Obtaining Gene-Modified HLA-E-Expressing Feeder Cells for Stimulation of Natural Killer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Construction of HLA-E-Containing Vector
2.2. Cell Lines
2.3. Peptide Synthesis
- -
- PIP (30% solution in NMP, 25 mL for 1 g of resin), 2 × 10 min;
- -
- DMF (20 mL for 1 g of resin), 5 × 5 min;
- -
- Fmoc-AA(PG)-OH (8 eq., 0.5M solution in NMP)/HATU (8 eq., 0.5M solution in NMP)/DIPEA (16 eq.), 40 min;
2.4. NK Cell Isolation and Magnetic Separation of CD57− Subset
2.5. Incubation of HLA-E Modified Cells with Peptide
2.6. HCMV IgG Detection
2.7. Cell Staining and Flow Cytometry Analysis
2.8. NK Cell Expansion
2.9. Cell Proliferation
2.10. Degranulation Assay
2.11. Cytokine-Dependent IFNγ Production Assay
2.12. Statistical Analysis
3. Results
3.1. Obtaining HLA-E-Expressing Feeder Cells on the Basis of K562 Cell Line and Their Characterization
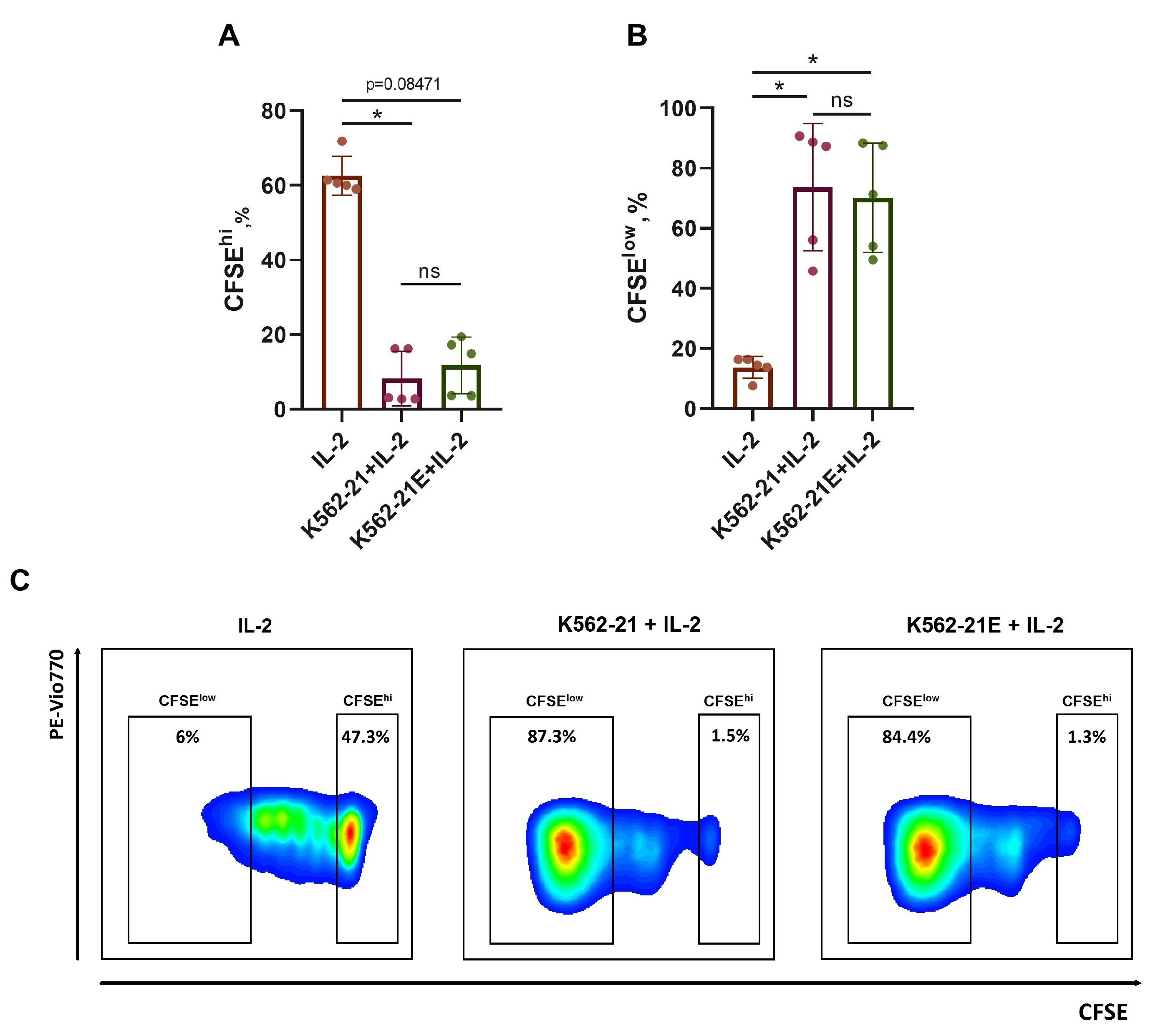
3.2. HLA-E-Presenting K562-21E Cells Induce Active NK Cell Proliferation
3.3. Analysis of CD57-Negative NK Cell Expansion Induced with the Use of HLA-E-Expressing K562 Feeder Cells
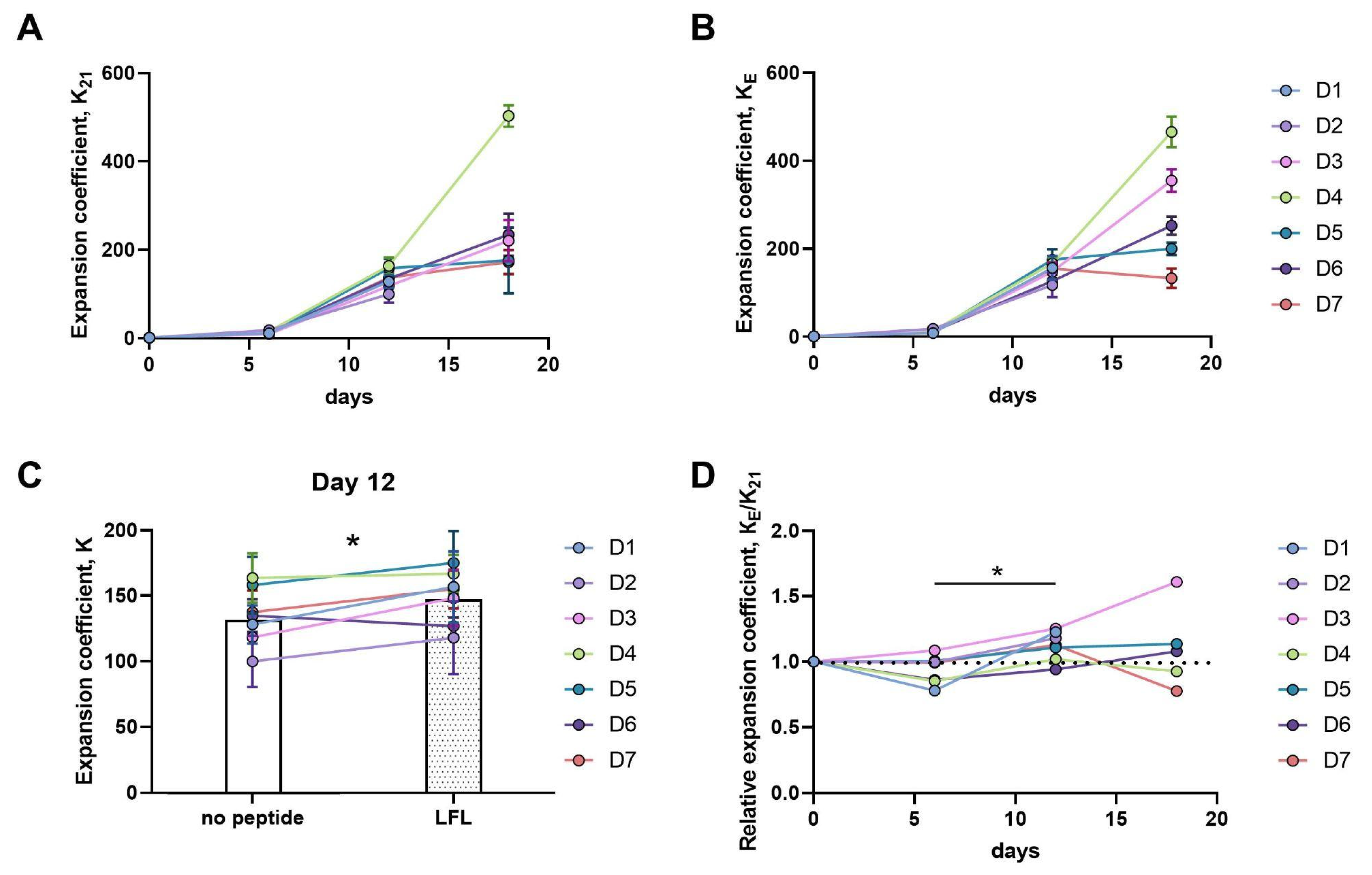
3.3.1. Twelve-Day Expansion of CD57− NK Cells Is Greater upon Presentation of the HLA-E/LFL Complex, Regardless of HCMV Serological Status
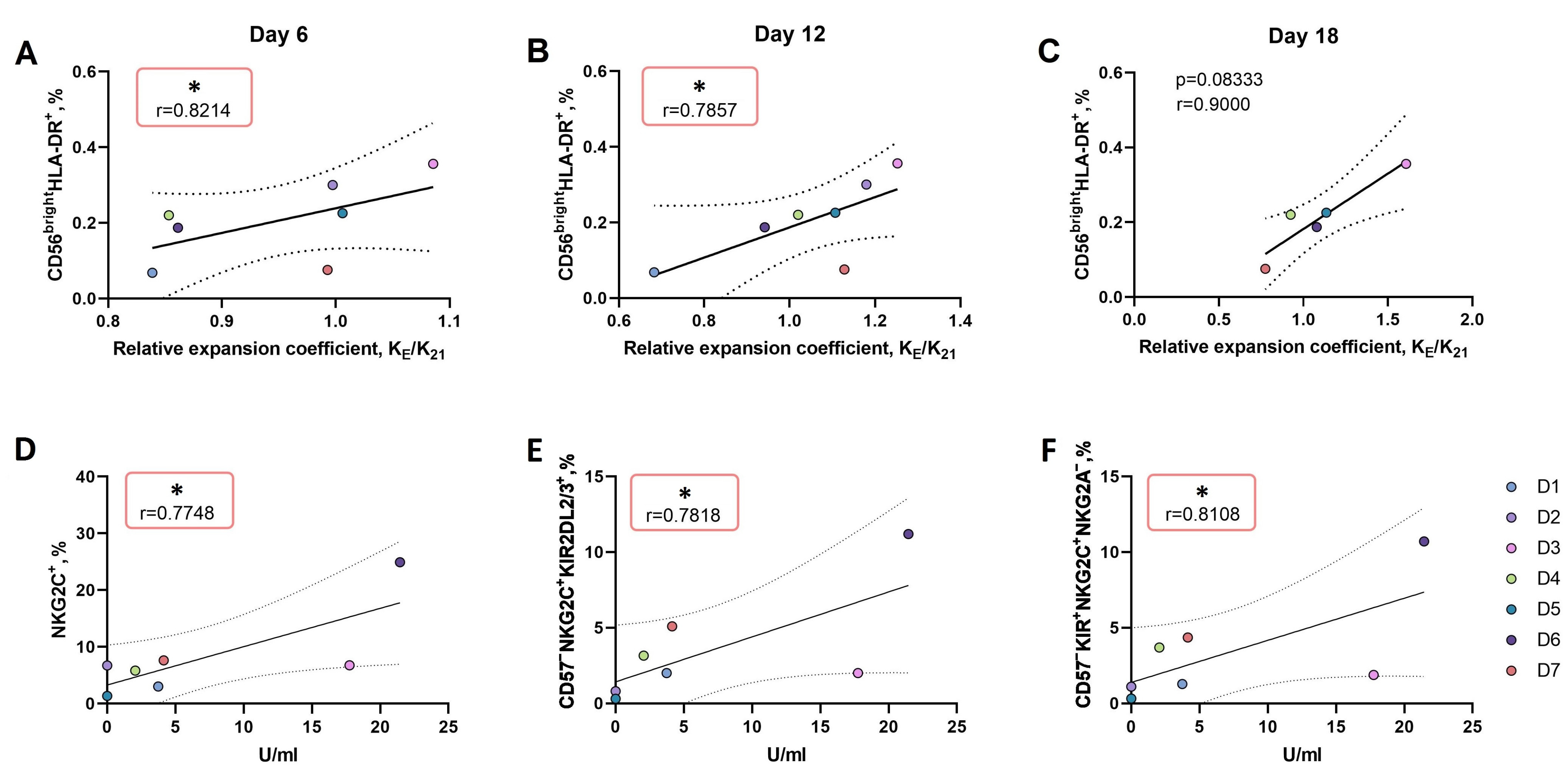
3.3.2. An Increase in the Proportion of CD56brightHLA-DR+ NK Cell Subset Ex Vivo Is Associated with More Intense Proliferation of NK Cells in Response to the LFL Peptide Presentation by K562-21E Feeder Cells
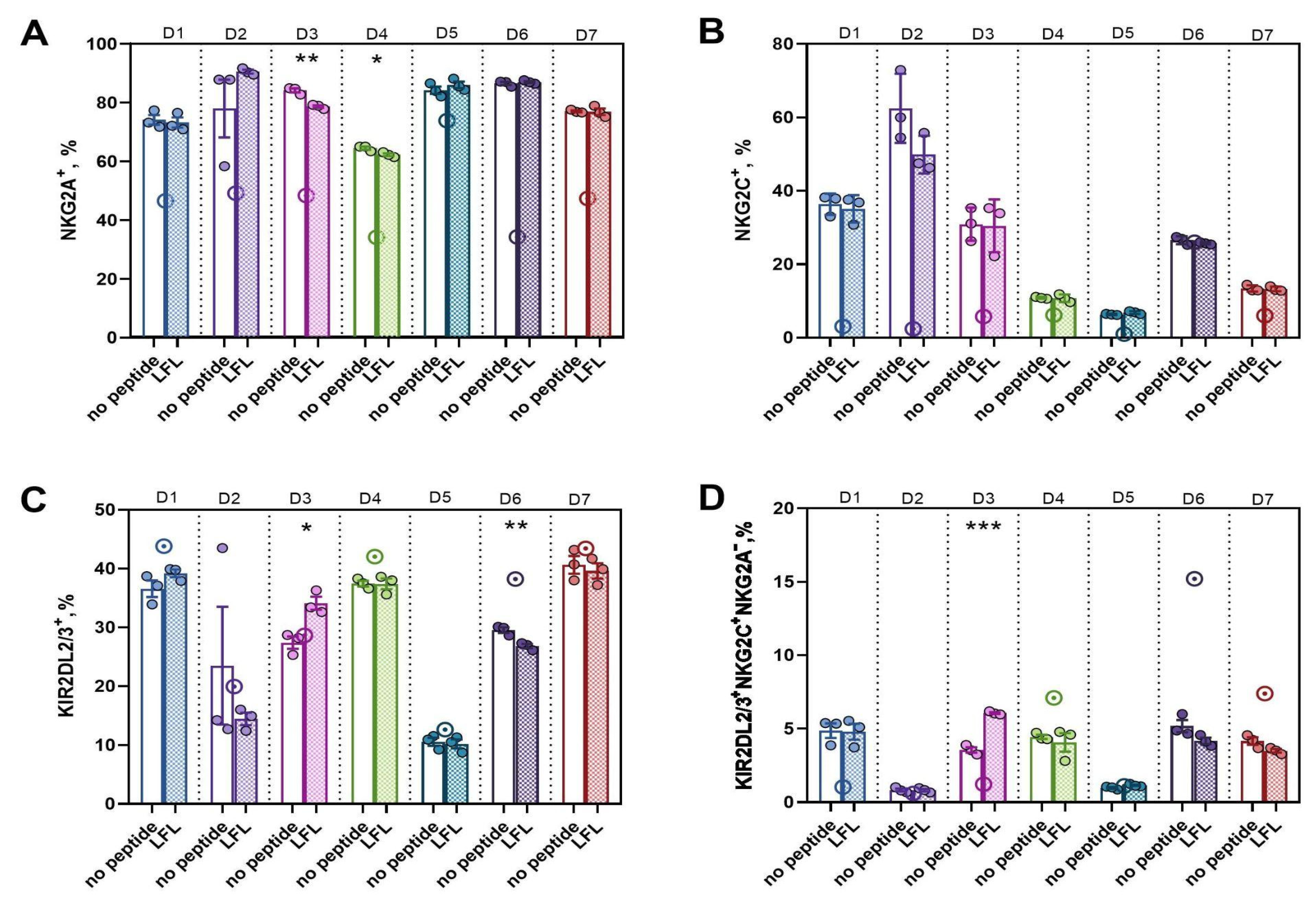
3.3.3. In CD57− NK Cell Cultures Actively Proliferating in Response to the Presentation of the HCMV Peptide, a High Proportion of KIR2DL2/3+NKG2C+NKG2A− Presumable Progenitors of Adaptive NK Cells was Observed
3.3.4. Active Proliferation of CD57− NK Cells in Response to the LFL Peptide Presentation Is Accompanied by Positive Functional Shifts
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, H.; Yang, L.; Wang, T.; Li, Z. NK Cell-Based Tumor Immunotherapy. Bioact. Mater. 2023, 31, 63–86. [Google Scholar] [CrossRef]
- Prager, I.; Watzl, C. Mechanisms of Natural Killer Cell-Mediated Cellular Cytotoxicity. J. Leukoc. Biol. 2019, 105, 1319–1329. [Google Scholar] [CrossRef]
- Laskowski, T.J.; Biederstädt, A.; Rezvani, K. Natural Killer Cells in Antitumour Adoptive Cell Immunotherapy. Nat. Rev. Cancer 2022, 22, 557–575. [Google Scholar] [CrossRef] [PubMed]
- de Rham, C.; Ferrari-Lacraz, S.; Jendly, S.; Schneiter, G.; Dayer, J.-M.; Villard, J. The Proinflammatory Cytokines IL-2, IL-15 and IL-21 Modulate the Repertoire of Mature Human Natural Killer Cell Receptors. Arthritis. Res. Ther. 2007, 9, R125. [Google Scholar] [CrossRef] [PubMed]
- Terrén, I.; Orrantia, A.; Mosteiro, A.; Vitallé, J.; Zenarruzabeitia, O.; Borrego, F. Metabolic Changes of Interleukin-12/15/18-Stimulated Human NK Cells. Sci. Rep. 2021, 11, 6472. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.; Li, L.; McCarty, J.; Kaur, I.; Yvon, E.; Shaim, H.; Muftuoglu, M.; Liu, E.; Orlowski, R.Z.; Cooper, L.; et al. Phase I Study of Cord Blood-Derived Natural Killer Cells Combined with Autologous Stem Cell Transplantation in Multiple Myeloma. Br. J. Haematol. 2017, 177, 457–466. [Google Scholar] [CrossRef]
- Childs, R.W.; Berg, M. Bringing Natural Killer Cells to the Clinic: Ex Vivo Manipulation. Hematol. Am. Soc. Hematol. Educ. Program 2013, 2013, 234–246. [Google Scholar] [CrossRef]
- Granzin, M.; Stojanovic, A.; Miller, M.; Childs, R.; Huppert, V.; Cerwenka, A. Highly Efficient IL-21 and Feeder Cell-Driven Ex Vivo Expansion of Human NK Cells with Therapeutic Activity in a Xenograft Mouse Model of Melanoma. Oncoimmunology 2016, 5, e1219007. [Google Scholar] [CrossRef]
- Imai, C.; Iwamoto, S.; Campana, D. Genetic Modification of Primary Natural Killer Cells Overcomes Inhibitory Signals and Induces Specific Killing of Leukemic Cells. Blood 2005, 106, 376–383. [Google Scholar] [CrossRef]
- Fujisaki, H.; Kakuda, H.; Imai, C.; Mullighan, C.G.; Campana, D. Replicative Potential of Human Natural Killer Cells. Br. J. Haematol. 2009, 145, 606–613. [Google Scholar] [CrossRef]
- Li, X.; He, C.; Liu, C.; Ma, J.; Ma, P.; Cui, H.; Tao, H.; Gao, B. Expansion of NK Cells from PBMCs Using Immobilized 4-1BBL and Interleukin-21. Int. J. Oncol. 2015, 47, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Denman, C.J.; Senyukov, V.V.; Somanchi, S.S.; Phatarpekar, P.V.; Kopp, L.M.; Johnson, J.L.; Singh, H.; Hurton, L.; Maiti, S.N.; Huls, M.H.; et al. Membrane-Bound IL-21 Promotes Sustained Ex Vivo Proliferation of Human Natural Killer Cells. PLoS ONE 2012, 7, e30264. [Google Scholar] [CrossRef]
- Cooper, M.A.; Elliott, J.M.; Keyel, P.A.; Yang, L.; Carrero, J.A.; Yokoyama, W.M. Cytokine-Induced Memory-like Natural Killer Cells. Proc. Natl. Acad. Sci. USA 2009, 106, 1915–1919. [Google Scholar] [CrossRef] [PubMed]
- Parrish-Novak, J.; Dillon, S.R.; Nelson, A.; Hammond, A.; Sprecher, C.; Gross, J.A.; Johnston, J.; Madden, K.; Xu, W.; West, J.; et al. Interleukin 21 and Its Receptor Are Involved in NK Cell Expansion and Regulation of Lymphocyte Function. Nature 2000, 408, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Skak, K.; Frederiksen, K.S.; Lundsgaard, D. Interleukin-21 Activates Human Natural Killer Cells and Modulates Their Surface Receptor Expression. Immunology 2008, 123, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Streltsova, M.A.; Erokhina, S.A.; Kanevskiy, L.M.; Lee, D.A.; Telford, W.G.; Sapozhnikov, A.M.; Kovalenko, E.I. Analysis of NK Cell Clones Obtained Using Interleukin-2 and Gene-Modified K562 Cells Revealed the Ability of “Senescent” NK Cells to Lose CD57 Expression and Start Expressing NKG2A. PLoS ONE 2018, 13, e0208469. [Google Scholar] [CrossRef] [PubMed]
- Bjorkstrom, N.K.; Riese, P.; Heuts, F.; Andersson, S.; Fauriat, C.; Ivarsson, M.A.; Bjorklund, A.T.; Flodstrom-Tullberg, M.; Michaelsson, J.; Rottenberg, M.E.; et al. Expression Patterns of NKG2A, KIR, and CD57 Define a Process of CD56dim NK-Cell Differentiation Uncoupled from NK-Cell Education. Blood 2010, 116, 3853–3864. [Google Scholar] [CrossRef]
- Gumá, M.; Budt, M.; Sáez, A.; Brckalo, T.; Hengel, H.; Angulo, A.; López-Botet, M. Expansion of CD94/NKG2C+ NK Cells in Response to Human Cytomegalovirus-Infected Fibroblasts. Blood 2006, 107, 3624–3631. [Google Scholar] [CrossRef]
- He, W.; Gea-Mallorquí, E.; Colin-York, H.; Fritzsche, M.; Gillespie, G.M.; Brackenridge, S.; Borrow, P.; McMichael, A.J. Intracellular Trafficking of HLA-E and Its Regulation. J. Exp. Med. 2023, 220, e20221941. [Google Scholar] [CrossRef]
- Llano, M.; Lee, N.; Navarro, F.; García, P.; Albar, J.P.; Geraghty, D.E.; López-Botet, M. HLA-E-Bound Peptides Influence Recognition by Inhibitory and Triggering CD94/NKG2 Receptors: Preferential Response to an HLA-G-Derived Nonamer. Eur. J. Immunol. 1998, 28, 2854–2863. [Google Scholar] [CrossRef]
- Hammer, Q.; Rückert, T.; Borst, E.M.; Dunst, J.; Haubner, A.; Durek, P.; Heinrich, F.; Gasparoni, G.; Babic, M.; Tomic, A.; et al. Peptide-Specific Recognition of Human Cytomegalovirus Strains Controls Adaptive Natural Killer Cells Article. Nat. Immunol. 2018, 19, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Davis, Z.B.; Cooley, S.A.; Cichocki, F.; Felices, M.; Wangen, R.; Luo, X.; DeFor, T.E.; Bryceson, Y.T.; Diamond, D.J.; Brunstein, C.; et al. Adaptive Natural Killer Cell and Killer Cell Immunoglobulin-Like Receptor-Expressing T Cell Responses Are Induced by Cytomegalovirus and Are Associated with Protection against Cytomegalovirus Reactivation after Allogeneic Donor Hematopoietic Cell Transplantation. Biol. Blood Marrow Transpl. 2015, 21, 1653–1662. [Google Scholar] [CrossRef]
- Yu, X.X.; Shang, Q.N.; Liu, X.F.; He, M.; Pei, X.Y.; Mo, X.D.; Lv, M.; Han, T.T.; Huo, M.R.; Zhao, X.S.; et al. Donor NKG2C Homozygosity Contributes to CMV Clearance after Haploidentical Transplantation. JCI Insight 2022, 7, e149120. [Google Scholar] [CrossRef] [PubMed]
- Kheav, V.D.; Busson, M.; Scieux, C.; de Latour, R.P.; Maki, G.; Haas, P.; Mazeron, M.C.; Carmagnat, M.; Masso, E.; Xhaard, A.; et al. Favorable Impact of Natural Killer Cell Reconstitution on Chronic Graft-versus-Host Disease and Cytomegalovirus Reactivation after Allogeneic Hematopoietic Stem Cell Transplantation. Haematologica 2014, 99, 1860–1867. [Google Scholar] [CrossRef]
- Cichocki, F.; Cooley, S.; Davis, Z.; DeFor, T.E.; Schlums, H.; Zhang, B.; Brunstein, C.G.; Blazar, B.R.; Wagner, J.; Diamond, D.J.; et al. CD56dimCD57+NKG2C+ NK Cell Expansion Is Associated with Reduced Leukemia Relapse after Reduced Intensity HCT. Leukemia 2016, 30, 456–463. [Google Scholar] [CrossRef]
- Russo, A.; Oliveira, G.; Berglund, S.; Greco, R.; Gambacorta, V.; Cieri, N.; Toffalori, C.; Zito, L.; Lorentino, F.; Piemontese, S.; et al. NK Cell Recovery after Haploidentical HSCT with Posttransplant Cyclophosphamide: Dynamics and Clinical Implications. Blood 2018, 131, 247–262. [Google Scholar] [CrossRef]
- Meazza, R.; Falco, M.; Loiacono, F.; Canevali, P.; Della Chiesa, M.; Bertaina, A.; Pagliara, D.; Merli, P.; Indio, V.; Galaverna, F.; et al. Phenotypic and Functional Characterization of NK Cells in AβT-Cell and B-Cell Depleted Haplo-HSCT to Cure Pediatric Patients with Acute Leukemia. Cancers 2020, 12, 2187. [Google Scholar] [CrossRef]
- Costa-Garcia, M.; Vera, A.; Moraru, M.; Vilches, C.; López-Botet, M.; Muntasell, A. Antibody-Mediated Response of NKG2C Bright NK Cells against Human Cytomegalovirus. J. Immunol. 2015, 194, 2715–2724. [Google Scholar] [CrossRef]
- Strong, R.K.; Holmes, M.A.; Li, P.; Braun, L.; Lee, N.; Geraghty, D.E. HLA-E Allelic Variants. Correlating Differential Expression, Peptide Affinities, Crystal Structures, and Thermal Stabilities. J. Biol. Chem. 2003, 278, 5082–5090. [Google Scholar] [CrossRef]
- Hosseini, E.; Schwarer, A.P.; Ghasemzadeh, M. Do Human Leukocyte Antigen E Polymorphisms Influence Graft-versus-Leukemia after Allogeneic Hematopoietic Stem Cell Transplantation? Exp. Hematol. 2015, 43, 149–157. [Google Scholar] [CrossRef]
- Lopez-Vergès, S.; Milush, J.M.; Pandey, S.; York, V.A.; Arakawa-Hoyt, J.; Pircher, H.; Norris, P.J.; Nixon, D.F.; Lanier, L.L. CD57 Defines a Functionally Distinct Population of Mature NK Cells in the Human CD56dimCD16+ NK-Cell Subset. Blood 2010, 116, 3865–3874. [Google Scholar] [CrossRef] [PubMed]
- Kobyzeva, P.A.; Streltsova, M.A.; Erokhina, S.A.; Kanevskiy, L.M.; Telford, W.G.; Sapozhnikov, A.M.; Kovalenko, E.I. CD56dimCD57−NKG2C+ NK Cells Retaining Proliferative Potential Are Possible Precursors of CD57+NKG2C+ Memory-like NK Cells. J. Leukoc. Biol. 2020, 108, 1379–1395. [Google Scholar] [CrossRef] [PubMed]
- Streltsova, M.A.; Barsov, E.V.; Erokhina, S.A.; Kovalenko, E.I. Retroviral Gene Transfer into Primary Human NK Cells Activated by IL-2 and K562 Feeder Cells Expressing Membrane-Bound IL-21. J. Immunol. Methods 2017, 450, 90–94. [Google Scholar] [CrossRef]
- Streltsova, M.A.; Erokhina, S.A.; Kanevskiy, L.M.; Grechikhina, M.V.; Kobyzeva, P.A.; Lee, D.A.; Telford, W.G.; Sapozhnikov, A.M.; Kovalenko, E.I. Recurrent Stimulation of Natural Killer Cell Clones with K562 Expressing Membrane-Bound Interleukin-21 Affects Their Phenotype, Interferon-γ Production, and Lifespan. Int. J. Mol. Sci. 2019, 20, 443. [Google Scholar] [CrossRef] [PubMed]
- Senko, D.A.; Timofeev, N.D.; Kasheverov, I.E.; Ivanov, I.A. Scope and Limitations of Pseudoprolines as Individual Amino Acids in Peptide Synthesis. Amino Acids 2021, 53, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Bland, F.A.; Lemberg, M.K.; McMichael, A.J.; Martoglio, B.; Braud, V.M. Requirement of the Proteasome for the Trimming of Signal Peptide-Derived Epitopes Presented by the Nonclassical Major Histocompatibility Complex Class I Molecule HLA-E. J. Biol. Chem. 2003, 278, 33747–33752. [Google Scholar] [CrossRef]
- Ustiuzhanina, M.O.; Vavilova, J.D.; Alekseeva, N.A.; Lutsenko, G.V.; Chudakov, D.M.; Kovalenko, E.I. Coordination of NK Cell Markers Expression and IgG Response in HCMV Infection. Med. Immunol. 2023, 25, 573–580. [Google Scholar] [CrossRef]
- Muntasell, A.; Vilches, C.; Angulo, A.; López-Botet, M. Adaptive Reconfiguration of the Human NK-Cell Compartment in Response to Cytomegalovirus: A Different Perspective of the Host-Pathogen Interaction. Eur. J. Immunol. 2013, 43, 1133–1141. [Google Scholar] [CrossRef]
- Gumá, M.; Angulo, A.; Vilches, C.; Gómez-Lozano, N.; Malats, N.; López-Botet, M. Imprint of Human Cytomegalovirus Infection on the NK Cell Receptor Repertoire. Blood 2004, 104, 3664–3671. [Google Scholar] [CrossRef]
- Rölle, A.; Pollmann, J.; Ewen, E.M.; Le, V.T.K.; Halenius, A.; Hengel, H.; Cerwenka, A. IL-12-Producing Monocytes and HLA-E Control HCMV-Driven NKG2C+ NK Cell Expansion. J. Clin. Investig. 2014, 124, 5305–5316. [Google Scholar] [CrossRef]
- Bayard, C.; Lepetitcorps, H.; Roux, A.; Larsen, M.; Fastenackels, S.; Salle, V.; Vieillard, V.; Marchant, A.; Stern, M.; Boddaert, J.; et al. Coordinated Expansion of Both Memory T Cells and NK Cells in Response to CMV Infection in Humans. Eur. J. Immunol. 2016, 46, 1168–1179. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.A.; Fehniger, T.A. Human Adaptive Natural Killer Cells: Beyond NKG2C. Trends. Immunol. 2016, 37, 351–353. [Google Scholar] [CrossRef]
- Phan, M.T.T.; Kim, J.; Koh, S.K.; Lim, Y.; Yu, H.; Lee, M.; Lee, J.M.; Kang, E.S.; Kim, H.Y.; Kim, S.K.; et al. Selective Expansion of NKG2C+ Adaptive NK Cells Using K562 Cells Expressing HLA-E. Int. J. Mol. Sci. 2022, 23, 9426. [Google Scholar] [CrossRef]
- Rölle, A.; Meyer, M.; Calderazzo, S.; Jäger, D.; Momburg, F. Distinct HLA-E Peptide Complexes Modify Antibody-Driven Effector Functions of Adaptive NK Cells. Cell Rep. 2018, 24, 1967–1976.e4. [Google Scholar] [CrossRef] [PubMed]
- Davis, Z.B.; Cogswell, A.; Scott, H.; Mertsching, A.; Boucau, J.; Wambua, D.; Le Gall, S.; Planelles, V.; Campbell, K.S.; Barker, E. A Conserved HIV-1-Derived Peptide Presented by HLA-E Renders Infected T-Cells Highly Susceptible to Attack by NKG2A/CD94-Bearing Natural Killer Cells. PLoS Pathog. 2016, 12, e1005421. [Google Scholar] [CrossRef] [PubMed]
- Haroun-Izquierdo, A.; Vincenti, M.; Netskar, H.; Van Ooijen, H.; Zhang, B.; Bendzick, L.; Kanaya, M.; Momayyezi, P.; Li, S.; Wiiger, M.T.; et al. Adaptive Single-KIR+NKG2C+ NK Cells Expanded from Select Superdonors Show Potent Missing-Self Reactivity and Efficiently Control HLA-Mismatched Acute Myeloid Leukemia. J. Immunother. Cancer 2022, 10, e005577. [Google Scholar] [CrossRef]
- Palamarchuk, A.I.; Alekseeva, N.A.; Streltsova, M.A.; Ustiuzhanina, M.O.; Kobyzeva, P.A.; Kust, S.A.; Grechikhina, M.V.; Boyko, A.A.; Shustova, O.A.; Sapozhnikov, A.M.; et al. Increased Susceptibility of the CD57—NK Cells Expressing KIR2DL2/3 and NKG2C to ICasp9 Gene Retroviral Transduction and the Relationships with Proliferative Potential, Activation Degree, and Death Induction Response. Int. J. Mol. Sci. 2021, 22, 13326. [Google Scholar] [CrossRef]
- Goodridge, J.P.; Jacobs, B.; Saetersmoen, M.L.; Clement, D.; Hammer, Q.; Clancy, T.; Skarpen, E.; Brech, A.; Landskron, J.; Grimm, C.; et al. Remodeling of Secretory Lysosomes during Education Tunes Functional Potential in NK Cells. Nat. Commun. 2019, 10, 514. [Google Scholar] [CrossRef]
- Evans, J.H.; Horowitz, A.; Mehrabi, M.; Wise, E.L.; Pease, J.E.; Riley, E.M.; Davis, D.M. A Distinct Subset of Human NK Cells Expressing HLA-DR Expand in Response to IL-2 and Can Aid Immune Responses to BCG. Eur. J. Immunol. 2011, 41, 1924–1933. [Google Scholar] [CrossRef]
- Erokhina, S.A.; Streltsova, M.A.; Kanevskiy, L.M.; Telford, W.G.; Sapozhnikov, A.M.; Kovalenko, E.I. HLA-DR+ NK Cells Are Mostly Characterized by Less Mature Phenotype and High Functional Activity. Immunol. Cell Biol. 2018, 96, 212–228. [Google Scholar] [CrossRef]
- Costa-García, M.; Ataya, M.; Moraru, M.; Vilches, C.; López-Botet, M.; Muntasell, A. Human Cytomegalovirus Antigen Presentation by HLA-DR+ NKG2C+ Adaptive NK Cells Specifically Activates Polyfunctional Effector Memory CD4+ T Lymphocytes. Front. Immunol. 2019, 10, 687. [Google Scholar] [CrossRef] [PubMed]
- Kovalenko, E.I.; Streltsova, M.A.; Kanevskiy, L.M.; Erokhina, S.A.; Telford, W.G. Identification of Human Memory-like NK Cells. Curr. Protoc. Cytom. 2017, 2017, 9.50.1–9.50.11. [Google Scholar] [CrossRef] [PubMed]
- Hanna, J.; Gonen-Gross, T.; Fitchett, J.; Rowe, T.; Daniels, M.; Arnon, T.I.; Gazit, R.; Joseph, A.; Schjetne, K.W.; Steinle, A.; et al. Novel APC-like Properties of Human NK Cells Directly Regulate T Cell Activation. J. Clin. Investig. 2004, 114, 1612–1623. [Google Scholar] [CrossRef] [PubMed]
- Caligiuri, M.A.; Zmuidzinas, A.; Manley, T.J.; Levine, H.; Smith, K.A.; Ritz, J. Functional Consequences of Interleukin 2 Receptor Expression on Resting Human Lymphocytes: Identification of a Novel Natural Killer Cell Subset with High Affinity Receptors. J. Exp. Med. 1990, 171, 1509–1526. [Google Scholar] [CrossRef]
- O’Sullivan, T.; Johnson, L.R.; Kang, H.H.; Sun, J.C. BNIP3- and BNIP3L-Mediated Mitophagy Promotes the Generation of Natural Killer Cell Memory. Immunity 2015, 43, 331–342. [Google Scholar] [CrossRef]
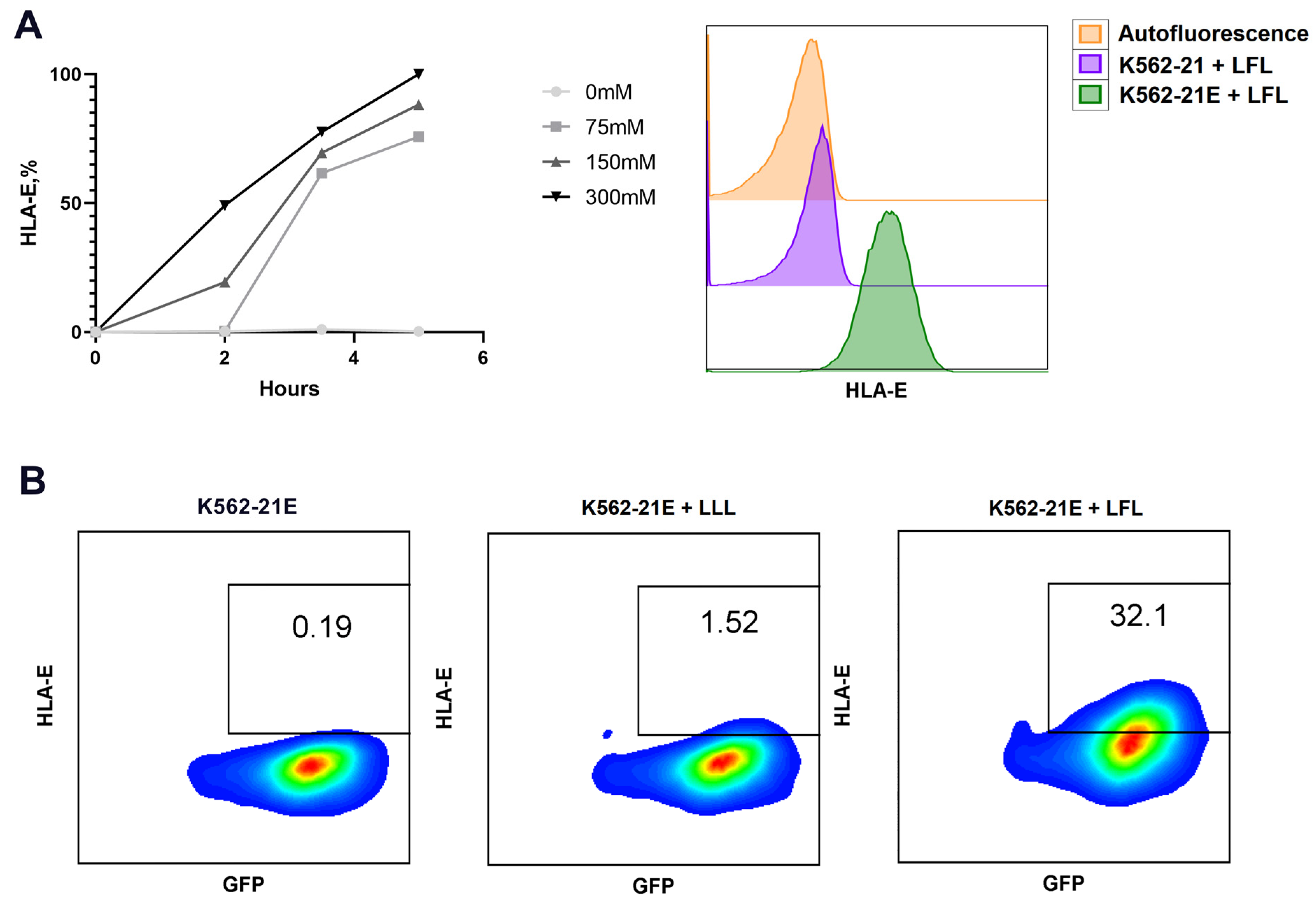

| K562-21E, 103/mL | Peptide Concentration, µM | Increase in the Level of HLA-E Expression Compared to Control, Arbitrary Units | Cell Mortality during 24 h Incubation, % | ||||
|---|---|---|---|---|---|---|---|
| 1 h | 2 h | 3.5 h | 5 h | 24 h | |||
| 200 | 0 | 1 | 1 | 1 | 1 | 1 | |
| 75 | 1.01 | 1.01 | 2.59 | 3.77 | |||
| 150 | 1.02 | 2.61 | 2.8 | 4.37 | |||
| 300 | 1.02 | 3.93 | 3.03 | 5.01 | 3.58 | 11.8 | |
| 500 | 300 | 3.44 | 28 | ||||
| 700 | 3.31 | 33 | |||||
| 2000 | 1.15 | 70 | |||||
| Donor Number | Age | Gender | HCMV Titer Range (U/mL) |
|---|---|---|---|
| 1 | 24 | Male | 3.7 |
| 2 | 25 | Female | 0 |
| 3 | 59 | Female | 17.8 |
| 4 | 40 | Male | 2.1 |
| 5 | 26 | Male | 0 |
| 6 | 64 | Female | 21.4 |
| 7 | 29 | Female | 4.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alekseeva, N.A.; Streltsova, M.A.; Vavilova, J.D.; Ustiuzhanina, M.O.; Palamarchuk, A.I.; Boyko, A.A.; Timofeev, N.D.; Popodko, A.I.; Kovalenko, E.I. Obtaining Gene-Modified HLA-E-Expressing Feeder Cells for Stimulation of Natural Killer Cells. Pharmaceutics 2024, 16, 133. https://doi.org/10.3390/pharmaceutics16010133
Alekseeva NA, Streltsova MA, Vavilova JD, Ustiuzhanina MO, Palamarchuk AI, Boyko AA, Timofeev ND, Popodko AI, Kovalenko EI. Obtaining Gene-Modified HLA-E-Expressing Feeder Cells for Stimulation of Natural Killer Cells. Pharmaceutics. 2024; 16(1):133. https://doi.org/10.3390/pharmaceutics16010133
Chicago/Turabian StyleAlekseeva, Nadezhda A., Maria A. Streltsova, Julia D. Vavilova, Maria O. Ustiuzhanina, Anastasia I. Palamarchuk, Anna A. Boyko, Nikita D. Timofeev, Alexey I. Popodko, and Elena I. Kovalenko. 2024. "Obtaining Gene-Modified HLA-E-Expressing Feeder Cells for Stimulation of Natural Killer Cells" Pharmaceutics 16, no. 1: 133. https://doi.org/10.3390/pharmaceutics16010133
APA StyleAlekseeva, N. A., Streltsova, M. A., Vavilova, J. D., Ustiuzhanina, M. O., Palamarchuk, A. I., Boyko, A. A., Timofeev, N. D., Popodko, A. I., & Kovalenko, E. I. (2024). Obtaining Gene-Modified HLA-E-Expressing Feeder Cells for Stimulation of Natural Killer Cells. Pharmaceutics, 16(1), 133. https://doi.org/10.3390/pharmaceutics16010133








