1, 45–74. [Google Scholar] [CrossRef]
- Rasheed, K.; Qayyum, A.; Ghaly, M.; Al-Fuqaha, A.; Razi, A.; Qadir, J. Explainable, Trustworthy, and Ethical Machine Learning for Healthcare: A Survey. Comput. Biol. Med. 2022, 149, 106043. [Google Scholar] [CrossRef]
- McCarron, T.L.; Moffat, K.; Wilkinson, G.; Zelinsky, S.; Boyd, J.M.; White, D.; Hassay, D.; Lorenzetti, D.L.; Marlett, N.J.; Noseworthy, T. Understanding Patient Engagement in Health System Decision-Making: A Co-Designed Scoping Review. Syst. Rev. 2019, 8, 97. [Google Scholar] [CrossRef] [PubMed]
- Becker, C.; Gross, S.; Gamp, M.; Beck, K.; Amacher, S.A.; Mueller, J.; Bohren, C.; Blatter, R.; Schaefert, R.; Schuetz, P.; et al. Patients’ Preference for Participation in Medical Decision-Making: Secondary Analysis of the BEDSIDE-OUTSIDE Trial. J. Gen. Intern. Med. 2023, 38, 1180–1189. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.C.; Swisher, C.L.; Chung, C.; Jaffray, D.; Sidey-Gibbons, C. On the Importance of Interpretable Machine Learning Predictions to Inform Clinical Decision Making in Oncology. Front. Oncol. 2023, 13, 1129380. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Pérez, R.; Bajorath, J. Interpretation of Machine Learning Models Using Shapley Values: Application to Compound Potency and Multi-Target Activity Predictions. J. Comput. Aided Mol. Des. 2020, 34, 1013–1026. [Google Scholar] [CrossRef] [PubMed]
- Tajgardoon, M.; Samayamuthu, M.J.; Calzoni, L.; Visweswaran, S. Patient-Specific Explanations for Predictions of Clinical Outcomes. ACI Open 2019, 3, e88–e97. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Depraetere, K.; Meesseman, L.; Silva, P.C.; Szymanowsky, R.; Fliegenschmidt, J.; Hulde, N.; Von Dossow, V.; Vanbiervliet, M.; De Baerdemaeker, J.; et al. Machine Learning-Based Prediction Models for Different Clinical Risks in Different Hospitals: Evaluation of Live Performance. J. Med. Internet Res. 2022, 24, e34295. [Google Scholar] [CrossRef]
- Petersson, L.; Larsson, I.; Nygren, J.M.; Nilsen, P.; Neher, M.; Reed, J.E.; Tyskbo, D.; Svedberg, P. Challenges to Implementing Artificial Intelligence in Healthcare: A Qualitative Interview Study with Healthcare Leaders in Sweden. BMC Health Serv. Res. 2022, 22, 850. [Google Scholar] [CrossRef]
- Nugent, B.M.; Madabushi, R.; Buch, B.; Peiris, V.; Crentsil, V.; Miller, V.M.; Bull, J.R.; Jenkins, M. Heterogeneity in Treatment Effects across Diverse Populations. Pharm. Stat. 2021, 20, 929–938. [Google Scholar] [CrossRef]
- He, Z.; Tang, X.; Yang, X.; Guo, Y.; George, T.J.; Charness, N.; Quan Hem, K.B.; Hogan, W.; Bian, J. Clinical Trial Generalizability Assessment in the Big Data Era: A Review. Clin. Transl. Sci. 2020, 13, 675–684. [Google Scholar] [CrossRef]
- Norori, N.; Hu, Q.; Aellen, F.M.; Faraci, F.D.; Tzovara, A. Addressing Bias in Big Data and AI for Health Care: A Call for Open Science. Patterns 2021, 2, 100347. [Google Scholar] [CrossRef]
- Drabiak, K. Leveraging Law and Ethics to Promote Safe and Reliable AI/ML in Healthcare. Front. Nucl. Med. 2022, 2, 983340. [Google Scholar] [CrossRef]
- Koppad, S.; Gkoutos, G.V.; Acharjee, A. Cloud Computing Enabled Big Multi-Omics Data Analytics. Bioinform. Biol. Insights 2021, 15, 11779322211035921. [Google Scholar] [CrossRef]
- Hofer, I.S.; Burns, M.; Kendale, S.; Wanderer, J.P. Realistically Integrating Machine Learning Into Clinical Practice: A Road Map of Opportunities, Challenges, and a Potential Future. Anesth. Analg. 2020, 130, 1115–1118. [Google Scholar] [CrossRef]
- Digital Health—StatPearls—NCBI Bookshelf. Available online: https://www.ncbi.nlm.nih.gov/books/NBK470260/ (accessed on 11 October 2023).
- Dunn, P.; Hazzard, E. Technology Approaches to Digital Health Literacy. Int. J. Cardiol. 2019, 293, 294–296. [Google Scholar] [CrossRef]
- Jandoo, T. WHO Guidance for Digital Health: What It Means for Researchers. Digit. Health 2020, 6, 2055207619898984. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.B.; Wei, W.Q.; Weeraratne, D.; Frisse, M.E.; Misulis, K.; Rhee, K.; Zhao, J.; Snowdon, J.L. Precision Medicine, AI, and the Future of Personalized Health Care. Clin. Transl. Sci. 2021, 14, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Luque, L.; Al Herbish, A.; Al Shammari, R.; Argente, J.; Bin-Abbas, B.; Deeb, A.; Dixon, D.; Zary, N.; Koledova, E.; Savage, M.O. Digital Health for Supporting Precision Medicine in Pediatric Endocrine Disorders: Opportunities for Improved Patient Care. Front. Pediatr. 2021, 9, 715705. [Google Scholar] [CrossRef] [PubMed]
- What Is Digital Health (Digital Healthcare) and Why Is It Important? Available online: https://www.techtarget.com/searchhealthit/definition/digital-health-digital-healthcare (accessed on 11 October 2023).
- Woods, L.; Dendere, R.; Eden, R.; Grantham, B.; Krivit, J.; Pearce, A.; McNeil, K.; Green, D.; Sullivan, C. Perceived Impact of Digital Health Maturity on Patient Experience, Population Health, Health Care Costs, and Provider Experience: Mixed Methods Case Study. J. Med. Internet Res. 2023, 25, e4586. [Google Scholar] [CrossRef] [PubMed]
- Kulynych, J.; Greely, H.T. Clinical Genomics, Big Data, and Electronic Medical Records: Reconciling Patient Rights with Research When Privacy and Science Collide. J. Law. Biosci. 2017, 4, 94–132. [Google Scholar] [CrossRef] [PubMed]
- Syed, R.; Eden, R.; Makasi, T.; Chukwudi, I.; Mamudu, A.; Kamalpour, M.; Kapugama Geeganage, D.; Sadeghianasl, S.; Leemans, S.J.J.; Goel, K.; et al. Digital Health Data Quality Issues: Systematic Review. J. Med. Internet Res. 2023, 25, e42615. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.; Maglaras, L.; Ferrag, M.A.; Almomani, I. Digitization of Healthcare Sector: A Study on Privacy and Security Concerns. ICT Express 2023, 9, 571–588. [Google Scholar] [CrossRef]
- Subbiah, V. The next Generation of Evidence-Based Medicine. Nat. Med. 2023, 29, 49–58. [Google Scholar] [CrossRef]
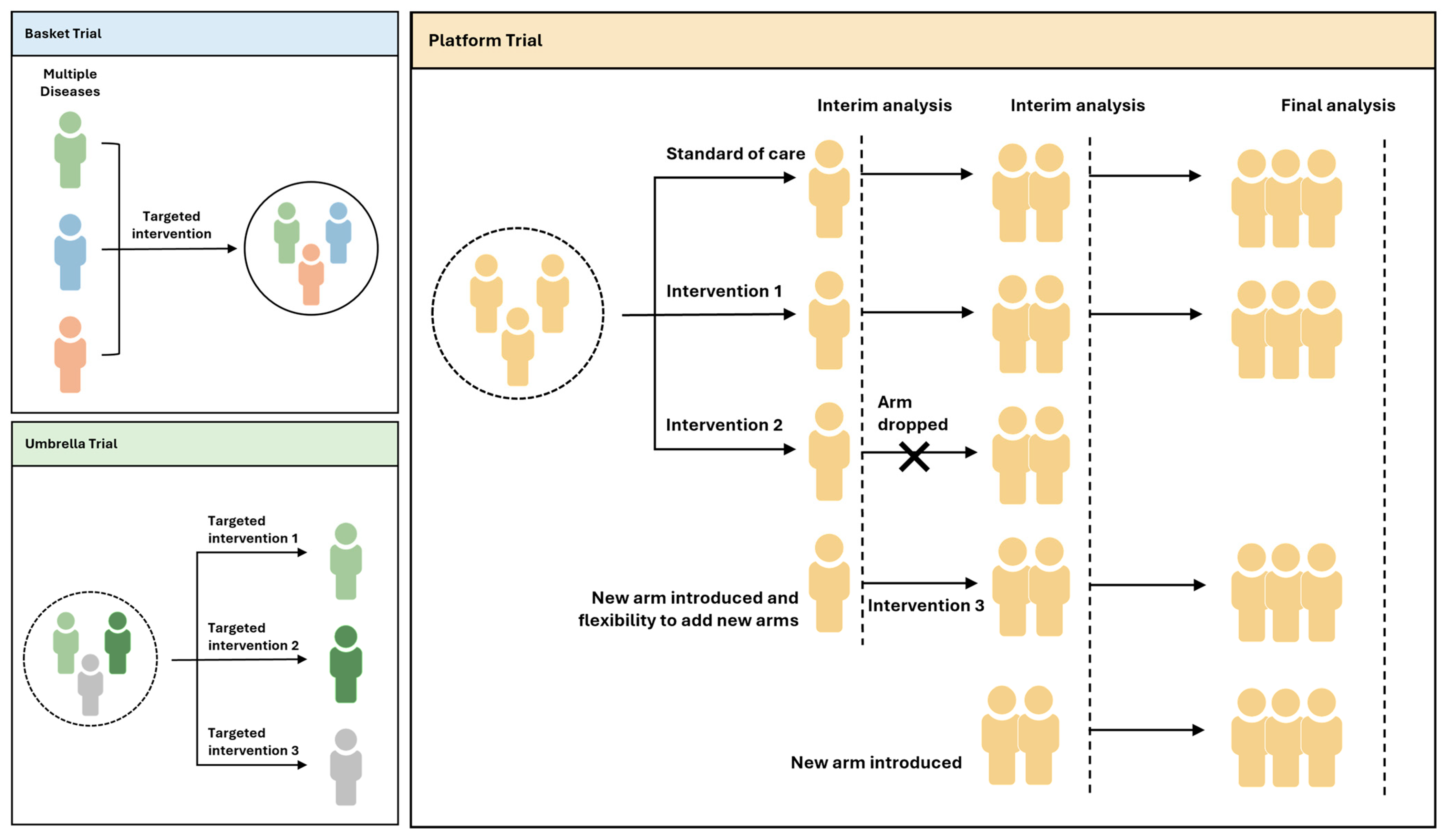
- Fountzilas, E.; Tsimberidou, A.M.; Vo, H.H.; Kurzrock, R. Clinical Trial Design in the Era of Precision Medicine. Genome Med. 2022, 14, 101. [Google Scholar] [CrossRef]
- Hirakawa, A.; Asano, J.; Sato, H.; Teramukai, S. Master Protocol Trials in Oncology: Review and New Trial Designs. Contemp. Clin. Trials Commun. 2018, 12, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Redman, M.W.; Allegra, C.J. The Master Protocol Concept. Semin. Oncol. 2015, 42, 724–730. [Google Scholar] [CrossRef] [PubMed]
- Woodcock, J.; LaVange, L.M. Master Protocols to Study Multiple Therapies, Multiple Diseases, or Both. N. Engl. J. Med. 2017, 377, 62–70. [Google Scholar] [CrossRef]
- Renfro, L.A.; Sargent, D.J. Statistical Controversies in Clinical Research: Basket Trials, Umbrella Trials, and Other Master Protocols: A Review and Examples. Ann. Oncol. 2017, 28, 34–43. [Google Scholar] [CrossRef]
- Food and Drug Administration. FDA Modernizes Clinical Trials with Master Protocols; CDER SBIA Chronicles: Silver Spring, MD, USA, 2019; pp. 1–2. [Google Scholar]
- Basket Clinical Trial Designs: The Key to Testing Innovative Therapies Is Innovation in Study Design and Conduct—ACRP. Available online: https://www.acrpnet.org/2020/02/basket-clinical-trial-designs-the-key-to-testing-innovative-therapies-is-innovation-in-study-design-and-conduct/ (accessed on 11 October 2023).
- Park, J.J.H.; Siden, E.; Zoratti, M.J.; Dron, L.; Harari, O.; Singer, J.; Lester, R.T.; Thorlund, K.; Mills, E.J. Systematic Review of Basket Trials, Umbrella Trials, and Platform Trials: A Landscape Analysis of Master Protocols. Trials 2019, 20, 572. [Google Scholar] [CrossRef] [PubMed]
- Home-Based Clinical Studies—A Paradigm Shift?—Clinical Trials Arena. Available online: https://www.clinicaltrialsarena.com/comment/home-based-clinical-studies-a-paradigm-shift-6094192-2/ (accessed on 11 October 2023).
- Franklin, M.; Thorn, J. Self-Reported and Routinely Collected Electronic Healthcare Resource-Use Data for Trial-Based Economic Evaluations: The Current State of Play in England and Considerations for the Future. BMC Med. Res. Methodol. 2019, 19, 8. [Google Scholar] [CrossRef]
- Virtual Clinical Trials|ObvioHealth. Available online: https://www.obviohealth.com/resources/how-virtual-clinical-trials-are-revolutionizing-health-research (accessed on 11 October 2023).
- FDA Grants Accelerated Approval to Pembrolizumab for First Tissue/Site Agnostic Indication|FDA. Available online: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-pembrolizumab-first-tissuesite-agnostic-indication (accessed on 11 October 2023).
- Wedam, S.; Fashoyin-Aje, L.; Bloomquist, E.; Tang, S.; Sridhara, R.; Goldberg, K.B.; Theoret, M.R.; Amiri-Kordestani, L.; Pazdur, R.; Beaver, J.A. FDA Approval Summary: Palbociclib for Male Patients with Metastatic Breast Cancer. Clin. Cancer Res. 2020, 26, 1208–1212. [Google Scholar] [CrossRef]
- Nice, E.C. The Omics Revolution: Beyond Genomics. A Meeting Report. Clin. Proteomics 2020, 17, 1. [Google Scholar] [CrossRef] [PubMed]
- Ochoa, D.; Karim, M.; Ghoussaini, M.; Hulcoop, D.G.; McDonagh, E.M.; Dunham, I. Human Genetics Evidence Supports Two-Thirds of the 2021 FDA-Approved Drugs. Nat. Rev. Drug Discov. 2022, 21, 551. [Google Scholar] [CrossRef] [PubMed]
- Abul-Husn, N.S.; Kenny, E.E. Personalized Medicine and the Power of Electronic Health Records. Cell 2019, 177, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Sitapati, A.; Kim, H.; Berkovich, B.; Marmor, R.; Singh, S.; El-Kareh, R.; Clay, B.; Ohno-Machado, L. Integrated Precision Medicine: The Role of Electronic Health Records in Delivering Personalized Treatment. Wiley Interdiscip. Rev. Syst. Biol. Med. 2017, 9, e1378. [Google Scholar] [CrossRef]
| Products | Therapeutic Indication |
| Cancer | |
| Abecma (multiple myeloma) | |
| Exkivity (lung cancer) | |
| Lumakras (lung cancer) | |
| Jemperli (endometrial cancer) | |
| Rybrevant (lung cancer) | |
| Scemblix (myeloid leukaemia) | |
| Tepmetko (lung cancer) | |
| Truseltiq (cholangiocarcinoma) | |
| Rare Diseases | |
| Amondys (muscular dystrophy) | |
| Evkeeza (homozygous familial hypercholesterolaemia) | |
| Nexviazyme (Pompe disease) | |
| Nulibry (molybdenum cofactor deficiency) | |
| Vyvgart (Myasthenia Gravis) | |
| Welireg (von Hippel–Lindau) | |
| Other Diseases | |
| Bylvay (progressive familial intrahepatic cholestasis) | |
| Cabenuva (HIV-1) | |
| Leqvio (hypercholesterolaemia) |
| Integration Points | Details |
|---|---|
| Data analysis | ML processes big data efficiently, improving patient outcomes in drug therapy. It identifies salient variables and delineates their interdependencies. |
| Predictive capabilities | ML algorithms excel in predictive capabilities, aiding pharmacometrics in understanding dose–exposure relationships (pharmacokinetics) and exposure marker effects (pharmacodynamics). |
| Complementing pharmacometric modelling | ML acts as a computational bridge, leveraging its flexibility to complement the complexity of principled pharmacometric modelling, resulting in synergistic effects in pharmacological applications. |
| Robustness of datasets | ML implementation in pharmacometrics requires robust datasets for training and testing, capturing the distribution of intrinsic and extrinsic factors of interest. |
| Overfitting | Evaluation data should not be used for training to prevent overfitting, ensuring the model generalizes well to unseen observations and doesn’t fit the training data perfectly. |
| Opportunities | How to Address Them? |
|---|---|
| PKPD model personalization | Developing ML techniques for efficient personalization of PK/PD models to individual patients using sparse data.
|
| Data integration for rare events | Designing models that integrate information from various sources (EHRs, social media, and wearable devices) to predict and manage rare adverse events not well-captured by traditional pharmacometrics models. Challenges: Scarcity of labeled data since rare events occur infrequently. |
| Adaptive clinical trials that can dynamically adjust treatment regimens based on real-time data analysis | Using ML as an assisted tool for clinical trial oversight, providing efficient ways to protect patient safety, reduce trial duration, and lower costs in clinical trial oversight. Challenges: Ensuring data quality and integrity when incorporating data from multiple sources. |
| Real-world evidence analysis | Using real-world evidence data to refine pharmacometrics models, accounting for patient heterogeneity, treatment variability, and long-term outcomes not adequately captured in controlled clinical trials. Challenges: Ensuring data quality and consistency. |
| Interpretable AI for decision Support | Developing interpretable ML models for transparent clinical decision-making. Challenges: Balancing model complexity and transparency; difficult interpretation potentially hindering their acceptance in clinical settings. |
| Uncertainty quantification | Enhancing pharmacometric models by incorporating uncertainty estimation techniques from ML, providing clinicians with confidence intervals for predictions and allowing for better risk assessment. |
| Multi-modal data fusion | Investigating methods to effectively fuse data from diverse modalities, such as genomics, proteomics, and imaging data, to create comprehensive patient profiles that can better inform treatment decisions. Breakdown of the multi-modal data fusion process: (1) data collection; (2) data preprocessing; (3) feature extraction and selection; (4) data fusion; (5) model development; (6) model validation; (7) clinical application; and (8) continuous learning (as new data becomes available, the models can be updated and refined, embodying the principles of continuous learning and improvement). |
| Longitudinal data analysis | Developing models for analyzing longitudinal data over extended periods to capture changes in patient response to treatments. |
| Ethical and regulatory Considerations | Addressing ethical implications and regulatory challenges of incorporating ML into pharmacometrics, including issues related to data privacy, bias, and validation. |
| Optimization of drug combination | Exploring ML algorithms to optimize drug combinations by predicting synergistic effects, potential adverse interactions, and tailoring treatments for individual patients. |
| EHR Benefits | Integration of EHR in Healthcare |
|---|---|
| Information access and sharing | EHRs facilitate quick and secure access to patients’ medical information, allowing healthcare professionals to make informed decisions and order care. |
| Better care management | EHRs help you better manage the care of chronic patients by enabling continuous monitoring and adjustment of treatment plans based on real-time data. |
| Integration and coordination | The integration of RSE (remote sensing and earth observation) into healthcare systems allows for more efficient coordination between different healthcare providers, improving continuity of care. |
| Clinical research | RSE data can be used in clinical research to identify health trends, evaluate the effectiveness of treatments, and improve evidence-based medicine. Furthermore, omics data, which encompasses genomic, transcriptomic, proteomic, and metabolomic information, plays a crucial role in precision medicine. This data enables the personalization of treatments based on the genetics and individual characteristics of each patient, improving the effectiveness of care. Omics data analysis also helps identify genetic markers of diseases, enabling early prevention and diagnosis. |
| Wearable Devices | Properties, Capabilities, and Applications |
|---|---|
| Smartwatches | Monitor heart rate, measure blood pressure, track physical activity, count steps, monitor sleep quality, and send reminders to move, drink water or perform exercises |
| Fitness trackers | Monitor steps, distance traveled, calories burned, heart rate, and even track specific exercises like running and swimming |
| Glucose-monitoring devices | For people with diabetes, devices such as continuous glucose monitors (CGM) offer the ability to monitor blood glucose levels in real-time. They can send alerts when glucose levels are out of ideal range |
| Portable electrocardiogram (ECG) devices | Some smartwatches can perform ECGs. They can detect abnormal heart rhythms, such as atrial fibrillation |
| Sleep-monitoring devices | These devices record sleep patterns, duration, and quality. They provide insights into improving sleep habits |
| Breath-monitoring devices | Can monitor respiratory rate and blood oxygen saturation. This is useful for monitoring breathing problems such as sleep apnea |
| Virtual and augmented reality (VR/AR) Devices | In rehabilitation areas and therapy, VR and AR devices create virtual environments for therapeutic purposes, such as rehabilitation after injuries or strokes |
| Smart glasses | These are used in medical settings for access to clinical information, real-time documentation, and telehealth |
| Physiological activity-monitoring devices | In addition to the most well-known devices, some wearables monitor specific physiological activities, such as body temperature, exposure to UV light, hydration, and much more |
| Wearable sensors for clinical research | In clinical research, wearable sensors are used to collect objective and accurate data about the health of patients in clinical studies, enabling a deeper understanding of different medical conditions |
| Augmented reality glasses for surgery | In medicine, augmented reality glasses are used by surgeons to provide real-time information during surgical procedures, making them more accurate and safer |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marques, L.; Costa, B.; Pereira, M.; Silva, A.; Santos, J.; Saldanha, L.; Silva, I.; Magalhães, P.; Schmidt, S.; Vale, N. Advancing Precision Medicine: A Review of Innovative In Silico Approaches for Drug Development, Clinical Pharmacology and Personalized Healthcare. Pharmaceutics 2024, 16, 332. https://doi.org/10.3390/pharmaceutics16030332
Marques L, Costa B, Pereira M, Silva A, Santos J, Saldanha L, Silva I, Magalhães P, Schmidt S, Vale N. Advancing Precision Medicine: A Review of Innovative In Silico Approaches for Drug Development, Clinical Pharmacology and Personalized Healthcare. Pharmaceutics. 2024; 16(3):332. https://doi.org/10.3390/pharmaceutics16030332
Chicago/Turabian StyleMarques, Lara, Bárbara Costa, Mariana Pereira, Abigail Silva, Joana Santos, Leonor Saldanha, Isabel Silva, Paulo Magalhães, Stephan Schmidt, and Nuno Vale. 2024. "Advancing Precision Medicine: A Review of Innovative In Silico Approaches for Drug Development, Clinical Pharmacology and Personalized Healthcare" Pharmaceutics 16, no. 3: 332. https://doi.org/10.3390/pharmaceutics16030332
APA StyleMarques, L., Costa, B., Pereira, M., Silva, A., Santos, J., Saldanha, L., Silva, I., Magalhães, P., Schmidt, S., & Vale, N. (2024). Advancing Precision Medicine: A Review of Innovative In Silico Approaches for Drug Development, Clinical Pharmacology and Personalized Healthcare. Pharmaceutics, 16(3), 332. https://doi.org/10.3390/pharmaceutics16030332











